KCF-S cluster No. 1494 (7 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| asterids | 9 |
| eudicotyledons | 9 |
| Liliopsida | 8 |
| rosids | 7 |
| Spermatophyta | 1 |
Cumulative family count (Top 20)
| class name | count |
|---|---|
|
Paeoniaceae
|
6 |
|
Apocynaceae
|
3 |
|
Iridaceae
|
2 |
|
Cactaceae
|
2 |
|
Xanthorrhoeaceae
|
2 |
|
Amaryllidaceae
|
2 |
|
Primulaceae
|
2 |
|
Rosaceae
|
2 |
|
Asteraceae
|
1 |
|
Posidoniaceae
|
1 |
|
Malvaceae
|
1 |
|
Poaceae
|
1 |
|
Viscaceae
|
1 |
|
Pinaceae
|
1 |
|
Aquifoliaceae
|
1 |
|
Rubiaceae
|
1 |
|
Betulaceae
|
1 |
|
Moraceae
|
1 |
|
Juglandaceae
|
1 |
|
Plantaginaceae
|
1 |
KEGG BRITE br08003
Categories (0)
| br08003 Category | # of metabolite |
|---|
metabolites link (0)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|
Metabolite list (7)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
C00000171
|
3-Acethyl-6-methoxybenzaldehyde
|
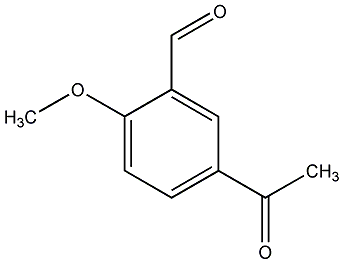
|
||||
|
C00002689
|
Apocynin
/ Acetovanillone |
CHEMBL346919
|
C056165
|
2 / 0 / 0 | 43 / 10 |
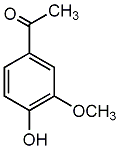
|
|
C00002693
|
3',4'-Dihydroxyacetophenone
|
CHEMBL243161
|
C032940
|
1 / 0 |
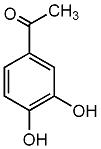
|
|
|
C00002704
|
Paeonol
|
CHEMBL1079227
|
C013638
|
4 / 2 / 3 |
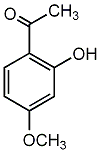
|
|
|
C00032617
|
2,5-Dihydroxy-4-methylacetophenone
|
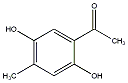
|
||||
|
C00032695
|
Acetoisovanillone
/ Isoacetovanillone |
C085119
|
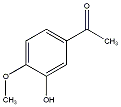
|
|||
|
C00033340
|
Resacetophenone
|
CHEMBL243374
|
C078634
|
1 / 0 / 0 |
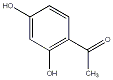
|
Human Protein / Gene in interactions
7 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q9HAS3 | Solute carrier family 28 member 3 | Unclassified protein | C00033340 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002704 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00002689 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002689 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00002704 | 0 / 0 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00002704 | 2 / 2 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002704 | 0 / 1 |
44 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1312 | COMT | catechol-O-methyltransferase (EC:2.1.1.6) |
C00002693
|
| 183 | AGT, ANHU, SERPINA8 | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
C00002689
|
| 1386 | ATF2, CRE-BP1, CREB2, HB16, TREB7 | activating transcription factor 2 (EC:2.3.1.48) |
C00002689
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00002689
|
| 6356 | CCL11, SCYA11 | chemokine (C-C motif) ligand 11 |
C00002689
|
| 6362 | CCL18, AMAC-1, AMAC1, CKb7, DC-CK1, DCCK1, MIP-4, PARC, SCYA18 | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
C00002689
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002689
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00002689
|
| 6354 | CCL7, FIC, MARC, MCP-3, MCP3, NC28, SCYA6, SCYA7 | chemokine (C-C motif) ligand 7 |
C00002689
|
| 948 | CD36, BDPLT10, CHDS7, FAT, GP3B, GP4, GPIV, PASIV, SCARB3 | CD36 molecule (thrombospondin receptor) |
C00002689
|
| 1437 | CSF2, GMCSF | colony stimulating factor 2 (granulocyte-macrophage) |
C00002689
|
| 2919 | CXCL1, FSP, GRO1, GROa, MGSA, MGSA-a, NAP-3, SCYB1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
C00002689
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00002689
|
| 1536 | CYBB, AMCBX2, CGD, GP91-1, GP91-PHOX, GP91PHOX, NOX2, p91-PHOX | cytochrome b-245, beta polypeptide |
C00002689
|
| 2033 | EP300, KAT3B, RSTS2, p300 | E1A binding protein p300 (EC:2.3.1.48) |
C00002689
|
| 2335 | FN1, CIG, ED-B, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | fibronectin 1 |
C00002689
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00002689
|
| 27429 | HTRA2, OMI, PARK13, PRSS25 | HtrA serine peptidase 2 (EC:3.4.21.108) |
C00002689
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00002689
|
| 3456 | IFNB1, IFB, IFF, IFNB | interferon, beta 1, fibroblast |
C00002689
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00002689
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002689
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002689
|
| 3684 | ITGAM, CD11B, CR3A, MAC-1, MAC1A, MO1A, SLEB6 | integrin, alpha M (complement component 3 receptor 3 subunit) |
C00002689
|
| 3717 | JAK2, JTK10, THCYT3 | Janus kinase 2 (EC:2.7.10.2) |
C00002689
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00002689
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002689
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002689
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00002689
|
| 5601 | MAPK9, JNK-55, JNK2, JNK2A, JNK2ALPHA, JNK2B, JNK2BETA, PRKM9, SAPK, SAPK1a, p54a, p54aSAPK | mitogen-activated protein kinase 9 (EC:2.7.11.24) |
C00002689
|
| 4282 | MIF, GIF, GLIF, MMIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) (EC:5.3.3.12 5.3.2.1) |
C00002689
|
| 4322 | MMP13, CLG3, MANDP1 | matrix metallopeptidase 13 (collagenase 3) (EC:3.4.24.-) |
C00002689
|
| 4481 | MSR1, CD204, SCARA1, SR-A, SRA, phSR1, phSR2 | macrophage scavenger receptor 1 |
C00002689
|
| 653361 | NCF1, NCF1A, NOXO2, SH3PXD1A, p47phox | neutrophil cytosolic factor 1 |
C00002689
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00002689
|
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00002689
|
| 4973 | OLR1, CLEC8A, LOX1, LOXIN, SCARE1, SLOX1 | oxidized low density lipoprotein (lectin-like) receptor 1 |
C00002689
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002689
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002689
|
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00002689
|
| 6772 | STAT1, CANDF7, ISGF-3, STAT91 | signal transducer and activator of transcription 1, 91kDa |
C00002689
|
| 7082 | TJP1, ZO-1 | tight junction protein 1 |
C00002689
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002689
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002689
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
Diseases related to CTD interactions
10 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D000419 | Albuminuria |
C00002689
|
| D001919 | Bradycardia |
C00002689
|
| D020520 | Brain Infarction |
C00002689
|
| D002310 | Cardiomyopathy, Alcoholic |
C00002689
|
| D003928 | Diabetic Nephropathies |
C00002689
|
| D006331 | Heart Diseases |
C00002689
|
| D006973 | Hypertension |
C00002689
|
| D007022 | Hypotension |
C00002689
|
| D009410 | Nerve Degeneration |
C00002689
|
| D011014 | Pneumonia |
C00002689
|