KCF-S cluster No. 208 (29 metabolites)
Corresponding Phytochemical cluster No. 48
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| Spermatophyta | 32 |
| asterids | 10 |
| rosids | 8 |
| Liliopsida | 7 |
| Magnoliophyta | 3 |
| Embryophyta | 1 |
Cumulative family count
| class name | count |
|---|---|
| Cupressaceae | 18 |
| Pinaceae | 11 |
| Lamiaceae | 5 |
| Poaceae | 4 |
| Meliaceae | 4 |
| Calceolariaceae | 3 |
| Zingiberaceae | 2 |
| Annonaceae | 2 |
| Cistaceae | 2 |
| Araucariaceae | 1 |
| Podocarpaceae | 1 |
| Euphorbiaceae | 1 |
| Brassicaceae | 1 |
| Araliaceae | 1 |
| Plagiochilaceae | 1 |
| Asteraceae | 1 |
| Taxaceae | 1 |
| Alismataceae | 1 |
| Schisandraceae | 1 |
Metabolite list (29)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
C00000867
|
9beta-Pimara-7,15-diene
|
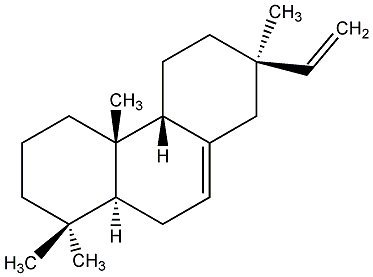
|
||||
|
C00000883
|
ent-Sandaracopimaradiene
|
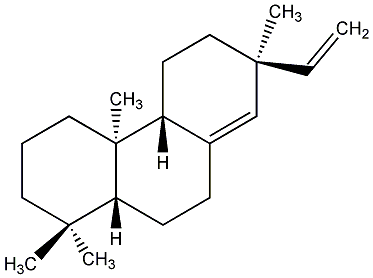
|
||||
|
C00000904
|
Virescenol B
|
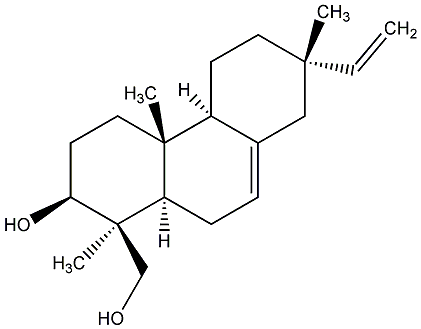
|
||||
|
C00003444
|
miropinic acid
/ Isopimaric acid / isodextropimaric acid |
CHEMBL512164
|
C115138
|
3 / 1 / 1 |
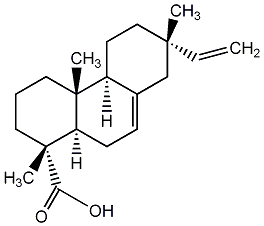
|
|
|
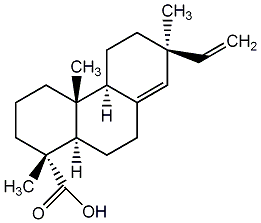
C00003468
|
(+)-Pimaric acid
|
CHEMBL513197
CHEMBL1397211 CHEMBL1410398 CHEMBL1735595 |
C008911
|
14 / 13 / 7 | 9 / 0 |
|
|
C00031283
|
Sandaracopimaric acid
/ (-)-Sandaracopimaric acid |
CHEMBL513197
CHEMBL1397211 CHEMBL1410398 CHEMBL1735595 |
C082072
|
14 / 13 / 7 |

|
|
|
C00031284
|
Sandaracopimarinal
|
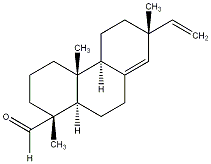
|
||||
|
C00031285
|
Sandaracopimarinol
|
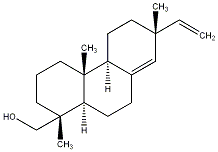
|
||||
|
C00031286
|
Sandaracopimarinol acetate
|

|
||||
|
C00032668
|
6,7-Dehydrosandaracopimaric acid
/ (+)-6,7-Dehydrosandaracopimaric acid |
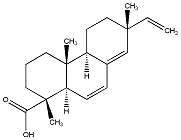
|
||||
|
C00034092
|
Oryzalexin D
/ (+)-Oryzalexin D |
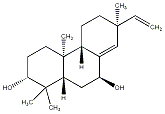
|
||||
|
C00034094
|
Oryzalexin F
|
CHEMBL254637
CHEMBL463903 |
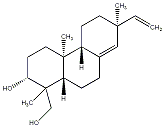
|
|||
|
C00034863
|
Isopimara-7,15-dien-2alpha-ol
/ (+)-Isopimara-7,15-dien-2alpha-ol |

|
||||
|
C00034864
|
Isopimara-7,15-dien-3beta-ol
/ (-)-Isopimara-7,15-dien-3beta-ol |
CHEMBL225866
|
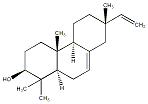
|
|||
|
C00034866
|
Isopimara-7,15-diene
|

|
||||
|
C00035989
|
Acanthoic acid
/ (-)-Acanthoic acid |
CHEMBL152325
CHEMBL512490 CHEMBL523295 |
4 / 0 / 0 |

|
||
|
C00036008
|
3beta-Hydroxyisopimaric acid
|
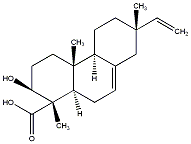
|
||||
|
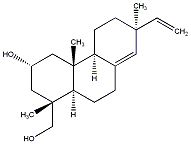
C00037101
|
ent-8(14),15-sandaracopimaradiene-2alpha,18-diol
/ (+)-ent-8(14),15-sandaracopimaradiene-2alpha,18-diol |
|
||||
|
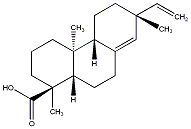
C00037105
|
ent-Pimara-8(14),15-dien-18-oic acid
|
CHEMBL513197
CHEMBL1397211 CHEMBL1410398 CHEMBL1735595 |
14 / 13 / 7 |
|
||
|
C00040774
|
17-Hydroxy-9-epi-ent-7,15-isopimaradiene
|

|
||||
|
C00040775
|
19-Hydroxy-9-epi-ent-7,15-isopimaradiene
|

|
||||
|
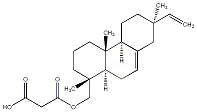
C00040777
|
19-Malonyl-9-epi-ent-7,15-isopimaradiene
|
|
||||
|
C00041813
|
Sagittine A
|
CHEMBL447656
|

|
|||
|
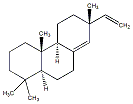
C00042933
|
Sandaracopimaradiene
/ (-)-Sandaracopimaradiene |
|
||||
|
C00048284
|
3beta-Acetoxy-7,15-isopimaradiene
|
|
||||
|
C00048309
|
Akhdarenol
|

|
||||
|
C00048379
|
ent-Pimara-8(14),15-diene
/ (-)-ent-Pimara-8(14),15-diene |
CHEMBL513197
CHEMBL1397211 CHEMBL1410398 CHEMBL1735595 |
14 / 13 / 7 |
|
||
|
C00048512
|
Pimara-8(14),15-diene
|
|
||||
|
C00049953
|
4-epi-Sandaracopimaric acid
/ (+)-4-epi-Sandaracopimaric acid |
CHEMBL513197
CHEMBL1397211 CHEMBL1410398 CHEMBL1735595 |
14 / 13 / 7 |
|
Human Protein / Gene in interactions
19 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P47870 | Gamma-aminobutyric acid receptor subunit beta-2 | GABA-A beta | C00003444 C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 0 |
| P14867 | Gamma-aminobutyric acid receptor subunit alpha-1 | GABA-A alpha | C00003444 C00003468 C00031283 C00037105 C00048379 C00049953 | 1 / 1 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00003468 C00031283 C00037105 C00048379 C00049953 | 1 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 1 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00003468 C00031283 C00037105 C00048379 C00049953 | 2 / 0 |
| O75496 | Geminin | Unclassified protein | C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 0 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00003468 C00031283 C00037105 C00048379 C00049953 | 7 / 3 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00003468 C00031283 C00037105 C00048379 C00049953 | 1 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00003468 C00031283 C00037105 C00048379 C00049953 | 1 / 1 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00003468 C00031283 C00037105 C00048379 C00049953 | 0 / 0 |
| P55055 | Oxysterols receptor LXR-beta | NR1H3 | C00035989 | 0 / 0 |
| Q13133 | Oxysterols receptor LXR-alpha | NR1H3 | C00035989 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00003444 | 0 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00035989 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00035989 | 0 / 0 |
9 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1386 | ATF2, CRE-BP1, CREB2, HB16, TREB7 | activating transcription factor 2 (EC:2.3.1.48) |
C00003468
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00003468
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00003468
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00003468
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00003468
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00003468
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00003468
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00003468
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00003468
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (13)
| OMIM | preferred title | UniProt |
|---|---|---|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #114500 | Colorectal cancer; crc |
P84022
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #611136 | Epilepsy, juvenile myoclonic, susceptibility to, 5; ejm5 |
P14867
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (7)
| KEGG | name | UniProt |
|---|---|---|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00808 | Idiopathic generalized epilepsies (IGEs) |
P14867
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|