Eucalyptus cypellocarpa
Species
KNApSAcK Entry
| Organism name | Eucalyptus cypellocarpa |
|---|---|
| Genus | Eucalyptus |
| Family | Myrtaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Eucalyptus |
|---|---|
| Linked NCBI taxonomy ID | 3932 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Myrtaceae |
|---|---|
| ID | 3931 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (15)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
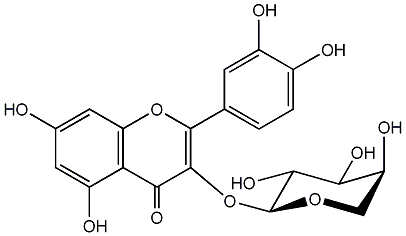
C00005368
|
Guaijaverin
/ Quercetin 3-O-alpha-L-arabinopyranoside |
CHEMBL183031
CHEMBL185461 CHEMBL488198 CHEMBL464507 CHEMBL520637 |
C088178
|
No. 2 | No. 15 |
|
||
|
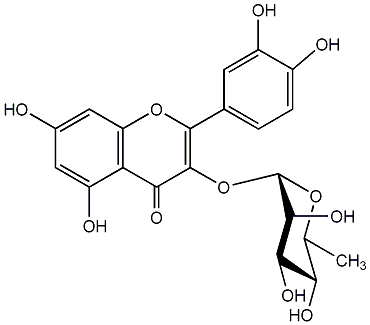
C00005374
|
Quercetin
|
CHEMBL82242
CHEMBL479232 CHEMBL1437696 |
C012526
|
14 / 2 / 2 | 2 / 1 | No. 2 | No. 15 |
|
|
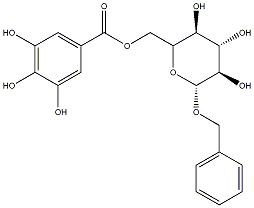
C00031624
|
Benzyl 6'-O-galloyl-beta-glucopyranoside
/ (-)-Benzyl 6'-O-galloyl-beta-glucopyranoside |
No. 532 |
|
|||||
|
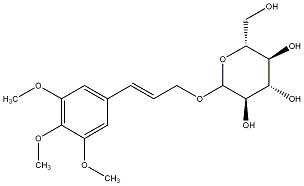
C00031878
|
Icariside H1
/ Juniperoside / (-)-Icariside H1 / (-)-Juniperoside |
No. 678 | No. 6 |
|
||||
|
C00033746
|
Cypellocarpin A
|
CHEMBL486175
|
No. 810 |

|
||||
|
C00046708
|
Cypellocarpin C
/ (-)-Cypellocarpin C |
CHEMBL247486
|
No. 810 |

|
||||
|
C00046707
|
Cypellocarpin B
/ (-)-Cypellocarpin B |
CHEMBL520205
|
No. 810 |

|
||||
|
C00010848
|
Oleuropeic acid
|
CHEMBL1077602
|
C417926
|
No. 983 | No. 35 |

|
||
|
C00031718
|
Cypellogin C
/ (+)-Cypellogin C |
No. 1511 |

|
|||||
|
C00031717
|
Cypellogin B
/ (+)-Cypellogin B |
No. 1511 |

|
|||||
|
C00031151
|
Resinoside B
|
No. 1511 |

|
|||||
|
C00031150
|
Resinoside A
|
No. 1511 |

|
|||||
|
C00031716
|
Cypellogin A
/ (-)-Cypellogin A |
No. 1511 |

|
|||||
|
C00011154
|
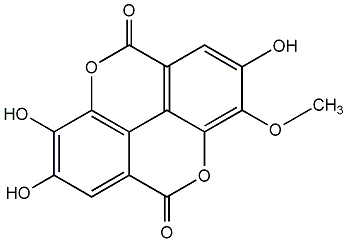
3-O-Methylellagic acid
|
CHEMBL564351
|
No. 2068 | No. 81 |
|
|||
|
C00048366
|
Cuniloside B
/ (-)-Cuniloside B |
No. 4168 |

|
Human Protein / Gene in interactions
14 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q9P1W9 | Serine/threonine-protein kinase pim-2 | Pim | C00005374 | 0 / 0 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00005374 | 1 / 1 |
| P07237 | Protein disulfide-isomerase | Enzyme | C00005374 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00005374 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00005374 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00005374 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00005374 | 1 / 1 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00005374 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00005374 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00005374 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00005374 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00005374 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00005374 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00005374 | 0 / 0 |
2 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00005374
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00005374
|