Epimedium membranaceum
Species
KNApSAcK Entry
| Organism name | Epimedium membranaceum |
|---|---|
| Genus | Epimedium |
| Family | Berberidaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Epimedium membranaceum |
|---|---|
| Linked NCBI taxonomy ID | 402159 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Berberidaceae |
|---|---|
| ID | 41773 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | eudicotyledons |
|---|---|
| ID | 71240 |
Metabolite list (4)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00004631
|
Quercetin
|
CHEMBL50
|
D011794
|
174 / 130 / 112 | 3132 / 63 | No. 3 | No. 15 |
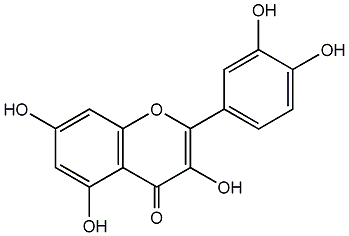
|
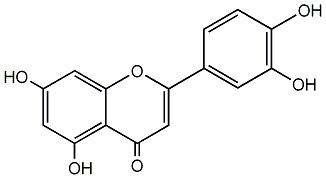
|
C00004565
|
Kaempferol
/ Nimbecetin / Rhamnolutein / Pelargidenolon / 5,7,4'-Trihydroxyflavonol / 3,5,7,4'-Tetrahydroxyflavone / 3,5,7-Trihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
CHEMBL150
|
C006552
|
76 / 69 / 87 | 86 / 8 | No. 71 | No. 15 |
|
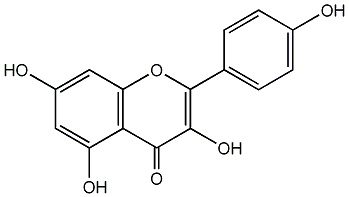
|
C00003817
|
Apigenin
/ 5,7,4'-Trihydroxyflavone |
CHEMBL28
|
D047310
|
164 / 119 / 105 | 68 / 8 | No. 71 | No. 15 |
|
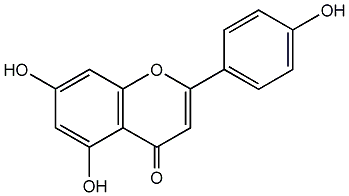
|
C00000674
|
Luteolin
|
CHEMBL151
|
D047311
|
99 / 91 / 95 | 76 / 6 | No. 71 | No. 15 |
|
Human Protein / Gene in interactions
249 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000674 C00003817 C00004565 C00004631 | 1 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| Q15078 | Cyclin-dependent kinase 5 activator 1 | REG serine/threonine protein kinase family | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P06276 | Cholinesterase | Hydrolase | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P49840 | Glycogen synthase kinase-3 alpha | Gsk | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00000674 C00003817 C00004565 C00004631 | 7 / 37 |
| Q9NPH5 | NADPH oxidase 4 | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| Q00535 | Cyclin-dependent kinase 5 | Cdk5 | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| Q8WWL7 | G2/mitotic-specific cyclin-B3 | Other cytosolic protein | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| O95067 | G2/mitotic-specific cyclin-B2 | Other cytosolic protein | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| Q16678 | Cytochrome P450 1B1 | Cytochrome P450 1B1 | C00000674 C00003817 C00004565 C00004631 | 4 / 4 |
| P14635 | G2/mitotic-specific cyclin-B1 | Other cytosolic protein | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00000674 C00003817 C00004565 C00004631 | 4 / 3 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P04150 | Glucocorticoid receptor | NR3C1 | C00000674 C00003817 C00004565 C00004631 | 0 / 1 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| O00255 | Menin | Unclassified protein | C00000674 C00003817 C00004565 C00004631 | 2 / 5 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00000674 C00003817 C00004565 C00004631 | 0 / 1 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00000674 C00003817 C00004565 C00004631 | 0 / 3 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00000674 C00003817 C00004565 C00004631 | 1 / 1 |
| P06493 | Cyclin-dependent kinase 1 | Cdc2 | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00000674 C00003817 C00004565 C00004631 | 1 / 2 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00000674 C00003817 C00004565 C00004631 | 3 / 3 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00000674 C00003817 C00004565 C00004631 | 1 / 1 |
| Q9UNQ0 | ATP-binding cassette sub-family G member 2 | ATP binding cassette | C00000674 C00003817 C00004565 C00004631 | 2 / 0 |
| P04062 | Glucosylceramidase | Enzyme | C00000674 C00003817 C00004565 C00004631 | 6 / 4 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00000674 C00003817 C00004565 C00004631 | 2 / 2 |
| P08183 | Multidrug resistance protein 1 | drug | C00000674 C00003817 C00004565 C00004631 | 1 / 0 |
| P04798 | Cytochrome P450 1A1 | Cytochrome P450 1A1 | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00000674 C00003817 C00004565 C00004631 | 1 / 1 |
| P39748 | Flap endonuclease 1 | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P11309 | Serine/threonine-protein kinase pim-1 | Pim | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| Q00534 | Cyclin-dependent kinase 6 | CMGC serine/threonine protein kinase family | C00000674 C00003817 C00004565 C00004631 | 1 / 0 |
| P22303 | Acetylcholinesterase | Hydrolase | C00000674 C00003817 C00004565 C00004631 | 1 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P36888 | Receptor-type tyrosine-protein kinase FLT3 | Pdgfr | C00000674 C00003817 C00004565 C00004631 | 1 / 1 |
| P10275 | Androgen receptor | NR3C4 | C00000674 C00003817 C00004565 C00004631 | 3 / 4 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000674 C00003817 C00004565 C00004631 | 0 / 1 |
| Q9Y3R4 | Sialidase-2 | Enzyme | C00000674 C00003817 C00004565 C00004631 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00000674 C00003817 C00004565 C00004631 | 1 / 1 |
| P02545 | Prelamin-A/C | Unclassified protein | C00000674 C00003817 C00004565 C00004631 | 11 / 10 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00000674 C00003817 C00004565 C00004631 | 1 / 1 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00000674 C00003817 C00004631 | 1 / 1 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00000674 C00003817 C00004631 | 3 / 1 |
| P37059 | Estradiol 17-beta-dehydrogenase 2 | Enzyme | C00003817 C00004565 C00004631 | 0 / 0 |
| O15296 | Arachidonate 15-lipoxygenase B | Enzyme | C00000674 C00004565 C00004631 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00000674 C00003817 C00004631 | 0 / 0 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00000674 C00003817 C00004631 | 3 / 2 |
| P06239 | Tyrosine-protein kinase Lck | Src | C00000674 C00003817 C00004631 | 0 / 1 |
| Q04760 | Lactoylglutathione lyase | Enzyme | C00000674 C00004565 C00004631 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00000674 C00003817 C00004631 | 2 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00000674 C00003817 C00004631 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00000674 C00003817 C00004565 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00000674 C00003817 C00004631 | 0 / 0 |
| P24941 | Cyclin-dependent kinase 2 | Cdc2 | C00000674 C00003817 C00004631 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00000674 C00003817 C00004631 | 0 / 0 |
| P27338 | Amine oxidase [flavin-containing] B | Oxidoreductase | C00000674 C00003817 C00004631 | 0 / 0 |
| P14679 | Tyrosinase | Oxidoreductase | C00000674 C00004565 C00004631 | 4 / 2 |
| Q99700 | Ataxin-2 | Unclassified protein | C00000674 C00003817 C00004565 | 1 / 1 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00003817 C00004565 C00004631 | 5 / 3 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00000674 C00003817 C00004631 | 4 / 1 |
| P33527 | Multidrug resistance-associated protein 1 | drugs | C00003817 C00004565 C00004631 | 0 / 0 |
| P14061 | Estradiol 17-beta-dehydrogenase 1 | Enzyme | C00003817 C00004565 C00004631 | 0 / 0 |
| P04745 | Alpha-amylase 1 | Enzyme | C00000674 C00004565 C00004631 | 0 / 0 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00003817 C00004565 C00004631 | 1 / 8 |
| Q15046 | Lysine--tRNA ligase | Enzyme | C00000674 C00003817 C00004631 | 2 / 1 |
| P38398 | Breast cancer type 1 susceptibility protein | Enzyme | C00000674 C00003817 | 4 / 2 |
| P51570 | Galactokinase | Enzyme | C00000674 C00004631 | 1 / 1 |
| Q9HC97 | G-protein coupled receptor 35 | Kynurenic acid receptor | C00000674 C00004631 | 0 / 0 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00000674 C00004631 | 1 / 0 |
| Q6W5P4 | Neuropeptide S receptor | Neuropeptide receptor | C00003817 C00004631 | 1 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00000674 C00004565 | 0 / 0 |
| P11387 | DNA topoisomerase 1 | Isomerase | C00000674 C00004631 | 0 / 0 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | C00003817 C00004631 | 5 / 1 |
| P23443 | Ribosomal protein S6 kinase beta-1 | p70 | C00003817 C00004631 | 0 / 0 |
| Q99549 | M-phase phosphoprotein 8 | Unclassified protein | C00003817 C00004631 | 0 / 0 |
| P68400 | Casein kinase II subunit alpha | Ck2 | C00003817 C00004631 | 0 / 0 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00003817 C00004565 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00003817 C00004631 | 0 / 0 |
| O00141 | Serine/threonine-protein kinase Sgk1 | AGC serine/threonine protein kinase SGK subfamily | C00003817 C00004631 | 0 / 0 |
| O75582 | Ribosomal protein S6 kinase alpha-5 | CAMK serine/threonine protein kinase MSKB subfamily | C00003817 C00004631 | 0 / 0 |
| O95271 | Tankyrase-1 | Enzyme | C00000674 C00003817 | 0 / 0 |
| P31749 | RAC-alpha serine/threonine-protein kinase | Akt | C00003817 C00004631 | 4 / 1 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | C00003817 C00004565 | 0 / 0 |
| P28907 | ADP-ribosyl cyclase 1 | Enzyme | C00000674 C00004631 | 0 / 1 |
| O14757 | Serine/threonine-protein kinase Chk1 | Chk1 | C00003817 C00004631 | 0 / 0 |
| Q02880 | DNA topoisomerase 2-beta | Isomerase | C00003817 C00004631 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00003817 C00004631 | 0 / 0 |
| Q8IW41 | MAP kinase-activated protein kinase 5 | CAMK serine/threonine protein kinase MAPKAPK | C00003817 C00004631 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00000674 C00004631 | 2 / 2 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00003817 C00004565 | 0 / 0 |
| P39900 | Macrophage metalloelastase | M10A | C00000674 C00004631 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00003817 C00004631 | 0 / 0 |
| P45984 | Mitogen-activated protein kinase 9 | Jnk | C00003817 C00004631 | 0 / 0 |
| O76074 | cGMP-specific 3',5'-cyclic phosphodiesterase | PDE_5A | C00003817 C00004631 | 0 / 0 |
| Q9H2K2 | Tankyrase-2 | Enzyme | C00000674 C00003817 | 0 / 0 |
| P35869 | Aryl hydrocarbon receptor | Transcription Factor | C00003817 C00004565 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00000674 C00003817 | 0 / 0 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00003817 C00004631 | 0 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00000674 C00004631 | 1 / 2 |
| P54855 | UDP-glucuronosyltransferase 2B15 | Enzyme | C00003817 C00004631 | 0 / 0 |
| P08254 | Stromelysin-1 | M10A | C00000674 C00004631 | 1 / 0 |
| P51449 | Nuclear receptor ROR-gamma | Nuclear hormone receptor subfamily 1 group F member 3 | C00003817 C00004565 | 0 / 0 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00003817 C00004631 | 5 / 2 |
| P15121 | Aldose reductase | Enzyme | C00004565 C00004631 | 0 / 0 |
| P48147 | Prolyl endopeptidase | S9A | C00004565 C00004631 | 0 / 0 |
| Q92887 | Canalicular multispecific organic anion transporter 1 | Unclassified protein | C00003817 C00004631 | 1 / 1 |
| P53778 | Mitogen-activated protein kinase 12 | p38 | C00003817 C00004631 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00000674 C00004631 | 0 / 0 |
| O15264 | Mitogen-activated protein kinase 13 | p38 | C00003817 C00004631 | 0 / 0 |
| P27487 | Dipeptidyl peptidase 4 | S9B | C00004565 C00004631 | 0 / 1 |
| P09874 | Poly [ADP-ribose] polymerase 1 | Enzyme | C00000674 C00003817 | 0 / 0 |
| Q12791 | Calcium-activated potassium channel subunit alpha-1 | KCNM, KCa1.x | C00003817 C00004565 | 1 / 1 |
| P03956 | Interstitial collagenase | M10A | C00000674 C00004631 | 0 / 1 |
| P06746 | DNA polymerase beta | Enzyme | C00000674 C00004631 | 0 / 0 |
| Q16539 | Mitogen-activated protein kinase 14 | p38 | C00003817 C00004631 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00000674 C00004631 | 0 / 0 |
| P08253 | 72 kDa type IV collagenase | M10A | C00000674 C00004631 | 1 / 3 |
| Q15759 | Mitogen-activated protein kinase 11 | p38 | C00003817 C00004631 | 0 / 0 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00003817 C00004631 | 0 / 0 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00004565 C00004631 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00000674 C00003817 | 0 / 1 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00003817 C00004631 | 2 / 2 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00003817 C00004631 | 2 / 0 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00003817 C00004631 | 0 / 0 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00000674 C00003817 | 1 / 4 |
| P45983 | Mitogen-activated protein kinase 8 | Jnk | C00003817 C00004631 | 0 / 0 |
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | C00003817 C00004631 | 3 / 0 |
| P56817 | Beta-secretase 1 | A1A | C00003817 C00004631 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00003817 C00004631 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00000674 C00003817 | 0 / 0 |
| P45452 | Collagenase 3 | M10A | C00000674 C00004631 | 1 / 1 |
| P49137 | MAP kinase-activated protein kinase 2 | CAMK serine/threonine protein kinase MAPKAPK | C00003817 C00004631 | 0 / 0 |
| P18054 | Arachidonate 12-lipoxygenase, 12S-type | Enzyme | C00000674 C00004631 | 2 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00003817 C00004565 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00004631 | 0 / 0 |
| P51679 | C-C chemokine receptor type 4 | CC chemokine receptor | C00004631 | 0 / 0 |
| P12931 | Proto-oncogene tyrosine-protein kinase Src | Src | C00004631 | 0 / 0 |
| Q9NPD5 | Solute carrier organic anion transporter family member 1B3 | Electrochemical transporter | C00004631 | 1 / 0 |
| P00390 | Glutathione reductase, mitochondrial | Oxidoreductase | C00004631 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00004631 | 0 / 1 |
| P06280 | Alpha-galactosidase A | Enzyme | C00000674 | 1 / 1 |
| Q9Y4K4 | Mitogen-activated protein kinase kinase kinase kinase 5 | STE serine/threonine protein kinase KHS subfamily | C00003817 | 0 / 0 |
| P42226 | Signal transducer and activator of transcription 6 | Unclassified protein | C00004631 | 0 / 0 |
| Q15418 | Ribosomal protein S6 kinase alpha-1 | Rskb | C00003817 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00004631 | 0 / 0 |
| P10145 | Interleukin-8 | Secreted protein | C00003817 | 0 / 0 |
| O15118 | Niemann-Pick C1 protein | Unclassified protein | C00003817 | 1 / 1 |
| Q92731 | Estrogen receptor beta | NR3A2 | C00003817 | 0 / 1 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00003817 | 0 / 0 |
| P21917 | D(4) dopamine receptor | Dopamine receptor | C00004631 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00004631 | 0 / 0 |
| Q9NWT6 | Hypoxia-inducible factor 1-alpha inhibitor | Enzyme | C00004631 | 0 / 0 |
| O14763 | Tumor necrosis factor receptor superfamily member 10B | Unclassified protein | C00000674 | 1 / 0 |
| Q00613 | Heat shock factor protein 1 | Unclassified protein | C00004631 | 0 / 0 |
| P16152 | Carbonyl reductase [NADPH] 1 | Enzyme | C00004631 | 0 / 0 |
| P09769 | Tyrosine-protein kinase Fgr | Src | C00003817 | 0 / 0 |
| P30518 | Vasopressin V2 receptor | Vasopressin and oxytocin receptor | C00004631 | 2 / 2 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00003817 | 0 / 0 |
| P53350 | Serine/threonine-protein kinase PLK1 | PLK serine/threonine protein kinase subfamily | C00004631 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00004631 | 1 / 2 |
| O75116 | Rho-associated protein kinase 2 | Rock | C00003817 | 0 / 0 |
| Q00796 | Sorbitol dehydrogenase | Enzyme | C00004631 | 0 / 0 |
| P68871 | Hemoglobin subunit beta | Secreted protein | C00003817 | 4 / 4 |
| P08238 | Heat shock protein HSP 90-beta | Other cytosolic protein | C00004631 | 0 / 0 |
| P07948 | Tyrosine-protein kinase Lyn | Src | C00003817 | 0 / 0 |
| P28845 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | Enzyme | C00004631 | 1 / 1 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00004631 | 0 / 0 |
| P17538 | Chymotrypsinogen B | S1A | C00004631 | 0 / 0 |
| Q15118 | [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1, mitochondrial | Pdhk | C00004631 | 0 / 0 |
| P05164 | Myeloperoxidase | Enzyme | C00004631 | 1 / 2 |
| P27986 | Phosphatidylinositol 3-kinase regulatory subunit alpha | Enzyme | C00004631 | 2 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00004631 | 0 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00003817 | 0 / 0 |
| P98170 | E3 ubiquitin-protein ligase XIAP | Other cytosolic protein | C00004565 | 1 / 1 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00003817 | 2 / 3 |
| Q9GZU7 | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 | Enzyme | C00004565 | 0 / 0 |
| P04792 | Heat shock protein beta-1 | Unclassified protein | C00004631 | 2 / 1 |
| O75828 | Carbonyl reductase [NADPH] 3 | Enzyme | C00004631 | 0 / 0 |
| P24385 | G1/S-specific cyclin-D1 | Other cytosolic protein | C00004631 | 1 / 8 |
| P11802 | Cyclin-dependent kinase 4 | CMGC serine/threonine protein kinase family | C00004631 | 1 / 3 |
| P08236 | Beta-glucuronidase | Enzyme | C00004631 | 1 / 2 |
| P25024 | C-X-C chemokine receptor type 1 | CXC chemokine receptor | C00004631 | 0 / 0 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00004631 | 0 / 0 |
| P49327 | Fatty acid synthase | Transferase | C00000674 | 0 / 0 |
| P08908 | 5-hydroxytryptamine receptor 1A | Serotonin receptor | C00004631 | 1 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00004631 | 1 / 2 |
| P62158 | Calmodulin | Unclassified protein | C00004631 | 1 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00004631 | 0 / 0 |
| Q99895 | Chymotrypsin-C | S1A | C00004631 | 0 / 2 |
| P27361 | Mitogen-activated protein kinase 3 | Erk | C00004631 | 0 / 0 |
| Q13554 | Calcium/calmodulin-dependent protein kinase type II subunit beta | Camk2 | C00004631 | 0 / 0 |
| P54646 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Ampk | C00003817 | 0 / 0 |
| P42336 | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | Enzyme | C00004631 | 9 / 1 |
| P43405 | Tyrosine-protein kinase SYK | Syk | C00003817 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00004631 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00004631 | 0 / 0 |
| Q13627 | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | CMGC dual-specificity kinase DYRK1 | C00003817 | 1 / 0 |
| P04054 | Phospholipase A2 | Enzyme | C00004631 | 0 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00004631 | 1 / 1 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00004631 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00000674 | 1 / 1 |
| P41240 | Tyrosine-protein kinase CSK | Csk | C00003817 | 0 / 0 |
| O15530 | 3-phosphoinositide-dependent protein kinase 1 | Pdk1 | C00003817 | 0 / 0 |
| P40225 | Thrombopoietin | Unclassified protein | C00003817 | 1 / 1 |
| P10632 | Cytochrome P450 2C8 | Cytochrome P450 2C8 | C00004631 | 0 / 0 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00004565 | 2 / 2 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00003817 | 1 / 0 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00004631 | 0 / 0 |
| P14618 | Pyruvate kinase PKM | Enzyme | C00003817 | 0 / 0 |
| O94956 | Solute carrier organic anion transporter family member 2B1 | Unclassified protein | C00004631 | 0 / 0 |
| Q9UN88 | Gamma-aminobutyric acid receptor subunit theta | GABA-A theta | C00003817 | 0 / 0 |
| P28472 | Gamma-aminobutyric acid receptor subunit beta-3 | GABA-A beta | C00003817 | 1 / 0 |
| P14867 | Gamma-aminobutyric acid receptor subunit alpha-1 | GABA-A alpha | C00003817 | 1 / 1 |
| P48169 | Gamma-aminobutyric acid receptor subunit alpha-4 | GABA-A alpha | C00003817 | 0 / 0 |
| O14764 | Gamma-aminobutyric acid receptor subunit delta | GABA-A delta | C00003817 | 1 / 1 |
| P31644 | Gamma-aminobutyric acid receptor subunit alpha-5 | GABA-A alpha | C00003817 | 0 / 0 |
| Q99928 | Gamma-aminobutyric acid receptor subunit gamma-3 | GABA-A gamma | C00003817 | 0 / 0 |
| P78334 | Gamma-aminobutyric acid receptor subunit epsilon | GABA-A epsilon | C00003817 | 0 / 0 |
| Q8N1C3 | Gamma-aminobutyric acid receptor subunit gamma-1 | GABA-A gamma | C00003817 | 0 / 0 |
| P34903 | Gamma-aminobutyric acid receptor subunit alpha-3 | GABA-A alpha | C00003817 | 0 / 0 |
| O00591 | Gamma-aminobutyric acid receptor subunit pi | GABA-A pi | C00003817 | 0 / 0 |
| P47870 | Gamma-aminobutyric acid receptor subunit beta-2 | GABA-A beta | C00003817 | 0 / 0 |
| P18507 | Gamma-aminobutyric acid receptor subunit gamma-2 | GABA-A gamma | C00003817 | 4 / 2 |
| P18505 | Gamma-aminobutyric acid receptor subunit beta-1 | GABA-A beta | C00003817 | 0 / 0 |
| Q16445 | Gamma-aminobutyric acid receptor subunit alpha-6 | GABA-A alpha | C00003817 | 0 / 0 |
| P47869 | Gamma-aminobutyric acid receptor subunit alpha-2 | GABA-A alpha | C00003817 | 1 / 0 |
| P53779 | Mitogen-activated protein kinase 10 | Jnk | C00003817 | 0 / 1 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00000674 | 0 / 0 |
| Q9Y6L6 | Solute carrier organic anion transporter family member 1B1 | Electrochemical transporter | C00004631 | 1 / 0 |
| P22694 | cAMP-dependent protein kinase catalytic subunit beta | Pka | C00003817 | 0 / 0 |
| P17612 | cAMP-dependent protein kinase catalytic subunit alpha | Pka | C00003817 | 0 / 0 |
| P22612 | cAMP-dependent protein kinase catalytic subunit gamma | Pka | C00003817 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00004631 | 0 / 0 |
| Q9HCP0 | Casein kinase I isoform gamma-1 | Ck1 | C00003817 | 0 / 0 |
| P48730 | Casein kinase I isoform delta | Ck1 | C00003817 | 1 / 0 |
| P49674 | Casein kinase I isoform epsilon | Ck1 | C00003817 | 0 / 0 |
| P78368 | Casein kinase I isoform gamma-2 | Ck1 | C00003817 | 0 / 0 |
| P48729 | Casein kinase I isoform alpha | Ck1 | C00003817 | 0 / 0 |
| Q02750 | Dual specificity mitogen-activated protein kinase kinase 1 | Ste7 | C00003817 | 1 / 1 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00004565 | 0 / 0 |
| Q14191 | Werner syndrome ATP-dependent helicase | Enzyme | C00000674 | 2 / 1 |
3155 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00000674
C00003817
C00004565
C00004631
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00000674
C00003817
C00004565
C00004631
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00000674
C00003817
C00004565
C00004631
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00000674
C00003817
C00004565
C00004631
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00000674
C00003817
C00004565
C00004631
|
| 9540 | TP53I3, PIG3 | tumor protein p53 inducible protein 3 |
C00000674
C00003817
C00004565
C00004631
|
| 8626 | TP63, AIS, B(p51A), B(p51B), EEC3, KET, LMS, NBP, OFC8, RHS, SHFM4, TP53CP, TP53L, TP73L, p40, p51, p53CP, p63, p73H, p73L | tumor protein p63 |
C00000674
C00003817
C00004565
C00004631
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00000674
C00003817
C00004565
C00004631
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00000674
C00003817
C00004565
C00004631
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00000674
C00003817
C00004565
C00004631
|
| 7161 | TP73, P73 | tumor protein p73 |
C00000674
C00003817
C00004565
C00004631
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00000674
C00003817
C00004565
C00004631
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00000674
C00003817
C00004565
C00004631
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00000674
C00003817
C00004565
C00004631
|
| 891 | CCNB1, CCNB | cyclin B1 |
C00000674
C00003817
C00004565
C00004631
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00000674
C00003817
C00004565
C00004631
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00000674
C00003817
C00004565
C00004631
|
| 1031 | CDKN2C, INK4C, p18, p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
C00000674
C00003817
C00004565
C00004631
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00000674
C00003817
C00004565
C00004631
|
| 3565 | IL4, BCGF-1, BCGF1, BSF-1, BSF1, IL-4 | interleukin 4 |
C00000674
C00003817
C00004565
C00004631
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00000674
C00003817
C00004565
C00004631
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00000674
C00003817
C00004565
C00004631
|
| 2203 | FBP1, FBP | fructose-1,6-bisphosphatase 1 (EC:3.1.3.11) |
C00000674
C00003817
C00004565
C00004631
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00000674
C00003817
C00004565
C00004631
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00000674
C00003817
C00004565
C00004631
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00000674
C00003817
C00004565
C00004631
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00000674
C00003817
C00004565
C00004631
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00000674
C00003817
C00004565
C00004631
|
| 8000 | PSCA, PRO232 | prostate stem cell antigen |
C00000674
C00003817
C00004565
C00004631
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00003817
C00004565
C00004631
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00003817
C00004565
C00004631
|
| 54205 | CYCS, CYC, HCS, THC4 | cytochrome c, somatic |
C00003817
C00004565
C00004631
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00000674
C00003817
C00004631
|
| 1437 | CSF2, GMCSF | colony stimulating factor 2 (granulocyte-macrophage) |
C00000674
C00004565
C00004631
|
| 4616 | GADD45B, GADD45BETA, MYD118 | growth arrest and DNA-damage-inducible, beta |
C00000674
C00004565
C00004631
|
| 2729 | GCLC, GCL, GCS, GLCL, GLCLC | glutamate-cysteine ligase, catalytic subunit (EC:6.3.2.2) |
C00003817
C00004565
C00004631
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00000674
C00003817
C00004631
|
| 54600 | UGT1A9, HLUGP4, LUGP4, UDPGT, UDPGT_1-9, UGT-1I, UGT1-09, UGT1-9, UGT1.9, UGT1AI, UGT1I | UDP glucuronosyltransferase 1 family, polypeptide A9 (EC:2.4.1.17) |
C00003817
C00004565
C00004631
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00003817
C00004565
C00004631
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00000674
C00003817
C00004631
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00003817
C00004565
C00004631
|
| 54659 | UGT1A3, UDPGT, UDPGT_1-3, UGT-1C, UGT1-03, UGT1.3, UGT1C | UDP glucuronosyltransferase 1 family, polypeptide A3 (EC:2.4.1.17) |
C00000674
C00004565
C00004631
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00000674
C00003817
C00004631
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00000674
C00004565
C00004631
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00000674
C00003817
C00004631
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00003817
C00004565
C00004631
|
| 1080 | CFTR, ABC35, ABCC7, CF, CFTR/MRP, MRP7, TNR-CFTR, dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) (EC:3.6.3.49) |
C00003817
C00004565
C00004631
|
| 4312 | MMP1, CLG, CLGN | matrix metallopeptidase 1 (interstitial collagenase) (EC:3.4.24.7) |
C00000674
C00003817
C00004631
|
| 598 | BCL2L1, BCL-XL/S, BCL2L, BCLX, BCLXL, BCLXS, Bcl-X, PPP1R52, bcl-xL, bcl-xS | BCL2-like 1 |
C00000674
C00004565
C00004631
|
| 958 | CD40, Bp50, CDW40, TNFRSF5, p50 | CD40 molecule, TNF receptor superfamily member 5 |
C00000674
C00003817
C00004631
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00000674
C00004565
C00004631
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00000674
C00003817
C00004631
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00003817
C00004565
C00004631
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00003817
C00004565
C00004631
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00000674
C00003817
C00004631
|
| 2810 | SFN, YWHAS | stratifin |
C00000674
C00004565
C00004631
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00000674
C00004565
C00004631
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00000674
C00003817
C00004631
|
| 8839 | WISP2, CCN5, CT58, CTGF-L | WNT1 inducible signaling pathway protein 2 |
C00000674
C00003817
C00004565
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00000674
C00004631
|
| 9970 | NR1I3, CAR, CAR1, MB67 | nuclear receptor subfamily 1, group I, member 3 |
C00004565
C00004631
|
| 5241 | PGR, NR3C3, PR | progesterone receptor |
C00003817
C00004565
|
| 10891 | PPARGC1A, LEM6, PGC-1(alpha), PGC-1v, PGC1, PGC1A, PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
C00004565
C00004631
|
| 4843 | NOS2, HEP-NOS, INOS, NOS, NOS2A | nitric oxide synthase 2, inducible (EC:1.14.13.39) |
C00004565
C00004631
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00004565
C00004631
|
| 6119 | RPA3, REPA3 | replication protein A3, 14kDa |
C00004565
C00004631
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00003817
C00004631
|
| 6513 | SLC2A1, DYT17, DYT18, DYT9, EIG12, GLUT, GLUT1, GLUT1DS, HTLVR, PED | solute carrier family 2 (facilitated glucose transporter), member 1 |
C00004565
C00004631
|
| 6773 | STAT2, ISGF-3, P113, STAT113 | signal transducer and activator of transcription 2, 113kDa |
C00004565
C00004631
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00000674
C00004631
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00004565
C00004631
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00004565
C00004631
|
| 4586 | MUC5AC, MUC5, TBM, leB | mucin 5AC, oligomeric mucus/gel-forming |
C00004565
C00004631
|
| 4502 | MT2A, MT2 | metallothionein 2A |
C00004565
C00004631
|
| 110357 |
C00004565
C00004631
|
||
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00004565
C00004631
|
| 4297 | KMT2A, ALL-1, CXXC7, HRX, HTRX1, MLL, MLL/GAS7, MLL1A, TET1-MLL, TRX1, WDSTS | lysine (K)-specific methyltransferase 2A (EC:2.1.1.43) |
C00004565
C00004631
|
| 29110 | TBK1, NAK, T2K | TANK-binding kinase 1 (EC:2.7.11.1) |
C00000674
C00004631
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00004565
C00004631
|
| 3567 | IL5, EDF, IL-5, TRF | interleukin 5 (colony-stimulating factor, eosinophil) |
C00004565
C00004631
|
| 3082 | HGF, DFNB39, F-TCF, HGFB, HPTA, SF | hepatocyte growth factor (hepapoietin A; scatter factor) |
C00004565
C00004631
|
| 3043 | HBB, CD113t-C, beta-globin | hemoglobin, beta |
C00004565
C00004631
|
| 2950 | GSTP1, DFN7, FAEES3, GST3, GSTP, PI | glutathione S-transferase pi 1 (EC:2.5.1.18) |
C00004565
C00004631
|
| 2938 | GSTA1, GST2, GSTA1-1, GTH1 | glutathione S-transferase alpha 1 (EC:2.5.1.18) |
C00004565
C00004631
|
| 7402 | UTRN, DMDL, DRP, DRP1 | utrophin |
C00003817
C00004565
|
| 1734 | DIO2, 5DII, D2, DIOII, SelY, TXDI2 | deiodinase, iodothyronine, type II (EC:1.97.1.10) |
C00004565
C00004631
|
| 6783 | SULT1E1, EST, EST-1, ST1E1, STE | sulfotransferase family 1E, estrogen-preferring, member 1 (EC:2.8.2.4) |
C00003817
C00004631
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00003817
C00004631
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00004565
C00004631
|
| 37618 |
C00004565
C00004631
|
||
| 1401 | CRP, PTX1 | C-reactive protein, pentraxin-related |
C00004565
C00004631
|
| 5747 | PTK2, FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK | protein tyrosine kinase 2 (EC:2.7.10.2) |
C00000674
C00004631
|
| 5347 | PLK1, PLK, STPK13 | polo-like kinase 1 (EC:2.7.11.21) |
C00000674
C00004631
|
| 4790 | NFKB1, EBP-1, KBF1, NF-kB1, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
C00000674
C00004631
|
| 1147 | CHUK, IKBKA, IKK-alpha, IKK1, IKKA, NFKBIKA, TCF16 | conserved helix-loop-helix ubiquitous kinase (EC:2.7.11.10) |
C00004565
C00004631
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00004565
C00004631
|
| 5601 | MAPK9, JNK-55, JNK2, JNK2A, JNK2ALPHA, JNK2B, JNK2BETA, PRKM9, SAPK, SAPK1a, p54a, p54aSAPK | mitogen-activated protein kinase 9 (EC:2.7.11.24) |
C00000674
C00004631
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00000674
C00004631
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00000674
C00004631
|
| 3726 | JUNB, AP-1 | jun B proto-oncogene |
C00000674
C00004631
|
| 3091 | HIF1A, HIF-1A, HIF-1alpha, HIF1, HIF1-ALPHA, MOP1, PASD8, bHLHe78 | hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
C00003817
C00004631
|
| 9429 | ABCG2, ABC15, ABCP, BCRP, BCRP1, BMDP, CD338, CDw338, EST157481, GOUT1, MRX, MXR, MXR1, UAQTL1 | ATP-binding cassette, sub-family G (WHITE), member 2 |
C00000674
C00004631
|
| 2064 | ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU, NGL, TKR1 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (EC:2.7.10.1) |
C00003817
C00004631
|
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00000674
C00004631
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00000674
C00004631
|
| 1950 | EGF, HOMG4, URG | epidermal growth factor |
C00000674
C00004631
|
| 1577 | CYP3A5, CP35, CYPIIIA5, P450PCN3, PCN3 | cytochrome P450, family 3, subfamily A, polypeptide 5 (EC:1.14.14.1) |
C00000674
C00004631
|
| 1588 | CYP19A1, ARO, ARO1, CPV1, CYAR, CYP19, CYPXIX, P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00003817
C00004631
|
| 1584 | CYP11B1, CPN1, CYP11B, FHI, P450C11 | cytochrome P450, family 11, subfamily B, polypeptide 1 (EC:1.14.15.4) |
C00003817
C00004631
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00000674
C00004631
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00000674
C00004631
|
| 972 | CD74, DHLAG, HLADG, II, Ia-GAMMA | CD74 molecule, major histocompatibility complex, class II invariant chain |
C00000674
C00004631
|
| 331 | XIAP, API3, BIRC4, IAP-3, ILP1, MIHA, XLP2, hIAP-3, hIAP3 | X-linked inhibitor of apoptosis |
C00000674
C00004631
|
| 890 | CCNA2, CCN1, CCNA | cyclin A2 |
C00000674
C00004631
|
| 4793 | NFKBIB, IKBB, TRIP9 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
C00000674
C00003817
|
| 239 | ALOX12, 12-LOX, 12S-LOX, LOG12 | arachidonate 12-lipoxygenase (EC:1.13.11.31) |
C00004565
C00004631
|
| 5243 | ABCB1, ABC20, CD243, CLCS, GP170, MDR1, P-GP, PGY1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 (EC:3.6.3.44) |
C00004565
C00004631
|
| 330 | BIRC3, AIP1, API2, CIAP2, HAIP1, HIAP1, MALT2, MIHC, RNF49, c-IAP2 | baculoviral IAP repeat containing 3 |
C00003817
C00004631
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00000674
C00004631
|
| 7048 | TGFBR2, AAT3, FAA3, LDS1B, LDS2B, MFS2, RIIC, TAAD2, TGFR-2, TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) (EC:2.7.11.30) |
C00004631
|
| 8309 | ACOX2, BCOX, BRCACOX, BRCOX, THCCox | acyl-CoA oxidase 2, branched chain (EC:1.17.99.3) |
C00004631
|
| 8310 | ACOX3 | acyl-CoA oxidase 3, pristanoyl (EC:1.3.3.6) |
C00004631
|
| 92370 | ACPL2 | acid phosphatase-like 2 (EC:3.1.3.2) |
C00004631
|
| 55 | ACPP, 5'-NT, ACP-3, ACP3, PAP | acid phosphatase, prostate (EC:3.1.3.2 3.1.3.5) |
C00004631
|
| 93953 | ACRC, NAAR1 | acidic repeat containing |
C00004631
|
| 80221 | ACSF2, ACSMW, AVYV493 | acyl-CoA synthetase family member 2 (EC:6.2.1.-) |
C00004631
|
| 51703 | ACSL5, ACS2, ACS5, FACL5 | acyl-CoA synthetase long-chain family member 5 (EC:6.2.1.3) |
C00004631
|
| 23305 | ACSL6, ACS2, FACL6, LACS_6, LACS2, LACS5 | acyl-CoA synthetase long-chain family member 6 (EC:6.2.1.3) |
C00004631
|
| 6296 | ACSM3, SA, SAH | acyl-CoA synthetase medium-chain family member 3 (EC:6.2.1.2) |
C00004631
|
| 54988 | ACSM5 | acyl-CoA synthetase medium-chain family member 5 (EC:6.2.1.2) |
C00004631
|
| 79611 | ACSS3 | acyl-CoA synthetase short-chain family member 3 (EC:6.2.1.1) |
C00004631
|
| 58 | ACTA1, ACTA, ASMA, CFTD, CFTD1, CFTDM, MPFD, NEM1, NEM2, NEM3 | actin, alpha 1, skeletal muscle |
C00004631
|
| 59 | ACTA2, AAT6, ACTSA, MYMY5 | actin, alpha 2, smooth muscle, aorta |
C00004631
|
| 60 | ACTB, BRWS1, PS1TP5BP1 | actin, beta |
C00004631
|
| 10121 | ACTR1A, ARP1, CTRN1 | ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
C00004631
|
| 10120 | ACTR1B, ARP1B, CTRN2, PC3 | ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
C00004631
|
| 64431 | ACTR6, ARP6, HSPC281, MSTP136, hARP6, hARPX | ARP6 actin-related protein 6 homolog (yeast) |
C00004631
|
| 95 | ACY1, ACY-1, ACY1D | aminoacylase 1 (EC:3.5.1.14) |
C00004631
|
| 97 | ACYP1, ACYPE | acylphosphatase 1, erythrocyte (common) type (EC:3.6.1.7) |
C00004631
|
| 100 | ADA | adenosine deaminase (EC:3.5.4.4) |
C00004631
|
| 161823 | ADAL | adenosine deaminase-like (EC:3.5.4.-) |
C00004631
|
| 8038 | ADAM12, MCMP, MCMPMltna, MLTN, MLTNA | ADAM metallopeptidase domain 12 |
C00004631
|
| 8751 | ADAM15, MDC15 | ADAM metallopeptidase domain 15 |
C00004631
|
| 8754 | ADAM9, CORD9, MCMP, MDC9, Mltng | ADAM metallopeptidase domain 9 |
C00004631
|
| 54507 | ADAMTSL4, ECTOL2, TSRC1 | ADAMTS-like 4 |
C00004631
|
| 120 | ADD3, ADDL | adducin 3 (gamma) |
C00004631
|
| 127 | ADH4, ADH-2 | alcohol dehydrogenase 4 (class II), pi polypeptide (EC:1.1.1.1) |
C00004631
|
| 128 | ADH5, ADH-3, ADHX, FALDH, FDH, GSH-FDH, GSNOR | alcohol dehydrogenase 5 (class III), chi polypeptide (EC:1.1.1.1 1.1.1.284) |
C00004631
|
| 130 | ADH6, ADH-5 | alcohol dehydrogenase 6 (class V) (EC:1.1.1.1) |
C00004631
|
| 133 | ADM, AM | adrenomedullin |
C00004631
|
| 23394 | ADNP, ADNP1 | activity-dependent neuroprotector homeobox |
C00004631
|
| 135 | ADORA2A, A2aR, ADORA2, RDC8 | adenosine A2a receptor |
C00004631
|
| 56985 | ADPRM, C17orf48, NBLA03831 | ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent (EC:3.6.1.13 3.6.1.16 3.6.1.53) |
C00004631
|
| 11047 | ADRM1, ARM-1, ARM1, GP110 | adhesion regulating molecule 1 |
C00004631
|
| 122622 | ADSSL1 | adenylosuccinate synthase like 1 (EC:6.3.4.4) |
C00004631
|
| 64782 | AEN, ISG20L1, pp12744 | apoptosis enhancing nuclease |
C00004631
|
| 114896 |
C00004631
|
||
| 79841 | AGBL2 | ATP/GTP binding protein-like 2 |
C00004631
|
| 59272 | ACE2, ACEH | angiotensin I converting enzyme 2 (EC:3.4.17.23) |
C00004631
|
| 178 | AGL, GDE | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase (EC:2.4.1.25 3.2.1.33) |
C00004631
|
| 79814 | AGMAT | agmatine ureohydrolase (agmatinase) (EC:3.5.3.11) |
C00004631
|
| 27161 | AGO2, EIF2C2, Q10 | argonaute RISC catalytic component 2 |
C00004631
|
| 84803 | AGPAT9, AGPAT_10, AGPAT8, GPAT3, LPAAT-theta, MAG1 | 1-acylglycerol-3-phosphate O-acyltransferase 9 (EC:2.3.1.51 2.3.1.15) |
C00004631
|
| 10551 | AGR2, AG2, GOB-4, HAG-2, PDIA17, XAG-2 | anterior gradient 2 |
C00004631
|
| 183 | AGT, ANHU, SERPINA8 | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
C00004631
|
| 23287 | AGTPBP1, CCP1, NNA1 | ATP/GTP binding protein 1 |
C00004631
|
| 185 | AGTR1, AG2S, AGTR1A, AGTR1B, AT1, AT1AR, AT1B, AT1BR, AT1R, AT2R1, AT2R1A, AT2R1B, HAT1R | angiotensin II receptor, type 1 |
C00004631
|
| 189 | AGXT, AGT, AGT1, AGXT1, PH1, SPAT, SPT, TLH6 | alanine-glyoxylate aminotransferase (EC:2.6.1.44 2.6.1.51) |
C00004631
|
| 57491 | AHRR, AHH, AHHR, bHLHe77 | aryl-hydrocarbon receptor repressor |
C00004631
|
| 197 | AHSG, A2HS, AHS, FETUA, HSGA | alpha-2-HS-glycoprotein |
C00004631
|
| 51390 | AIG1, AIG-1, dJ95L4.1 | androgen-induced 1 |
C00004631
|
| 202 | AIM1, CRYBG1, ST4 | absent in melanoma 1 |
C00004631
|
| 205 | AK4, AK_4, AK3, AK3L1, AK3L2 | adenylate kinase 4 (EC:2.7.4.10 2.7.4.6) |
C00004631
|
| 9590 | AKAP12, AKAP250, SSeCKS | A kinase (PRKA) anchor protein 12 |
C00004631
|
| 79647 | AKIRIN1, C1orf108, RP11-781D11.2, STRF2 | akirin 1 |
C00004631
|
| 55122 | AKIRIN2, C6orf166, FBI1, dJ486L4.2 | akirin 2 |
C00004631
|
| 231 | AKR1B1, ADR, ALDR1, ALR2, AR | aldo-keto reductase family 1, member B1 (aldose reductase) (EC:1.1.1.21) |
C00004631
|
| 57016 | AKR1B10, AKR1B11, AKR1B12, ALDRLn, ARL-1, ARL1, HIS, HSI | aldo-keto reductase family 1, member B10 (aldose reductase) (EC:1.1.1.2) |
C00004631
|
| 1646 | AKR1C2, AKR1C-pseudo, BABP, DD, DD2, DDH2, HAKRD, HBAB, MCDR2, SRXY8 | aldo-keto reductase family 1, member C2 (EC:1.3.1.20 1.1.1.357) |
C00004631
|
| 8644 | AKR1C3, DD3, DDX, HA1753, HAKRB, HAKRe, HSD17B5, PGFS, hluPGFS | aldo-keto reductase family 1, member C3 (EC:1.3.1.20 1.1.1.188 1.1.1.239 1.1.1.64 1.1.1.112 1.1.1.357) |
C00004631
|
| 6718 | AKR1D1, 3o5bred, CBAS2, SRD5B1 | aldo-keto reductase family 1, member D1 (EC:1.3.1.3) |
C00004631
|
| 211 | ALAS1, ALAS, ALAS3, ALASH, MIG4 | aminolevulinate, delta-, synthase 1 (EC:2.3.1.37) |
C00004631
|
| 213 | ALB, PRO0883, PRO0903, PRO1341 | albumin |
C00004631
|
| 216 | ALDH1A1, ALDC, ALDH-E1, ALDH1, ALDH11, PUMB1, RALDH1 | aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) |
C00004631
|
| 219 | ALDH1B1, ALDH5, ALDHX | aldehyde dehydrogenase 1 family, member B1 (EC:1.2.1.3) |
C00004631
|
| 217 | ALDH2, ALDH-E2, ALDHI, ALDM | aldehyde dehydrogenase 2 family (mitochondrial) (EC:1.2.1.3) |
C00004631
|
| 224 | ALDH3A2, ALDH10, FALDH, SLS | aldehyde dehydrogenase 3 family, member A2 (EC:1.2.1.3) |
C00004631
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00004631
|
| 7915 | ALDH5A1, SSADH, SSDH | aldehyde dehydrogenase 5 family, member A1 (EC:1.2.1.24) |
C00004631
|
| 4329 | ALDH6A1, MMSADHA, MMSDH | aldehyde dehydrogenase 6 family, member A1 (EC:1.2.1.27 1.2.1.18) |
C00004631
|
| 226 | ALDOA, ALDA, GSD12 | aldolase A, fructose-bisphosphate (EC:4.1.2.13) |
C00004631
|
| 230 | ALDOC, ALDC | aldolase C, fructose-bisphosphate (EC:4.1.2.13) |
C00004631
|
| 84920 | ALG10, ALG10A, DIE2, KCR1 | ALG10, alpha-1,2-glucosyltransferase (EC:2.4.1.256) |
C00004631
|
| 79868 | ALG13, CDG1S, CXorf45, GLT28D1, MDS031, TDRD13, YGL047W | ALG13, UDP-N-acetylglucosaminyltransferase subunit (EC:3.4.19.12 2.4.1.141) |
C00004631
|
| 199857 | ALG14 | ALG14, UDP-N-acetylglucosaminyltransferase subunit |
C00004631
|
| 29929 | ALG6, CDG1C | ALG6, alpha-1,3-glucosyltransferase (EC:2.4.1.267) |
C00004631
|
| 7840 | ALMS1, ALSS | Alstrom syndrome 1 |
C00004631
|
| 241 | ALOX5AP, FLAP | arachidonate 5-lipoxygenase-activating protein |
C00004631
|
| 248 | ALPI, IAP | alkaline phosphatase, intestinal (EC:3.1.3.1) |
C00004631
|
| 115701 | ALPK2, HAK | alpha-kinase 2 |
C00004631
|
| 144193 | AMDHD1 | amidohydrolase domain containing 1 (EC:3.5.2.7) |
C00004631
|
| 9949 | AMMECR1, AMMERC1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
C00004631
|
| 196394 | AMN1 | antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
C00004631
|
| 51421 | AMOTL2, LCCP | angiomotin like 2 |
C00004631
|
| 271 | AMPD2 | adenosine monophosphate deaminase 2 (EC:3.5.4.6) |
C00004631
|
| 272 | AMPD3 | adenosine monophosphate deaminase 3 (EC:3.5.4.6) |
C00004631
|
| 275 | AMT, GCE, GCST, GCVT, NKH | aminomethyltransferase (EC:2.1.2.10) |
C00004631
|
| 10393 | ANAPC10, APC10, DOC1 | anaphase promoting complex subunit 10 |
C00004631
|
| 29945 | ANAPC4, APC4 | anaphase promoting complex subunit 4 |
C00004631
|
| 283 | ANG, ALS9, HEL168, RNASE4, RNASE5 | angiogenin, ribonuclease, RNase A family, 5 (EC:3.1.27.-) |
C00004631
|
| 51129 | ANGPTL4, ANGPTL2, ARP4, FIAF, HARP, HFARP, NL2, PGAR, UNQ171, pp1158 | angiopoietin-like 4 |
C00004631
|
| 287 | ANK2, ANK-2, LQT4, brank-2 | ankyrin 2, neuronal |
C00004631
|
| 63926 | ANKEF1, ANKRD5, dJ839B4.6 | ankyrin repeat and EF-hand domain containing 1 |
C00004631
|
| 56172 | ANKH, ANK, CCAL2, CMDJ, CPPDD, HANK, MANK | ANKH inorganic pyrophosphate transport regulator |
C00004631
|
| 54882 | ANKHD1, MASK, VBARP | ankyrin repeat and KH domain containing 1 |
C00004631
|
| 57037 | ANKMY2, ZMYND20 | ankyrin repeat and MYND domain containing 2 |
C00004631
|
| 57763 | ANKRA2, ANKRA | ankyrin repeat, family A (RFXANK-like), 2 |
C00004631
|
| 55608 | ANKRD10 | ankyrin repeat domain 10 |
C00004631
|
| 23253 | ANKRD12, ANCO-2, ANCO1, GAC-1, Nbla00144 | ankyrin repeat domain 12 |
C00004631
|
| 54522 | ANKRD16 | ankyrin repeat domain 16 |
C00004631
|
| 353322 | ANKRD37, Lrp2bp | ankyrin repeat domain 37 |
C00004631
|
| 257629 | ANKS4B, HARP | ankyrin repeat and sterile alpha motif domain containing 4B |
C00004631
|
| 54443 | ANLN, Scraps, scra | anillin, actin binding protein |
C00004631
|
| 81611 | ANP32E, LANP-L, LANPL | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
C00004631
|
| 290 | ANPEP, APN, CD13, GP150, LAP1, P150, PEPN | alanyl (membrane) aminopeptidase (EC:3.4.11.2) |
C00004631
|
| 118429 | ANTXR2, CMG-2, CMG2, HFS, ISH, JHF | anthrax toxin receptor 2 |
C00004631
|
| 301 | ANXA1, ANX1, LPC1 | annexin A1 |
C00004631
|
| 302 | ANXA2, ANX2, ANX2L4, CAL1H, LIP2, LPC2, LPC2D, P36, PAP-IV | annexin A2 |
C00004631
|
| 306 | ANXA3, ANX3 | annexin A3 (EC:3.1.4.43) |
C00004631
|
| 308 | ANXA5, ANX5, ENX2, PP4, RPRGL3 | annexin A5 |
C00004631
|
| 309 | ANXA6, ANX6, CBP68 | annexin A6 |
C00004631
|
| 314 | AOC2, DAO2, RAO | amine oxidase, copper containing 2 (retina-specific) (EC:1.4.3.21) |
C00004631
|
| 8639 | AOC3, HPAO, SSAO, VAP-1, VAP1 | amine oxidase, copper containing 3 (EC:1.4.3.21) |
C00004631
|
| 316 | AOX1, AO, AOH1 | aldehyde oxidase 1 (EC:1.2.3.1) |
C00004631
|
| 10053 | AP1M2, AP1-mu2, HSMU1B, MU-1B, MU1B, mu2 | adaptor-related protein complex 1, mu 2 subunit |
C00004631
|
| 1174 | AP1S1, AP19, CLAPS1, MEDNIK, SIGMA1A, WUGSC:H_DJ0747G18.2 | adaptor-related protein complex 1, sigma 1 subunit |
C00004631
|
| 10947 | AP3M2, AP47B, CLA20, P47B | adaptor-related protein complex 3, mu 2 subunit |
C00004631
|
| 55745 | AP5M1, C14orf108, MUDENG, Mu5, MuD | adaptor-related protein complex 5, mu 1 subunit |
C00004631
|
| 317 | APAF1, APAF-1, CED4 | apoptotic peptidase activating factor 1 |
C00004631
|
| 9546 | APBA3, MGC:15815, X11L2, mint3 | amyloid beta (A4) precursor protein-binding, family A, member 3 |
C00004631
|
| 323 | APBB2, FE65L, FE65L1 | amyloid beta (A4) precursor protein-binding, family B, member 2 |
C00004631
|
| 324 | APC, BTPS2, DP2, DP2.5, DP3, GS, PPP1R46 | adenomatous polyposis coli |
C00004631
|
| 333 | APLP1, APLP | amyloid beta (A4) precursor-like protein 1 |
C00004631
|
| 116519 | APOA5, APOAV, RAP3 | apolipoprotein A-V |
C00004631
|
| 338 | APOB, FLDB, LDLCQ4 | apolipoprotein B |
C00004631
|
| 9582 | APOBEC3B, A3B, APOBEC1L, ARCD3, ARP4, DJ742C19.2, PHRBNL, bK150C2.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
C00004631
|
| 60489 | APOBEC3G, A3G, ARCD, ARP-9, ARP9, CEM-15, CEM15, bK150C2.7, dJ494G10.1 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (EC:3.5.4.5) |
C00004631
|
| 341 | APOC1 | apolipoprotein C-I |
C00004631
|
| 344 | APOC2, APO-CII, APOC-II | apolipoprotein C-II |
C00004631
|
| 345 | APOC3, APOCIII, HALP2 | apolipoprotein C-III |
C00004631
|
| 348 | APOE, AD2, LDLCQ5, LPG | apolipoprotein E |
C00004631
|
| 319 | APOF, Apo-F, LTIP | apolipoprotein F |
C00004631
|
| 350 | APOH, B2G1, B2GP1, BG | apolipoprotein H (beta-2-glycoprotein I) |
C00004631
|
| 81575 | APOLD1, VERGE | apolipoprotein L domain containing 1 |
C00004631
|
| 55937 | APOM, G3a, HSPC336, NG20, apo-M | apolipoprotein M |
C00004631
|
| 351 | APP, AAA, ABETA, ABPP, AD1, APPI, CTFgamma, CVAP, PN-II, PN2 | amyloid beta (A4) precursor protein |
C00004631
|
| 55198 | APPL2, DIP13B | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
C00004631
|
| 282679 | AQP11, AQPX1 | aquaporin 11 |
C00004631
|
| 360 | AQP3, AQP-3, GIL | aquaporin 3 (Gill blood group) |
C00004631
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00004631
|
| 377 | ARF3 | ADP-ribosylation factor 3 |
C00004631
|
| 10565 | ARFGEF1, ARFGEP1, BIG1, P200 | ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
C00004631
|
| 383 | ARG1 | arginase 1 (EC:3.5.3.1) |
C00004631
|
| 384 | ARG2 | arginase 2 (EC:3.5.3.1) |
C00004631
|
| 9824 | ARHGAP11A, GAP_(1-12) | Rho GTPase activating protein 11A |
C00004631
|
| 55114 | ARHGAP17, MST066, MST110, MSTP038, MSTP066, MSTP110, NADRIN, PP367, PP4534, RICH1, RICH1B, WBP15 | Rho GTPase activating protein 17 |
C00004631
|
| 93663 | ARHGAP18, MacGAP, SENEX, bA307O14.2 | Rho GTPase activating protein 18 |
C00004631
|
| 84986 | ARHGAP19 | Rho GTPase activating protein 19 |
C00004631
|
| 9411 | ARHGAP29, PARG1, RP11-255E17.1 | Rho GTPase activating protein 29 |
C00004631
|
| 9912 | ARHGAP44, NPC-A-10, RICH2 | Rho GTPase activating protein 44 |
C00004631
|
| 84837 | ARHGAP5-AS1, C14orf128 | ARHGAP5 antisense RNA 1 (head to head) |
C00004631
|
| 397 | ARHGDIB, D4, GDIA2, GDID4, LYGDI, Ly-GDI, RAP1GN1, RhoGDI2 | Rho GDP dissociation inhibitor (GDI) beta |
C00004631
|
| 64283 | ARHGEF28, RGNEF, RIP2, p190RHOGEF | Rho guanine nucleotide exchange factor (GEF) 28 |
C00004631
|
| 50650 | ARHGEF3, GEF3, STA3, XPLN | Rho guanine nucleotide exchange factor (GEF) 3 |
C00004631
|
| 196528 | ARID2, BAF200, p200 | AT rich interactive domain 2 (ARID, RFX-like) |
C00004631
|
| 200894 | ARL13B, ARL2L1, JBTS8 | ADP-ribosylation factor-like 13B |
C00004631
|
| 54622 | ARL15, ARFRP2 | ADP-ribosylation factor-like 15 |
C00004631
|
| 379 | ARL4D, ARF4L, ARL6 | ADP-ribosylation factor-like 4D |
C00004631
|
| 84100 | ARL6, BBS3, RP55 | ADP-ribosylation factor-like 6 |
C00004631
|
| 10550 | ARL6IP5, DERP11, GTRAP3-18, JWA, PRAF3, addicsin, hp22, jmx | ADP-ribosylation-like factor 6 interacting protein 5 |
C00004631
|
| 64860 | ARMCX5 | armadillo repeat containing, X-linked 5 |
C00004631
|
| 406 | ARNTL, BMAL1, BMAL1c, JAP3, MOP3, PASD3, TIC, bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like |
C00004631
|
| 56938 | ARNTL2, BMAL2, CLIF, MOP9, PASD9, bHLHe6 | aryl hydrocarbon receptor nuclear translocator-like 2 |
C00004631
|
| 408 | ARRB1, ARB1, ARR1 | arrestin, beta 1 |
C00004631
|
| 27106 | ARRDC2, CLONE24945 | arrestin domain containing 2 |
C00004631
|
| 57561 | ARRDC3, TLIMP | arrestin domain containing 3 |
C00004631
|
| 410 | ARSA, MLD | arylsulfatase A (EC:3.1.6.8) |
C00004631
|
| 415 | ARSE, ASE, CDPX, CDPX1, CDPXR | arylsulfatase E (chondrodysplasia punctata 1) (EC:3.1.6.1) |
C00004631
|
| 22901 | ARSG | arylsulfatase G |
C00004631
|
| 420 | ART4, ARTC4, CD297, DO, DOK1 | ADP-ribosyltransferase 4 (EC:2.4.2.31) |
C00004631
|
| 421 | ARVCF | armadillo repeat gene deleted in velocardiofacial syndrome |
C00004631
|
| 427 | ASAH1, AC, ACDase, ASAH, PHP, PHP32, SMAPME | N-acylsphingosine amidohydrolase (acid ceramidase) 1 (EC:3.5.1.23) |
C00004631
|
| 8853 | ASAP2, AMAP2, CENTB3, DDEF2, PAG3, PAP, Pap-alpha, SHAG1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
C00004631
|
| 339201 | ASB16-AS1, C17orf65 | ASB16 antisense RNA 1 |
C00004631
|
| 51676 | ASB2, ASB-2 | ankyrin repeat and SOCS box containing 2 |
C00004631
|
| 140462 | ASB9 | ankyrin repeat and SOCS box containing 9 |
C00004631
|
| 10973 | ASCC3, ASC1p200, HELIC1, RNAH | activating signal cointegrator 1 complex subunit 3 (EC:3.6.4.12) |
C00004631
|
| 55723 | ASF1B, CIA-II | anti-silencing function 1B histone chaperone |
C00004631
|
| 432 | ASGR1, ASGPR, ASGPR1, CLEC4H1, HL-1 | asialoglycoprotein receptor 1 |
C00004631
|
| 444 | ASPH, AAH, BAH, CASQ2BP1, HAAH, JCTN, junctin | aspartate beta-hydroxylase (EC:1.14.11.16) |
C00004631
|
| 253982 | ASPHD1 | aspartate beta-hydroxylase domain containing 1 |
C00004631
|
| 259266 | ASPM, ASP, Calmbp1, MCPH5 | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
C00004631
|
| 151516 | ASPRV1, MUNO, SASP, SASPase, Taps | aspartic peptidase, retroviral-like 1 |
C00004631
|
| 79058 | ASPSCR1, ASPCR1, ASPL, ASPS, RCC17, TUG, UBXD9, UBXN9 | alveolar soft part sarcoma chromosome region, candidate 1 |
C00004631
|
| 80150 | ASRGL1, ALP, ALP1, CRASH | asparaginase like 1 (EC:3.5.1.1 3.4.19.5) |
C00004631
|
| 445 | ASS1, ASS, CTLN1 | argininosuccinate synthase 1 (EC:6.3.4.5) |
C00004631
|
| 28990 | ASTE1 | asteroid homolog 1 (Drosophila) |
C00004631
|
| 55726 | ASUN, C12orf11, GCT1, Mat89Bb, NET48, SPATA30 | asunder spermatogenesis regulator |
C00004631
|
| 467 | ATF3 | activating transcription factor 3 |
C00004631
|
| 55729 | ATF7IP, AM, ATF-IP, MCAF, MCAF1, p621 | activating transcription factor 7 interacting protein |
C00004631
|
| 89849 | ATG16L2, ATG16B, WDR80 | autophagy related 16-like 2 (S. cerevisiae) |
C00004631
|
| 23130 | ATG2A | autophagy related 2A |
C00004631
|
| 64422 | ATG3, APG3, APG3-LIKE, APG3L, PC3-96 | autophagy related 3 |
C00004631
|
| 51062 | ATL1, AD-FSP, FSP1, GBP3, HSN1D, SPG3, SPG3A, atlastin1 | atlastin GTPase 1 |
C00004631
|
| 64225 | ATL2, ARL3IP2, ARL6IP2, atlastin2 | atlastin GTPase 2 |
C00004631
|
| 57130 | ATP13A1, ATP13A | ATPase type 13A1 (EC:3.6.3.8) |
C00004631
|
| 476 | ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide (EC:3.6.3.9) |
C00004631
|
| 483 | ATP1B3, ATPB-3, CD298 | ATPase, Na+/K+ transporting, beta 3 polypeptide (EC:3.6.3.9) |
C00004631
|
| 487 | ATP2A1, ATP2A, SERCA1 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 (EC:3.6.3.8) |
C00004631
|
| 516 | ATP5G1, ATP5A, ATP5G | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
C00004631
|
| 533 | ATP6V0B, ATP6F, HATPL, VMA16 | ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (EC:3.6.3.14) |
C00004631
|
| 528 | ATP6V1C1, ATP6C, ATP6D, VATC, Vma5 | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 (EC:3.6.3.14) |
C00004631
|
| 90423 | ATP6V1E2, ATP6E1, ATP6EL2, ATP6V1EL2, VMA4 | ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
C00004631
|
| 538 | ATP7A, DSMAX, MK, MNK, SMAX3 | ATPase, Cu++ transporting, alpha polypeptide (EC:3.6.3.54) |
C00004631
|
| 540 | ATP7B, PWD, WC1, WD, WND | ATPase, Cu++ transporting, beta polypeptide (EC:3.6.3.54) |
C00004631
|
| 5205 | ATP8B1, ATPIC, BRIC, FIC1, ICP1, PFIC, PFIC1 | ATPase, aminophospholipid transporter, class I, type 8B, member 1 (EC:3.6.3.1) |
C00004631
|
| 10079 | ATP9A, ATPIIA | ATPase, class II, type 9A (EC:3.6.3.1) |
C00004631
|
| 545 | ATR, FCTCS, FRP1, MEC1, SCKL, SCKL1 | ataxia telangiectasia and Rad3 related (EC:2.7.11.1) |
C00004631
|
| 4287 | ATXN3, AT3, ATX3, JOS, MJD, MJD1, SCA3 | ataxin 3 (EC:3.4.19.12) |
C00004631
|
| 549 | AUH | AU RNA binding protein/enoyl-CoA hydratase (EC:4.2.1.18) |
C00004631
|
| 6790 | AURKA, AIK, ARK1, AURA, AURORA2, BTAK, PPP1R47, STK15, STK6, STK7 | aurora kinase A (EC:2.7.11.1) |
C00004631
|
| 9212 | AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, PPP1R48, STK12, STK5, aurkb-sv1, aurkb-sv2 | aurora kinase B (EC:2.7.11.1) |
C00004631
|
| 8313 | AXIN2, AXIL, ODCRCS | axin 2 |
C00004631
|
| 558 | AXL, JTK11, UFO | AXL receptor tyrosine kinase (EC:2.7.10.1) |
C00004631
|
| 145173 | B3GALTL, B3GLCT, B3GTL, B3Glc-T, Gal-T, beta3Glc-T | beta 1,3-galactosyltransferase-like |
C00004631
|
| 26229 | B3GAT3, GLCATI | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (EC:2.4.1.135) |
C00004631
|
| 146712 | B3GNTL1, B3GNT8 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (EC:2.4.1.-) |
C00004631
|
| 2583 | B4GALNT1, GALGT, GALNACT, GalNAc-T, SPG26 | beta-1,4-N-acetyl-galactosaminyl transferase 1 (EC:2.4.1.92) |
C00004631
|
| 8702 | B4GALT4, B4Gal-T4, beta4Gal-T4 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 (EC:2.4.1.90 2.4.1.275) |
C00004631
|
| 570 | BAAT, BACAT, BAT | bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) (EC:2.3.1.65 3.1.2.2) |
C00004631
|
| 572 | BAD, BBC2, BCL2L8 | BCL2-associated agonist of cell death |
C00004631
|
| 9532 | BAG2, BAG-2, dJ417I1.2 | BCL2-associated athanogene 2 |
C00004631
|
| 9530 | BAG4, BAG-4, SODD | BCL2-associated athanogene 4 |
C00004631
|
| 578 | BAK1, BAK, BAK-LIKE, BCL2L7, CDN1 | BCL2-antagonist/killer 1 |
C00004631
|
| 25805 | BAMBI, NMA | BMP and activin membrane-bound inhibitor |
C00004631
|
| 11177 | BAZ1A, ACF1, WALp1, WCRF180, hACF1 | bromodomain adjacent to zinc finger domain, 1A |
C00004631
|
| 29994 | BAZ2B, WALp4 | bromodomain adjacent to zinc finger domain, 2B |
C00004631
|
| 166379 | BBS12, C4orf24 | Bardet-Biedl syndrome 12 |
C00004631
|
| 583 | BBS2, BBS | Bardet-Biedl syndrome 2 |
C00004631
|
| 585 | BBS4 | Bardet-Biedl syndrome 4 |
C00004631
|
| 129880 | BBS5 | Bardet-Biedl syndrome 5 |
C00004631
|
| 27241 | BBS9, B1, C18, D1, PTHB1 | Bardet-Biedl syndrome 9 |
C00004631
|
| 4059 | BCAM, AU, CD239, LU, MSK19 | basal cell adhesion molecule (Lutheran blood group) |
C00004631
|
| 9564 | BCAR1, CAS, CAS1, CASS1, CRKAS, P130Cas | breast cancer anti-estrogen resistance 1 |
C00004631
|
| 8412 | BCAR3, NSP2, SH2D3B | breast cancer anti-estrogen resistance 3 |
C00004631
|
| 586 | BCAT1, BCATC, BCT1, ECA39, MECA39, PNAS121, PP18 | branched chain amino-acid transaminase 1, cytosolic (EC:2.6.1.42) |
C00004631
|
| 10295 | BCKDK, BCKDKD, BDK | branched chain ketoacid dehydrogenase kinase (EC:2.7.11.4) |
C00004631
|
| 79370 | BCL2L14, BCLG | BCL2-like 14 (apoptosis facilitator) |
C00004631
|
| 599 | BCL2L2, BCL-W, BCL2-L-2, BCLW, PPP1R51 | BCL2-like 2 |
C00004631
|
| 54880 | BCOR, ANOP2, MAA2, MCOPS2 | BCL6 corepressor |
C00004631
|
| 613 | BCR, ALL, BCR1, CML, D22S11, D22S662, PHL | breakpoint cluster region (EC:2.7.11.1) |
C00004631
|
| 622 | BDH1, BDH, SDR9C1 | 3-hydroxybutyrate dehydrogenase, type 1 (EC:1.1.1.30) |
C00004631
|
| 56898 | BDH2, DHRS6, EFA6R, PRO20933, SDR15C1, UCPA-OR, UNQ6308 | 3-hydroxybutyrate dehydrogenase, type 2 (EC:1.1.1.30) |
C00004631
|
| 55859 | BEX1, BEX2, HBEX2, HGR74-h | brain expressed, X-linked 1 |
C00004631
|
| 51283 | BFAR, BAR, RNF47 | bifunctional apoptosis regulator |
C00004631
|
| 632 | BGLAP, BGP, OC, OCN | bone gamma-carboxyglutamate (gla) protein |
C00004631
|
| 8553 | BHLHE40, BHLHB2, DEC1, HLHB2, SHARP-2, STRA13, Stra14 | basic helix-loop-helix family, member e40 |
C00004631
|
| 635 | BHMT, BHMT1 | betaine--homocysteine S-methyltransferase (EC:2.1.1.5) |
C00004631
|
| 23743 | BHMT2 | betaine--homocysteine S-methyltransferase 2 (EC:2.1.1.10) |
C00004631
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00004631
|
| 638 | BIK, BIP1, BP4, NBK | BCL2-interacting killer (apoptosis-inducing) |
C00004631
|
| 84320 | ACBD6 | acyl-CoA binding domain containing 6 |
C00004631
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00004631
|
| 54841 | BIVM | basic, immunoglobulin-like variable motif containing |
C00004631
|
| 641 | BLM, BS, RECQ2, RECQL2, RECQL3 | Bloom syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00004631
|
| 282991 | BLOC1S2, BLOS2, CEAP, CEAP11, RP11-316M21.4 | biogenesis of lysosomal organelles complex-1, subunit 2 |
C00004631
|
| 649 | BMP1, OI13, PCOLC, PCP, PCP2, TLD | bone morphogenetic protein 1 (EC:3.4.24.19) |
C00004631
|
| 650 | BMP2, BDA2, BMP2A | bone morphogenetic protein 2 |
C00004631
|
| 55589 | BMP2K, BIKE | BMP2 inducible kinase (EC:2.7.11.1) |
C00004631
|
| 652 | BMP4, BMP2B, BMP2B1, MCOPS6, OFC11, ZYME | bone morphogenetic protein 4 |
C00004631
|
| 353500 | BMP8A | bone morphogenetic protein 8a |
C00004631
|
| 657 | BMPR1A, 10q23del, ACVRLK3, ALK3, CD292, SKR5 | bone morphogenetic protein receptor, type IA (EC:2.7.11.30) |
C00004631
|
| 664 | BNIP3, NIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 |
C00004631
|
| 665 | BNIP3L, BNIP3a, NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like |
C00004631
|
| 79866 | BORA, C13orf34 | bora, aurora kinase A activator |
C00004631
|
| 80341 | BPIFB2, BPIL1, C20orf184, LPLUNC2, RYSR, dJ726C3.2 | BPI fold containing family B, member 2 |
C00004631
|
| 2186 | BPTF, FAC1, FALZ, NURF301 | bromodomain PHD finger transcription factor |
C00004631
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00004631
|
| 675 | BRCA2, BRCC2, BROVCA2, FACD, FAD, FAD1, FANCB, FANCD, FANCD1, GLM3, PNCA2 | breast cancer 2, early onset |
C00004631
|
| 6046 | BRD2, D6S113E, FSH, FSRG1, NAT, RING3, RNF3 | bromodomain containing 2 |
C00004631
|
| 29117 | BRD7, BP75, CELTIX1, NAG4 | bromodomain containing 7 |
C00004631
|
| 676 | BRDT, BRD6, CT9 | bromodomain, testis-specific |
C00004631
|
| 26580 | BSCL2, GNG3LG, HMN5, SPG17 | Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
C00004631
|
| 682 | BSG, 5F7, CD147, EMMPRIN, M6, OK, TCSF | basigin |
C00004631
|
| 8927 | BSN, ZNF231 | bassoon presynaptic cytomatrix protein |
C00004631
|
| 121551 | BTBD11, ABTB2B | BTB (POZ) domain containing 11 |
C00004631
|
| 55643 | BTBD2 | BTB (POZ) domain containing 2 |
C00004631
|
| 91408 | BTF3L4 | basic transcription factor 3-like 4 |
C00004631
|
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative |
C00004631
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00004631
|
| 699 | BUB1, BUB1A, BUB1L, hBUB1 | BUB1 mitotic checkpoint serine/threonine kinase (EC:2.7.11.1) |
C00004631
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00004631
|
| 9184 | BUB3, BUB3L, hBUB3 | BUB3 mitotic checkpoint protein |
C00004631
|
| 11067 | C10orf10, DEPP, FIG | chromosome 10 open reading frame 10 |
C00004631
|
| 83938 | C10orf11, OCA5, OCA7 | chromosome 10 open reading frame 11 |
C00004631
|
| 119032 | C10orf32, RP11-753C18.6 | chromosome 10 open reading frame 32 |
C00004631
|
| 91894 | C11orf52 | chromosome 11 open reading frame 52 |
C00004631
|
| 28970 | C11orf54, PTD012 | chromosome 11 open reading frame 54 |
C00004631
|
| 220042 | C11orf82, noxin | chromosome 11 open reading frame 82 |
C00004631
|
| 387763 | C11orf96, AG2 | chromosome 11 open reading frame 96 |
C00004631
|
| 90488 | C12orf23 | chromosome 12 open reading frame 23 |
C00004631
|
| 57103 | C12orf5, FR2BP, TIGAR | chromosome 12 open reading frame 5 (EC:3.1.3.46) |
C00004631
|
| 144608 | C12orf60 | chromosome 12 open reading frame 60 |
C00004631
|
| 387882 | C12orf75, AGD3, OCC-1, OCC1 | chromosome 12 open reading frame 75 |
C00004631
|
| 11161 | C14orf1, ERG28, NET51 | chromosome 14 open reading frame 1 |
C00004631
|
| 122525 | C14orf28, DRIP-1, DRIP1, c14_5270 | chromosome 14 open reading frame 28 |
C00004631
|
| 84419 | C15orf48, FOAP-11, NMES1 | chromosome 15 open reading frame 48 |
C00004631
|
| 89927 | C16orf45 | chromosome 16 open reading frame 45 |
C00004631
|
| 64755 | C16orf58, RUS | chromosome 16 open reading frame 58 |
C00004631
|
| 388272 | C16orf87 | chromosome 16 open reading frame 87 |
C00004631
|
| 388327 | C17orf100 | chromosome 17 open reading frame 100 |
C00004631
|
| 284018 | C17orf58 | chromosome 17 open reading frame 58 |
C00004631
|
| 79415 | C17orf62 | chromosome 17 open reading frame 62 |
C00004631
|
| 64149 | C17orf75, NJMU-R1 | chromosome 17 open reading frame 75 |
C00004631
|
| 125144 | C17orf76-AS1, C17orf45, NCRNA00188, TSAP19, FAM211A-AS1 | C17orf76 antisense RNA 1 |
C00004631
|
| 162681 | C18orf54, LAS2 | chromosome 18 open reading frame 54 |
C00004631
|
| 147685 | C19orf18 | chromosome 19 open reading frame 18 |
C00004631
|
| 148223 | C19orf25 | chromosome 19 open reading frame 25 |
C00004631
|
| 284422 | C19orf77 | chromosome 19 open reading frame 77 |
C00004631
|
| 55908 | C19orf80, ANGPTL8, PRO1185, PVPA599, RIFL, TD26 | chromosome 19 open reading frame 80 |
C00004631
|
| 56913 | C1GALT1, C1GALT, T-synthase | core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 (EC:2.4.1.122) |
C00004631
|
| 29071 | C1GALT1C1, C1GALT2, C38H2-L1, COSMC, MST143, TNPS | C1GALT1-specific chaperone 1 |
C00004631
|
| 79762 | C1orf115, RP11-322F10.4 | chromosome 1 open reading frame 115 |
C00004631
|
| 128346 | C1orf162 | chromosome 1 open reading frame 162 |
C00004631
|
| 127703 | C1orf216 | chromosome 1 open reading frame 216 |
C00004631
|
| 57035 | C1orf63, DJ465N24.2.1, NPD014, RP3-465N24.4 | chromosome 1 open reading frame 63 |
C00004631
|
| 149563 | C1orf64, ERRF, RP11-5P18.4 | chromosome 1 open reading frame 64 |
C00004631
|
| 112770 | C1orf85, NCU-G1 | chromosome 1 open reading frame 85 |
C00004631
|
| 56683 | C21orf59, C21orf48, CILD26, FBB18 | chromosome 21 open reading frame 59 |
C00004631
|
| 25966 | C2CD2, C21orf25, C21orf258, TMEM24L | C2 calcium-dependent domain containing 2 |
C00004631
|
| 145741 | C2CD4A, FAM148A, NLF1 | C2 calcium-dependent domain containing 4A |
C00004631
|
| 9847 | C2CD5, CDP138, KIAA0528 | C2 calcium-dependent domain containing 5 |
C00004631
|
| 257407 | C2orf72 | chromosome 2 open reading frame 72 |
C00004631
|
| 130355 | C2orf76, AIM29 | chromosome 2 open reading frame 76 |
C00004631
|
| 389084 | C2orf82, UNQ830 | chromosome 2 open reading frame 82 |
C00004631
|
| 285315 | C3orf33, AC3-33 | chromosome 3 open reading frame 33 |
C00004631
|
| 339883 | C3orf35, APRG1 | chromosome 3 open reading frame 35 |
C00004631
|
| 285237 | C3orf38 | chromosome 3 open reading frame 38 |
C00004631
|
| 79669 | C3orf52, TTMP | chromosome 3 open reading frame 52 |
C00004631
|
| 722 | C4BPA, C4BP, PRP | complement component 4 binding protein, alpha |
C00004631
|
| 725 | C4BPB, C4BP | complement component 4 binding protein, beta |
C00004631
|
| 54969 | C4orf27 | chromosome 4 open reading frame 27 |
C00004631
|
| 401152 | C4orf3 | chromosome 4 open reading frame 3 |
C00004631
|
| 401115 | C4orf48, CHR4_55 | chromosome 4 open reading frame 48 |
C00004631
|
| 727 | C5, C5a, C5b, CPAMD4 | complement component 5 |
C00004631
|
| 728 | C5AR1, C5A, C5AR, C5R1, CD88 | complement component 5a receptor 1 |
C00004631
|
| 27202 | C5AR2, C5L2, GPF77, GPR77 | complement component 5a receptor 2 |
C00004631
|
| 375444 | C5orf34 | chromosome 5 open reading frame 34 |
C00004631
|
| 63920 | C5orf54, Buster3, ZBED8 | chromosome 5 open reading frame 54 |
C00004631
|
| 221491 | C6orf1, LBH | chromosome 6 open reading frame 1 |
C00004631
|
| 79624 | C6orf211 | chromosome 6 open reading frame 211 |
C00004631
|
| 441150 | C6orf226 | chromosome 6 open reading frame 226 |
C00004631
|
| 50854 | C6orf48, D6S57, G8 | chromosome 6 open reading frame 48 |
C00004631
|
| 347744 | C6orf52 | chromosome 6 open reading frame 52 |
C00004631
|
| 135154 | C6orf57 | chromosome 6 open reading frame 57 |
C00004631
|
| 84310 | C7orf50, YCR016W | chromosome 7 open reading frame 50 |
C00004631
|
| 154791 | C7orf55, FMC1 | chromosome 7 open reading frame 55 |
C00004631
|
| 154743 | C7orf60 | chromosome 7 open reading frame 60 |
C00004631
|
| 731 | C8A | complement component 8, alpha polypeptide |
C00004631
|
| 732 | C8B, C82 | complement component 8, beta polypeptide |
C00004631
|
| 733 | C8G, C8C | complement component 8, gamma polypeptide |
C00004631
|
| 56892 | C8orf4, TC-1, TC1 | chromosome 8 open reading frame 4 |
C00004631
|
| 401466 | C8orf59 | chromosome 8 open reading frame 59 |
C00004631
|
| 401546 | C9orf152, bA470J20.2 | chromosome 9 open reading frame 152 |
C00004631
|
| 138199 | C9orf41 | chromosome 9 open reading frame 41 |
C00004631
|
| 138241 | C9orf85 | chromosome 9 open reading frame 85 |
C00004631
|
| 203197 | C9orf91, RP11-402G3.2 | chromosome 9 open reading frame 91 |
C00004631
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00004631
|
| 91768 | CABLES1, CABL1, CABLES, HsT2563, IK3-1 | Cdk5 and Abl enzyme substrate 1 |
C00004631
|
| 27101 | CACYBP, GIG5, RP1-102G20.6, S100A6BP, SIP | calcyclin binding protein |
C00004631
|
| 790 | CAD | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (EC:2.1.3.2 3.5.2.3 6.3.5.5) |
C00004631
|
| 23705 | CADM1, BL2, IGSF4, IGSF4A, NECL2, Necl-2, RA175, ST17, SYNCAM, TSLC1, sTSLC-1, sgIGSF, synCAM1 | cell adhesion molecule 1 |
C00004631
|
| 93664 | CADPS2 | Ca++-dependent secretion activator 2 |
C00004631
|
| 8536 | CAMK1, CAMKI | calcium/calmodulin-dependent protein kinase I (EC:2.7.11.17) |
C00004631
|
| 79823 | CAMKMT, C2orf34, CLNMT, CaM_KMT, Cam, KMT | calmodulin-lysine N-methyltransferase (EC:2.1.1.60) |
C00004631
|
| 57662 | CAMSAP3, KIAA1543, NEZHA | calmodulin regulated spectrin-associated protein family, member 3 |
C00004631
|
| 23125 | CAMTA2 | calmodulin binding transcription activator 2 |
C00004631
|
| 55832 | CAND1, TIP120, TIP120A | cullin-associated and neddylation-dissociated 1 |
C00004631
|
| 6650 | CAPN15, SOLH | calpain 15 |
C00004631
|
| 824 | CAPN2, CANP2, CANPL2, CANPml, mCANP | calpain 2, (m/II) large subunit (EC:3.4.22.53) |
C00004631
|
| 65981 | CAPRIN2, C1QDC1, EEG-1, EEG1, RNG140 | caprin family member 2 |
C00004631
|
| 399726 | CASC10, C10orf114, bA418C1.3 | cancer susceptibility candidate 10 |
C00004631
|
| 57082 | CASC5, AF15Q14, CT29, D40, KNL1, PPP1R55, Spc7, hKNL-1, hSpc105 | cancer susceptibility candidate 5 |
C00004631
|
| 64921 | CASD1, C7orf12, NBLA04196 | CAS1 domain containing 1 |
C00004631
|
| 834 | CASP1, ICE, IL1BC, P45 | caspase 1, apoptosis-related cysteine peptidase (EC:3.4.22.36) |
C00004631
|
| 839 | CASP6, MCH2 | caspase 6, apoptosis-related cysteine peptidase (EC:3.4.22.59) |
C00004631
|
| 39 | ACAT2 | acetyl-CoA acetyltransferase 2 (EC:2.3.1.9) |
C00004631
|
| 38 | ACAT1, ACAT, MAT, T2, THIL | acetyl-CoA acetyltransferase 1 (EC:2.3.1.9) |
C00004631
|
| 37 | ACADVL, ACAD6, LCACD, VLCAD | acyl-CoA dehydrogenase, very long chain (EC:1.3.8.9) |
C00004631
|
| 347732 | CATSPER3, CACRC | cation channel, sperm associated 3 |
C00004631
|
| 857 | CAV1, BSCL3, CGL3, MSTP085, PPH3, VIP21, CAV | caveolin 1, caveolae protein, 22kDa |
C00004631
|
| 873 | CBR1, CBR, SDR21C1, hCBR1 | carbonyl reductase 1 (EC:1.1.1.189 1.1.1.197 1.1.1.184) |
C00004631
|
| 84869 | CBR4, SDR45C1 | carbonyl reductase 4 (EC:1.1.1.-) |
C00004631
|
| 875 | CBS, HIP4 | cystathionine-beta-synthase (EC:4.2.1.22) |
C00004631
|
| 23468 | CBX5, HP1, HP1A | chromobox homolog 5 |
C00004631
|
| 23466 | CBX6 | chromobox homolog 6 |
C00004631
|
| 200014 | CC2D1B | coiled-coil and C2 domain containing 1B |
C00004631
|
| 56267 | CCBL2, KAT3, KATIII, RP4-531M19.2 | cysteine conjugate-beta lyase 2 (EC:2.6.1.7 4.4.1.13 2.6.1.63) |
C00004631
|
| 55013 | CCDC109B, MCUb | coiled-coil domain containing 109B |
C00004631
|
| 90693 | CCDC126 | coiled-coil domain containing 126 |
C00004631
|
| 81576 | CCDC130 | coiled-coil domain containing 130 |
C00004631
|
| 165055 | CCDC138 | coiled-coil domain containing 138 |
C00004631
|
| 80071 | CCDC15 | coiled-coil domain containing 15 |
C00004631
|
| 149483 | CCDC17 | coiled-coil domain containing 17 |
C00004631
|
| 51134 | CCDC41, Cep83, NY-REN-58 | coiled-coil domain containing 41 |
C00004631
|
| 100286684 |
C00004631
|
||
| 237859 |
C00004631
|
||
| 84660 | CCDC62, CT109, ERAP75, TSP-NY | coiled-coil domain containing 62 |
C00004631
|
| 80323 | CCDC68, SE57-1 | coiled-coil domain containing 68 |
C00004631
|
| 26112 | CCDC69 | coiled-coil domain containing 69 |
C00004631
|
| 60494 | CCDC81 | coiled-coil domain containing 81 |
C00004631
|
| 55704 | CCDC88A, APE, GIRDIN, GIV, GRDN, HkRP1, KIAA1212 | coiled-coil domain containing 88A |
C00004631
|
| 80212 | CCDC92 | coiled-coil domain containing 92 |
C00004631
|
| 54535 | CCHCR1, C6orf18, HCR, SBP | coiled-coil alpha-helical rod protein 1 |
C00004631
|
| 6359 | CCL15, HCC-2, HMRP-2B, LKN-1, LKN1, MIP-1_delta, MIP-1D, MIP-5, MRP-2B, NCC-3, NCC3, SCYA15, SCYL3, SY15 | chemokine (C-C motif) ligand 15 |
C00004631
|
| 6360 | CCL16, CKb12, HCC-4, ILINCK, LCC-1, LEC, LMC, Mtn-1, NCC-4, NCC4, SCYA16, SCYL4 | chemokine (C-C motif) ligand 16 |
C00004631
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00004631
|
| 36 | ACADSB, 2-MEBCAD, ACAD7, SBCAD | acyl-CoA dehydrogenase, short/branched chain (EC:1.3.8.5) |
C00004631
|
| 57820 | CCNB1IP1, C14orf18, HEI10 | cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
C00004631
|
| 9133 | CCNB2, HsT17299 | cyclin B2 |
C00004631
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00004631
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00004631
|
| 896 | CCND3 | cyclin D3 |
C00004631
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00004631
|
| 9134 | CCNE2, CYCE2 | cyclin E2 |
C00004631
|
| 899 | CCNF, FBX1, FBXO1 | cyclin F |
C00004631
|
| 901 | CCNG2 | cyclin G2 |
C00004631
|
| 9236 | CCPG1, CPR8 | cell cycle progression 1 |
C00004631
|
| 1235 | CCR6, BN-1, C-C_CKR-6, CC-CKR-6, CCR-6, CD196, CKR-L3, CKRL3, CMKBR6, DCR2, DRY6, GPR29, GPRCY4, STRL22 | chemokine (C-C motif) receptor 6 |
C00004631
|
| 1236 | CCR7, BLR2, CD197, CDw197, CMKBR7, EBI1 | chemokine (C-C motif) receptor 7 |
C00004631
|
| 135228 | CD109, CPAMD7, RP11-525G3.1 | CD109 molecule |
C00004631
|
| 977 | CD151, GP27, MER2, PETA-3, RAPH, SFA1, TSPAN24 | CD151 molecule (Raph blood group) |
C00004631
|
| 100133941 | CD24, CD24A | CD24 molecule |
C00004631
|
| 9936 | CD302, BIMLEC, CLEC13A, DCL-1, DCL1 | CD302 molecule |
C00004631
|
| 915 | CD3D, CD3-DELTA, T3D | CD3d molecule, delta (CD3-TCR complex) |
C00004631
|
| 84129 | ACAD11, ACAD-11 | acyl-CoA dehydrogenase family, member 11 |
C00004631
|
| 4179 | CD46, AHUS2, MCP, MIC10, TLX, TRA2.10 | CD46 molecule, complement regulatory protein |
C00004631
|
| 1604 | CD55, CR, CROM, DAF, TC | CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
C00004631
|
| 965 | CD58, LFA-3, LFA3, ag3 | CD58 molecule |
C00004631
|
| 966 | CD59, 16.3A5, 1F5, EJ16, EJ30, EL32, G344, HRF-20, HRF20, MAC-IP, MACIF, MEM43, MIC11, MIN1, MIN2, MIN3, MIRL, MSK21, p18-20 | CD59 molecule, complement regulatory protein |
C00004631
|
| 967 | CD63, LAMP-3, ME491, MLA1, OMA81H, TSPAN30 | CD63 molecule |
C00004631
|
| 924 | CD7, GP40, LEU-9, TP41, Tp40 | CD7 molecule |
C00004631
|
| 31 | ACACA, ACAC, ACACAD, ACC, ACC1, ACCA | acetyl-CoA carboxylase alpha (EC:6.4.1.2 6.3.4.14) |
C00004631
|
| 9308 | CD83, BL11, HB15 | CD83 molecule |
C00004631
|
| 928 | CD9, BTCC-1, DRAP-27, MIC3, MRP-1, TSPAN-29, TSPAN29 | CD9 molecule |
C00004631
|
| 22918 | CD93, C1QR1, C1qR(P), C1qRP, CDw93, ECSM3, MXRA4, dJ737E23.1 | CD93 molecule |
C00004631
|
| 976 | CD97, TM7LN1 | CD97 molecule |
C00004631
|
| 978 | CDA, CDD | cytidine deaminase (EC:3.5.4.5) |
C00004631
|
| 8881 | CDC16, ANAPC6, APC6, CUT9 | cell division cycle 16 |
C00004631
|
| 34411 |
C00004631
|
||
| 991 | CDC20, CDC20A, bA276H19.3, p55CDC | cell division cycle 20 |
C00004631
|
| 995 | CDC25C, CDC25, PPP1R60 | cell division cycle 25C (EC:3.1.3.48) |
C00004631
|
| 996 | CDC27, ANAPC3, APC3, CDC27Hs, D0S1430E, D17S978E, HNUC, NUC2 | cell division cycle 27 |
C00004631
|
| 11140 | CDC37, P50CDC37 | cell division cycle 37 |
C00004631
|
| 31052 |
C00004631
|
||
| 990 | CDC6, CDC18L, HsCDC18, HsCDC6 | cell division cycle 6 |
C00004631
|
| 8317 | CDC7, CDC7L1, HsCDC7, Hsk1, huCDC7 | cell division cycle 7 (EC:2.7.11.1) |
C00004631
|
| 79577 | CDC73, C1orf28, HPTJT, HRPT2, HYX | cell division cycle 73 |
C00004631
|
| 83461 | CDCA3, GRCC8, TOME-1 | cell division cycle associated 3 |
C00004631
|
| 113130 | CDCA5, SORORIN | cell division cycle associated 5 |
C00004631
|
| 55536 | CDCA7L, JPO2, R1, RAM2 | cell division cycle associated 7-like |
C00004631
|
| 999 | CDH1, Arc-1, CD324, CDHE, ECAD, LCAM, UVO | cadherin 1, type 1, E-cadherin (epithelial) |
C00004631
|
| 1009 | CDH11, CAD11, CDHOB, OB, OSF-4 | cadherin 11, type 2, OB-cadherin (osteoblast) |
C00004631
|
| 1000 | CDH2, CD325, CDHN, CDw325, NCAD | cadherin 2, type 1, N-cadherin (neuronal) |
C00004631
|
| 29965 | CDIP1, C16orf5, CDIP, I1, LITAFL | cell death-inducing p53 target 1 |
C00004631
|
| 10449 | ACAA2, DSAEC | acetyl-CoA acyltransferase 2 (EC:2.3.1.16) |
C00004631
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00004631
|
| 8851 | CDK5R1, CDK5P35, CDK5R, NCK5A, p23, p25, p35, p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
C00004631
|
| 55755 | CDK5RAP2, C48, Cep215, MCPH3 | CDK5 regulatory subunit associated protein 2 |
C00004631
|
| 1024 | CDK8, K35 | cyclin-dependent kinase 8 (EC:2.7.11.23 2.7.11.22) |
C00004631
|
| 1025 | CDK9, C-2k, CDC2L4, CTK1, PITALRE, TAK | cyclin-dependent kinase 9 (EC:2.7.11.23 2.7.11.22) |
C00004631
|
| 54901 | CDKAL1 | CDK5 regulatory subunit associated protein 1-like 1 |
C00004631
|
| 58527 | ABRACL, C6orf115, Costars, HSPC280, PRO2013 | ABRA C-terminal like |
C00004631
|
| 1028 | CDKN1C, BWCR, BWS, KIP2, WBS, p57, p57Kip2 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
C00004631
|
| 1029 | CDKN2A, ARF, CDK4I, CDKN2, CMM2, INK4, INK4A, MLM, MTS-1, MTS1, P14, P14ARF, P16, P16-INK4A, P16INK4, P16INK4A, P19, P19ARF, TP16 | cyclin-dependent kinase inhibitor 2A |
C00004631
|
| 1030 | CDKN2B, CDK4I, INK4B, MTS2, P15, TP15, p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
C00004631
|
| 1033 | CDKN3, CDI1, CIP2, KAP, KAP1 | cyclin-dependent kinase inhibitor 3 (EC:3.1.3.16 3.1.3.48) |
C00004631
|
| 1036 | CDO1 | cysteine dioxygenase type 1 (EC:1.13.11.20) |
C00004631
|
| 30850 | CDR2L, HUMPPA | cerebellar degeneration-related protein 2-like |
C00004631
|
| 284040 | CDRT4 | CMT1A duplicated region transcript 4 |
C00004631
|
| 1040 | CDS1, CDS | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 (EC:2.7.7.41) |
C00004631
|
| 81620 | CDT1, DUP, RIS2 | chromatin licensing and DNA replication factor 1 |
C00004631
|
| 1048 | CEACAM5, CD66e, CEA | carcinoembryonic antigen-related cell adhesion molecule 5 |
C00004631
|
| 1050 | CEBPA, C/EBP-alpha, CEBP | CCAAT/enhancer binding protein (C/EBP), alpha |
C00004631
|
| 1051 | CEBPB, C/EBP-beta, CRP2, IL6DBP, LAP, LIP, NF-IL6, TCF5 | CCAAT/enhancer binding protein (C/EBP), beta |
C00004631
|
| 1052 | CEBPD, C/EBP-delta, CELF, CRP3, NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta |
C00004631
|
| 1056 | CEL, BAL, BSDL, BSSL, CELL, CEase, FAP, FAPP, LIPA, MODY8 | carboxyl ester lipase (EC:3.1.1.13 3.1.1.3) |
C00004631
|
| 10658 | CELF1, BRUNOL2, CUG-BP, CUGBP, CUGBP1, EDEN-BP, NAB50, NAPOR, hNab50 | CUGBP, Elav-like family member 1 |
C00004631
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00004631
|
| 1060 | CENPC, CENP-C, CENPC1, MIF2, hcp-4 | centromere protein C |
C00004631
|
| 1062 | CENPE, CENP-E, KIF10, PPP1R61 | centromere protein E, 312kDa |
C00004631
|
| 1063 | CENPF, CENF, PRO1779, hcp-1 | centromere protein F, 350/400kDa |
C00004631
|
| 2491 | CENPI, CENP-I, FSHPRH1, LRPR1, Mis6 | centromere protein I |
C00004631
|
| 55835 | CENPJ, BM032, CENP-J, CPAP, LAP, LIP1, MCPH6, SASS4, SCKL4, Sas-4 | centromere protein J |
C00004631
|
| 64105 | CENPK, AF5alpha, CENP-K, P33, Solt | centromere protein K |
C00004631
|
| 387103 | CENPW, C6orf173, CENP-W, CUG2 | centromere protein W |
C00004631
|
| 26920 |
C00004631
|
||
| 145508 | CEP128, C14orf145, C14orf61, LEDP/132 | centrosomal protein 128kDa |
C00004631
|
| 9662 | CEP135, CEP4, KIAA0635, MCPH8 | centrosomal protein 135kDa |
C00004631
|
| 22995 | CEP152, MCPH4, MCPH9, SCKL5 | centrosomal protein 152kDa |
C00004631
|
| 283638 | CEP170B, CEP170R, FAM68C, KIAA0284 | centrosomal protein 170B |
C00004631
|
| 55165 | CEP55, C10orf3, CT111, URCC6 | centrosomal protein 55kDa |
C00004631
|
| 23177 | CEP68, KIAA0582 | centrosomal protein 68kDa |
C00004631
|
| 80321 | CEP70, BITE | centrosomal protein 70kDa |
C00004631
|
| 55722 | CEP72 | centrosomal protein 72kDa |
C00004631
|
| 84131 | CEP78, C9orf81, IP63 | centrosomal protein 78kDa |
C00004631
|
| 84902 | CEP89, CCDC123, CEP123 | centrosomal protein 89kDa |
C00004631
|
| 91012 | CERS5, LASS5, Trh4 | ceramide synthase 5 (EC:2.3.1.24) |
C00004631
|
| 8824 | CES2, CE-2, CES2A1, PCE-2, iCE | carboxylesterase 2 (EC:3.1.1.1 3.1.1.84 3.1.1.56) |
C00004631
|
| 629 | CFB, AHUS4, BF, BFD, CFAB, FB, FBI12, GBG, H2-Bf, PBF2 | complement factor B (EC:3.4.21.47) |
C00004631
|
| 3426 | CFI, AHUS3, ARMD13, C3BINA, C3b-INA, FI, IF, KAF | complement factor I (EC:3.4.21.45) |
C00004631
|
| 1072 | CFL1, CFL | cofilin 1 (non-muscle) |
C00004631
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00004631
|
| 84952 | CGNL1, JACOP | cingulin-like 1 |
C00004631
|
| 494143 | CHAC2 | ChaC, cation transport regulator homolog 2 (E. coli) |
C00004631
|
| 8208 | CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSPH7, MPP7 | chromatin assembly factor 1, subunit B (p60) |
C00004631
|
| 400916 | CHCHD10, C22orf16, N27C7-4 | coiled-coil-helix-coiled-coil-helix domain containing 10 |
C00004631
|
| 9557 | CHD1L, ALC1, CHDL | chromodomain helicase DNA binding protein 1-like (EC:3.6.4.12) |
C00004631
|
| 1106 | CHD2, EEOC | chromodomain helicase DNA binding protein 2 (EC:3.6.4.12) |
C00004631
|
| 11200 | CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53, hCds1 | checkpoint kinase 2 (EC:2.7.11.1) |
C00004631
|
| 1119 | CHKA, CHK, CK, CKI, EK | choline kinase alpha (EC:2.7.1.32 2.7.1.82) |
C00004631
|
| 1122 | CHML, REP2 | choroideremia-like (Rab escort protein 2) |
C00004631
|
| 1124 | CHN2, ARHGAP3, BCH, CHN2-3, RHOGAP3 | chimerin 2 |
C00004631
|
| 26973 | CHORDC1, CHP1 | cysteine and histidine-rich domain (CHORD) containing 1 |
C00004631
|
| 79586 | CHPF, CHSY2, CSS2 | chondroitin polymerizing factor (EC:2.4.1.175 2.4.1.226) |
C00004631
|
| 57053 | CHRNA10 | cholinergic receptor, nicotinic, alpha 10 (neuronal) |
C00004631
|
| 166012 | CHST13, C4ST3 | carbohydrate (chondroitin 4) sulfotransferase 13 (EC:2.8.2.5) |
C00004631
|
| 51363 | CHST15, BRAG, GALNAC4S-6ST, RP11-47G11.1 | carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 (EC:2.8.2.33) |
C00004631
|
| 9469 | CHST3, C6ST, C6ST1, HSD | carbohydrate (chondroitin 6) sulfotransferase 3 (EC:2.8.2.17) |
C00004631
|
| 83539 | CHST9, GALNAC4ST-2 | carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
C00004631
|
| 63922 | CHTF18, C16orf41, C321D2.2, C321D2.3, C321D2.4, CHL12, Ctf18, RUVBL | CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
C00004631
|
| 54921 | CHTF8, CTF8, DERPC | CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
C00004631
|
| 27141 | CIDEB | cell death-inducing DFFA-like effector b |
C00004631
|
| 63924 | CIDEC, CIDE-3, CIDE3, FPLD5, FSP27 | cell death-inducing DFFA-like effector c |
C00004631
|
| 1153 | CIRBP, CIRP | cold inducible RNA binding protein |
C00004631
|
| 11113 | CIT, CRIK, STK21 | citron (rho-interacting, serine/threonine kinase 21) (EC:2.7.11.1) |
C00004631
|
| 4435 | CITED1, MSG1 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
C00004631
|
| 10370 | CITED2, ASD8, MRG-1, MRG1, P35SRJ, VSD2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
C00004631
|
| 1152 | CKB, B-CK, CKBB | creatine kinase, brain (EC:2.7.3.2) |
C00004631
|
| 51192 | CKLF, C32, CKLF1, CKLF2, CKLF3, CKLF4, UCK-1 | chemokine-like factor |
C00004631
|
| 23122 | CLASP2 | cytoplasmic linker associated protein 2 |
C00004631
|
| 1182 | CLCN3, CLC3, ClC-3 | chloride channel, voltage-sensitive 3 |
C00004631
|
| 1184 | CLCN5, CLC5, CLCK2, ClC-5, DENTS, NPHL1, NPHL2, XLRH, XRN, hCIC-K2 | chloride channel, voltage-sensitive 5 |
C00004631
|
| 9076 | CLDN1, CLD1, ILVASC, SEMP1 | claudin 1 |
C00004631
|
| 9071 | CLDN10, CPETRL3, OSP-L | claudin 10 |
C00004631
|
| 23562 | CLDN14, DFNB29 | claudin 14 |
C00004631
|
| 1364 | CLDN4, CPE-R, CPER, CPETR, CPETR1, WBSCR8, hCPE-R | claudin 4 |
C00004631
|
| 1047 | CLGN | calmegin |
C00004631
|
| 7461 | CLIP2, CLIP, CLIP-115, CYLN2, WBSCR3, WBSCR4, WSCR3, WSCR4 | CAP-GLY domain containing linker protein 2 |
C00004631
|
| 1195 | CLK1, CLK, CLK/STY, STY | CDC-like kinase 1 (EC:2.7.12.1) |
C00004631
|
| 79789 | CLMN | calmin (calponin-like, transmembrane) |
C00004631
|
| 1203 | CLN5, NCL | ceroid-lipofuscinosis, neuronal 5 |
C00004631
|
| 10978 | CLP1, HEAB, hClp1 | cleavage and polyadenylation factor I subunit 1 (EC:2.7.1.78) |
C00004631
|
| 10845 | CLPX | caseinolytic mitochondrial matrix peptidase chaperone subunit |
C00004631
|
| 1212 | CLTB, LCB | clathrin, light chain B |
C00004631
|
| 171425 | CLYBL, CLB | citrate lyase beta like |
C00004631
|
| 134147 | CMBL, JS-1 | carboxymethylenebutenolidase homolog (Pseudomonas) (EC:3.1.-.-) |
C00004631
|
| 129607 | CMPK2, TMPK2, TYKi, UMP-CMPK2 | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial (EC:2.7.4.6 2.7.4.14) |
C00004631
|
| 112616 | CMTM7, CKLFSF7 | CKLF-like MARVEL transmembrane domain containing 7 |
C00004631
|
| 55783 | CMTR2, AFT, FTSJD1, HMTr2, MTr2 | cap methyltransferase 2 |
C00004631
|
| 9337 | CNOT8, CAF1, CALIF, Caf1b, POP2, hCAF1 | CCR4-NOT transcription complex, subunit 8 (EC:3.1.13.4) |
C00004631
|
| 22837 | COBLL1, COBLR1 | cordon-bleu WH2 repeat protein-like 1 |
C00004631
|
| 10466 | COG5, CDG2I, GOLTC1, GTC90 | component of oligomeric golgi complex 5 |
C00004631
|
| 57511 | COG6, CDG2L, COD2, SHNS | component of oligomeric golgi complex 6 |
C00004631
|
| 8161 | COIL, CLN80, p80-coilin | coilin |
C00004631
|
| 80781 | COL18A1, KNO, KNO1, KS | collagen, type XVIII, alpha 1 |
C00004631
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00004631
|
| 1280 | COL2A1, ANFH, AOM, COL11A3, SEDC, STL1 | collagen, type II, alpha 1 |
C00004631
|
| 1281 | COL3A1, EDS4A | collagen, type III, alpha 1 |
C00004631
|
| 1294 | COL7A1, EBD1, EBDCT, EBR1 | collagen, type VII, alpha 1 |
C00004631
|
| 120376 | COLCA2, C11orf93, CASC13 | colorectal cancer associated 2 |
C00004631
|
| 81035 | COLEC12, CLP1, NSR2, SCARA4, SRCL | collectin sub-family member 12 |
C00004631
|
| 150684 | COMMD1, C2orf5, MURR1 | copper metabolism (Murr1) domain containing 1 |
C00004631
|
| 51397 | COMMD10, PTD002 | COMM domain containing 10 |
C00004631
|
| 54951 | COMMD8 | COMM domain containing 8 |
C00004631
|
| 1312 | COMT | catechol-O-methyltransferase (EC:2.1.1.6) |
C00004631
|
| 11316 | COPE, epsilon-COP | coatomer protein complex, subunit epsilon |
C00004631
|
| 51138 | COPS4, CSN4 | COP9 signalosome subunit 4 |
C00004631
|
| 10987 | COPS5, CSN5, JAB1, MOV-34, SGN5 | COP9 signalosome subunit 5 |
C00004631
|
| 51805 | COQ3, DHHBMT, DHHBMTASE, UG0215E05, bA9819.1 | coenzyme Q3 methyltransferase (EC:2.1.1.114 2.1.1.64 2.1.1.222) |
C00004631
|
| 7464 | CORO2A, CLIPINB, IR10, WDR2 | coronin, actin binding protein, 2A |
C00004631
|
| 10391 | CORO2B, CLIPINC | coronin, actin binding protein, 2B |
C00004631
|
| 23406 | COTL1, CLP | coactosin-like 1 (Dictyostelium) |
C00004631
|
| 1353 | COX11, COX11P | cytochrome c oxidase assembly homolog 11 (yeast) |
C00004631
|
| 90639 | COX19 | cytochrome c oxidase assembly homolog 19 (S. cerevisiae) |
C00004631
|
| 116228 | COX20, FAM36A | COX20 cytochrome C oxidase assembly factor |
C00004631
|
| 1356 | CP, CP-2 | ceruloplasmin (ferroxidase) (EC:1.16.3.1) |
C00004631
|
| 1361 | CPB2, CPU, PCPB, TAFI | carboxypeptidase B2 (plasma) (EC:3.4.17.20) |
C00004631
|
| 79974 | CPED1, C7orf58 | cadherin-like and PC-esterase domain containing 1 |
C00004631
|
| 1369 | CPN1, CPN, SCPN | carboxypeptidase N, polypeptide 1 (EC:3.4.17.3) |
C00004631
|
| 1370 | CPN2, ACBP | carboxypeptidase N, polypeptide 2 (EC:3.4.17.3) |
C00004631
|
| 221184 | CPNE2, COPN2, CPN2 | copine II |
C00004631
|
| 10404 | CPQ, LDP, PGCP | carboxypeptidase Q (EC:3.4.17.-) |
C00004631
|
| 1374 | CPT1A, CPT1, CPT1-L, L-CPT1 | carnitine palmitoyltransferase 1A (liver) (EC:2.3.1.21) |
C00004631
|
| 1375 | CPT1B, CPT1-M, CPT1M, CPTI, CPTI-M, M-CPT1, MCCPT1, MCPT1 | carnitine palmitoyltransferase 1B (muscle) (EC:2.3.1.21) |
C00004631
|
| 1376 | CPT2, CPT1, CPTASE, IIAE4 | carnitine palmitoyltransferase 2 (EC:2.3.1.21) |
C00004631
|
| 54504 | CPVL, HVLP | carboxypeptidase, vitellogenic-like |
C00004631
|
| 1385 | CREB1, CREB | cAMP responsive element binding protein 1 |
C00004631
|
| 64764 | CREB3L2, BBF2H7 | cAMP responsive element binding protein 3-like 2 |
C00004631
|
| 84699 | CREB3L3, CREB-H, CREBH | cAMP responsive element binding protein 3-like 3 |
C00004631
|
| 1387 | CREBBP, CBP, KAT3A, RSTS | CREB binding protein (EC:2.3.1.48) |
C00004631
|
| 9419 | CRIPT | cysteine-rich PDZ-binding protein |
C00004631
|
| 9244 | CRLF1, CISS, CISS1, CLF, CLF-1, NR6, zcytor5 | cytokine receptor-like factor 1 |
C00004631
|
| 51379 | CRLF3, CREME-9, CREME9, CRLM9, CYTOR4, FRWS, p48.2 | cytokine receptor-like factor 3 |
C00004631
|
| 54675 | CRLS1, C20orf155, CLS, CLS1, GCD10, dJ967N21.6 | cardiolipin synthase 1 |
C00004631
|
| 1427 | CRYGS, CRYG8, CTRCT20 | crystallin, gamma S |
C00004631
|
| 51084 | CRYL1, GDH, lambda-CRY | crystallin, lambda 1 (EC:1.1.1.45) |
C00004631
|
| 1428 | CRYM, DFNA40, THBP | crystallin, mu (EC:1.5.1.25) |
C00004631
|
| 9946 | CRYZL1, 4P11, QOH-1 | crystallin, zeta (quinone reductase)-like 1 |
C00004631
|
| 1441 | CSF3R, CD114, GCSFR | colony stimulating factor 3 receptor (granulocyte) |
C00004631
|
| 55790 | CSGALNACT1, CSGalNAcT-1, ChGn, beta4GalNAcT | chondroitin sulfate N-acetylgalactosaminyltransferase 1 (EC:2.4.1.174) |
C00004631
|
| 55454 | CSGALNACT2, CHGN2, GALNACT-2, GALNACT2 | chondroitin sulfate N-acetylgalactosaminyltransferase 2 (EC:2.4.1.174) |
C00004631
|
| 1459 | CSNK2A2, CK2A2, CSNK2A1 | casein kinase 2, alpha prime polypeptide (EC:2.7.11.1) |
C00004631
|
| 1466 | CSRP2, CRP2, LMO5, SmLIM | cysteine and glycine-rich protein 2 |
C00004631
|
| 1475 | CSTA, AREI, STF1, STFA | cystatin A (stefin A) |
C00004631
|
| 1478 | CSTF2, CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
C00004631
|
| 4253 | CTAGE5, MEA6, MGEA, MGEA11, MGEA6 | CTAGE family, member 5 |
C00004631
|
| 80169 | CTC1, AAF-132, AAF132, C17orf68, CRMCC, tmp494178 | CTS telomere maintenance complex component 1 |
C00004631
|
| 1490 | CTGF, CCN2, HCS24, IGFBP8, NOV2 | connective tissue growth factor |
C00004631
|
| 115908 | CTHRC1 | collagen triple helix repeat containing 1 |
C00004631
|
| 1495 | CTNNA1, CAP102 | catenin (cadherin-associated protein), alpha 1, 102kDa |
C00004631
|
| 1499 | CTNNB1, CTNNB, MRD19, armadillo | catenin (cadherin-associated protein), beta 1, 88kDa |
C00004631
|
| 51797 |
C00004631
|
||
| 1506 | CTRL, CTRL1 | chymotrypsin-like (EC:3.4.21.-) |
C00004631
|
| 1508 | CTSB, APPS, CPSB | cathepsin B (EC:3.4.22.1) |
C00004631
|
| 1075 | CTSC, CPPI, DPP-I, DPP1, DPPI, HMS, JP, JPD, PALS, PDON1, PLS | cathepsin C (EC:3.4.14.1) |
C00004631
|
| 1509 | CTSD, CLN10, CPSD | cathepsin D (EC:3.4.23.5) |
C00004631
|
| 1519 | CTSO, CTSO1 | cathepsin O (EC:3.4.22.42) |
C00004631
|
| 2017 | CTTN, EMS1 | cortactin |
C00004631
|
| 404093 | CUEDC1 | CUE domain containing 1 |
C00004631
|
| 79004 | CUEDC2, C10orf66, bA18I14.5 | CUE domain containing 2 |
C00004631
|
| 8450 | CUL4B, MRXHF2, MRXS15, MRXSC, SFM2 | cullin 4B |
C00004631
|
| 51076 | CUTC, RP11-483F11.3 | cutC copper transporter |
C00004631
|
| 50624 | CUZD1, ERG-1, UO-44 | CUB and zona pellucida-like domains 1 |
C00004631
|
| 6376 | CX3CL1, ABCD-3, C3Xkine, CXC3, CXC3C, NTN, NTT, SCYD1, fractalkine, neurotactin | chemokine (C-X3-C motif) ligand 1 |
C00004631
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00004631
|
| 58191 | CXCL16, CXCLG16, SR-PSOX, SRPSOX | chemokine (C-X-C motif) ligand 16 |
C00004631
|
| 2920 | CXCL2, CINC-2a, GRO2, GROb, MGSA-b, MIP-2a, MIP2, MIP2A, SCYB2 | chemokine (C-X-C motif) ligand 2 |
C00004631
|
| 55086 | CXorf57 | chromosome X open reading frame 57 |
C00004631
|
| 1528 | CYB5A, CYB5, MCB5 | cytochrome b5 type A (microsomal) |
C00004631
|
| 1535 | CYBA, p22-PHOX | cytochrome b-245, alpha polypeptide |
C00004631
|
| 114757 | CYGB, HGB, STAP | cytoglobin |
C00004631
|
| 100270688 |
C00004631
|
||
| 22885 | ABLIM3 | actin binding LIM protein family, member 3 |
C00004631
|
| 57406 | ABHD6 | abhydrolase domain containing 6 (EC:3.1.1.23) |
C00004631
|
| 1591 | CYP24A1, CP24, CYP24, HCAI, P450-CC24 | cytochrome P450, family 24, subfamily A, polypeptide 1 (EC:1.14.13.126) |
C00004631
|
| 1594 | CYP27B1, CP2B, CYP1, CYP1alpha, CYP27B, P450c1, PDDR, VDD1, VDDR, VDDRI, VDR | cytochrome P450, family 27, subfamily B, polypeptide 1 (EC:1.14.13.13) |
C00004631
|
| 1556 | CYP2B7P, CYP2B, CYP2B7, CYP2B7P1 | cytochrome P450, family 2, subfamily B, polypeptide 7, pseudogene |
C00004631
|
| 1558 | CYP2C8, CPC8, CYPIIC8, MP-12/MP-20 | cytochrome P450, family 2, subfamily C, polypeptide 8 (EC:1.14.14.1) |
C00004631
|
| 1565 | CYP2D6, CPD6, CYP2D, CYP2D7AP, CYP2D7BP, CYP2D7P2, CYP2D8P2, CYP2DL1, CYPIID6, P450-DB1, P450C2D, P450DB1 | cytochrome P450, family 2, subfamily D, polypeptide 6 (EC:1.14.14.1) |
C00004631
|
| 1573 | CYP2J2, CPJ2 | cytochrome P450, family 2, subfamily J, polypeptide 2 (EC:1.14.14.1) |
C00004631
|
| 29785 | CYP2S1 | cytochrome P450, family 2, subfamily S, polypeptide 1 (EC:1.14.14.1) |
C00004631
|
| 51099 | ABHD5, CDS, CGI58, IECN2, NCIE2 | abhydrolase domain containing 5 (EC:2.3.1.51) |
C00004631
|
| 57834 | CYP4F11, CYPIVF11 | cytochrome P450, family 4, subfamily F, polypeptide 11 (EC:1.14.14.1) |
C00004631
|
| 1595 | CYP51A1, CP51, CYP51, CYPL1, LDM, P450-14DM, P450L1 | cytochrome P450, family 51, subfamily A, polypeptide 1 (EC:1.14.13.70) |
C00004631
|
| 1582 | CYP8B1, CP8B, CYP12 | cytochrome P450, family 8, subfamily B, polypeptide 1 (EC:1.14.13.95) |
C00004631
|
| 3491 | CYR61, CCN1, GIG1, IGFBP10 | cysteine-rich, angiogenic inducer, 61 |
C00004631
|
| 1601 | DAB2, DOC-2, DOC2 | Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
C00004631
|
| 26007 | DAK, NET45 | dihydroxyacetone kinase 2 homolog (S. cerevisiae) (EC:2.7.1.29 4.6.1.15 2.7.1.28) |
C00004631
|
| 1612 | DAPK1, DAPK | death-associated protein kinase 1 (EC:2.7.11.1) |
C00004631
|
| 1615 | DARS, HBSL | aspartyl-tRNA synthetase (EC:6.1.1.12) |
C00004631
|
| 10926 | DBF4, ASK, CHIF, DBF4A, ZDBF1 | DBF4 homolog (S. cerevisiae) |
C00004631
|
| 138948 | DBH-AS1, NCRNA00118 | DBH antisense RNA 1 |
C00004631
|
| 1627 | DBN1, D0S117E | drebrin 1 |
C00004631
|
| 51163 | DBR1 | debranching RNA lariats 1 |
C00004631
|
| 54876 | DCAF16, C4orf30 | DDB1 and CUL4 associated factor 16 |
C00004631
|
| 55827 | DCAF6, 1200006M05Rik, ARCAP, IQWD1, MSTP055, NRIP, PC326 | DDB1 and CUL4 associated factor 6 |
C00004631
|
| 131566 | DCBLD2, CLCP1, ESDN | discoidin, CUB and LCCL domain containing 2 |
C00004631
|
| 51473 | DCDC2, DCDC2A, RU2, RU2S | doublecortin domain containing 2 |
C00004631
|
| 1633 | DCK | deoxycytidine kinase (EC:2.7.1.74) |
C00004631
|
| 196513 | DCP1B, DCP1, hDcp1b | decapping mRNA 1B |
C00004631
|
| 28960 | DCPS, DCS1, HINT-5, HINT5, HSL1 | decapping enzyme, scavenger (EC:3.6.1.59) |
C00004631
|
| 55208 | DCUN1D2, C13orf17 | DCN1, defective in cullin neddylation 1, domain containing 2 |
C00004631
|
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00004631
|
| 1644 | DDC, AADC | dopa decarboxylase (aromatic L-amino acid decarboxylase) (EC:4.1.1.28) |
C00004631
|
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00004631
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00004631
|
| 1663 | DDX11, CHL1, CHLR1, KRG2, WABS | DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 (EC:3.6.4.13) |
C00004631
|
| 10521 | DDX17, P72, RH70 | DEAD (Asp-Glu-Ala-Asp) box helicase 17 (EC:3.6.4.13) |
C00004631
|
| 11218 | DDX20, DP103, GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 (EC:3.6.4.13) |
C00004631
|
| 68278 |
C00004631
|
||
| 11056 | DDX52, HUSSY19, ROK1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (EC:3.6.4.13) |
C00004631
|
| 23586 | DDX58, RIG-I, RIGI, RLR-1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (EC:3.6.4.13) |
C00004631
|
| 1666 | DECR1, DECR, NADPH, SDR18C1 | 2,4-dienoyl CoA reductase 1, mitochondrial (EC:1.3.1.34) |
C00004631
|
| 54849 | DEF8 | differentially expressed in FDCP 8 homolog (mouse) |
C00004631
|
| 1667 | DEFA1, DEF1, DEFA2, HNP-1, HP-1, HP1, MRS | defensin, alpha 1 |
C00004631
|
| 100294660 |
C00004631
|
||
| 7913 | DEK, D6S231E | DEK oncogene |
C00004631
|
| 57706 | DENND1A, FAM31A, KIAA1608, RP11-230L22.3 | DENN/MADD domain containing 1A |
C00004631
|
| 8562 | DENR, DRP, DRP1, SMAP-3 | density-regulated protein |
C00004631
|
| 55635 | DEPDC1, DEP.8, DEPDC1-V2, DEPDC1A, SDP35 | DEP domain containing 1 |
C00004631
|
| 55789 | DEPDC1B, BRCC3, XTP1 | DEP domain containing 1B |
C00004631
|
| 420357 |
C00004631
|
||
| 91614 | DEPDC7, TR2, dJ85M6.4 | DEP domain containing 7 |
C00004631
|
| 1676 | DFFA, DFF-45, DFF1, ICAD | DNA fragmentation factor, 45kDa, alpha polypeptide |
C00004631
|
| 1677 | DFFB, CAD, CPAN, DFF-40, DFF2, DFF40 | DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) |
C00004631
|
| 1687 | DFNA5, ICERE-1 | deafness, autosomal dominant 5 |
C00004631
|
| 84649 | DGAT2, ARAT, GS1999FULL | diacylglycerol O-acyltransferase 2 (EC:2.3.1.20 2.3.1.76) |
C00004631
|
| 54487 | DGCR8, C22orf12, DGCRK6, Gy1, pasha | DGCR8 microprocessor complex subunit |
C00004631
|
| 1606 | DGKA, DAGK, DAGK1, DGK-alpha | diacylglycerol kinase, alpha 80kDa (EC:2.7.1.107) |
C00004631
|
| 1718 | DHCR24, DCE, Nbla03646, SELADIN1, seladin-1 | 24-dehydrocholesterol reductase (EC:1.3.1.72) |
C00004631
|
| 1717 | DHCR7, SLOS | 7-dehydrocholesterol reductase (EC:1.3.1.21) |
C00004631
|
| 200895 | DHFRL1, DHFRP4 | dihydrofolate reductase-like 1 (EC:1.5.1.3) |
C00004631
|
| 147015 | DHRS13, SDR7C5 | dehydrogenase/reductase (SDR family) member 13 |
C00004631
|
| 51635 | DHRS7, SDR34C1, retDSR4, retSDR4 | dehydrogenase/reductase (SDR family) member 7 |
C00004631
|
| 54505 | DHX29, DDX29 | DEAH (Asp-Glu-Ala-His) box polypeptide 29 (EC:3.6.4.13) |
C00004631
|
| 79665 | DHX40, DDX40, PAD | DEAH (Asp-Glu-Ala-His) box polypeptide 40 (EC:3.6.4.13) |
C00004631
|
| 56616 | DIABLO, DFNA64, SMAC | diablo, IAP-binding mitochondrial protein |
C00004631
|
| 445162 |
C00004631
|
||
| 1733 | DIO1, 5DI, TXDI1 | deiodinase, iodothyronine, type I (EC:1.97.1.10) |
C00004631
|
| 23181 | DIP2A, C21orf106, DIP2 | DIP2 disco-interacting protein 2 homolog A (Drosophila) |
C00004631
|
| 84925 | DIRC2, RCC4 | disrupted in renal carcinoma 2 |
C00004631
|
| 22943 | DKK1, DKK-1, SK | dickkopf WNT signaling pathway inhibitor 1 |
C00004631
|
| 27121 | DKK4, DKK-4 | dickkopf WNT signaling pathway inhibitor 4 |
C00004631
|
| 1738 | DLD, DLDD, DLDH, E3, GCSL, LAD, PHE3 | dihydrolipoamide dehydrogenase (EC:1.8.1.4) |
C00004631
|
| 1739 | DLG1, DLGH1, SAP-97, SAP97, dJ1061C18.1.1, hdlg | discs, large homolog 1 (Drosophila) |
C00004631
|
| 9787 | DLGAP5, DLG7, HURP | discs, large (Drosophila) homolog-associated protein 5 |
C00004631
|
| 1746 | DLX2, TES-1, TES1 | distal-less homeobox 2 |
C00004631
|
| 1750 | DLX6 | distal-less homeobox 6 |
C00004631
|
| 1760 | DMPK, DM, DM1, DM1PK, DMK, MDPK, MT-PK | dystrophia myotonica-protein kinase (EC:2.7.11.1) |
C00004631
|
| 1657 | DMXL1 | Dmx-like 1 |
C00004631
|
| 23312 | DMXL2, RC3 | Dmx-like 2 |
C00004631
|
| 55172 | DNAAF2, C14orf104, CILD10, KTU, PF13 | dynein, axonemal, assembly factor 2 |
C00004631
|
| 3301 | DNAJA1, DJ-2, DjA1, HDJ2, HSDJ, HSJ2, HSPF4, NEDD7, hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 |
C00004631
|
| 3300 | DNAJB2, DSMA5, HSJ-1, HSJ1, HSPF3 | DnaJ (Hsp40) homolog, subfamily B, member 2 |
C00004631
|
| 11080 | DNAJB4, DNAJW, DjB4, HLJ1 | DnaJ (Hsp40) homolog, subfamily B, member 4 |
C00004631
|
| 56521 | DNAJC12, JDP1 | DnaJ (Hsp40) homolog, subfamily C, member 12 |
C00004631
|
| 27000 | DNAJC2, MPHOSPH11, MPP11, ZRF1, ZUO1 | DnaJ (Hsp40) homolog, subfamily C, member 2 |
C00004631
|
| 5611 | DNAJC3, ERdj6, HP58, P58, P58IPK, PRKRI | DnaJ (Hsp40) homolog, subfamily C, member 3 |
C00004631
|
| 1773 | DNASE1, DNL1, DRNI | deoxyribonuclease I (EC:3.1.21.1) |
C00004631
|
| 1774 | DNASE1L1, DNAS1L1, DNASEX, DNL1L, G4.8, XIB | deoxyribonuclease I-like 1 (EC:3.1.21.-) |
C00004631
|
| 1776 | DNASE1L3, DHP2, DNAS1L3, LSD, SLEB16 | deoxyribonuclease I-like 3 (EC:3.1.21.-) |
C00004631
|
| 144132 | DNHD1, C11orf47, CCDC35, DHCD1, DNHD1L | dynein heavy chain domain 1 |
C00004631
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00004631
|
| 30836 | DNTTIP2, ERBP, FCF2, HSU15552, LPTS-RP2, TdIF2 | deoxynucleotidyltransferase, terminal, interacting protein 2 |
C00004631
|
| 9732 | DOCK4 | dedicator of cytokinesis 4 |
C00004631
|
| 80005 | DOCK5 | dedicator of cytokinesis 5 |
C00004631
|
| 220164 | DOK6, DOK5L, HsT3226 | docking protein 6 |
C00004631
|
| 22845 | DOLK, CDG1M, DK, DK1, SEC59, TMEM15 | dolichol kinase (EC:2.7.1.108) |
C00004631
|
| 23033 | DOPEY1, KIAA1117, dJ202D23.2 | dopey family member 1 |
C00004631
|
| 285381 | DPH3, DELGIP, DELGIP1, DESR1, DPH3A, KTI11, ZCSL2 | diphthamide biosynthesis 3 |
C00004631
|
| 89978 | DPH6, ATPBD4 | diphthamine biosynthesis 6 (EC:6.3.1.14) |
C00004631
|
| 8818 | DPM2, CDG1U | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (EC:2.4.1.83) |
C00004631
|
| 54344 | DPM3, CDG1O | dolichyl-phosphate mannosyltransferase polypeptide 3 (EC:2.4.1.83) |
C00004631
|
| 1803 | DPP4, ADABP, ADCP2, CD26, DPPIV, TP103 | dipeptidyl-peptidase 4 (EC:3.4.14.5) |
C00004631
|
| 84661 | DPY30, Cps25, HDPY-30, Saf19 | dpy-30 homolog (C. elegans) |
C00004631
|
| 1806 | DPYD, DHP, DHPDHASE, DPD | dihydropyrimidine dehydrogenase (EC:1.3.1.2) |
C00004631
|
| 1808 | DPYSL2, CRMP-2, CRMP2, DHPRP2, DRP-2, DRP2, N2A3, ULIP-2, ULIP2 | dihydropyrimidinase-like 2 |
C00004631
|
| 10570 | DPYSL4, CRMP3, DRP-4, ULIP4 | dihydropyrimidinase-like 4 |
C00004631
|
| 55332 | DRAM1, DRAM | DNA-damage regulated autophagy modulator 1 |
C00004631
|
| 10589 | DRAP1, NC2-alpha | DR1-associated protein 1 (negative cofactor 2 alpha) |
C00004631
|
| 1819 | DRG2 | developmentally regulated GTP binding protein 2 |
C00004631
|
| 1824 | DSC2, ARVD11, CDHF2, DG2, DGII/III, DSC3 | desmocollin 2 |
C00004631
|
| 79075 | DSCC1, DCC1 | DNA replication and sister chromatid cohesion 1 |
C00004631
|
| 29940 | DSE, DSEP, DSEPI, SART-2, SART2 | dermatan sulfate epimerase (EC:5.1.3.19) |
C00004631
|
| 11034 | DSTN, ACTDP, ADF, bA462D18.2 | destrin (actin depolymerizing factor) |
C00004631
|
| 112487 | DTD2, C14orf126 | D-tyrosyl-tRNA deacylase 2 (putative) |
C00004631
|
| 51514 | DTL, CDT2, DCAF2, L2DTL, RAMP | denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
C00004631
|
| 56986 | DTWD1 | DTW domain containing 1 |
C00004631
|
| 1843 | DUSP1, CL100, HVH1, MKP-1, MKP1, PTPN10 | dual specificity phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00004631
|
| 285193 | DUSP28, DUSP26, VHP | dual specificity phosphatase 28 (EC:3.1.3.16 3.1.3.48) |
C00004631
|
| 1846 | DUSP4, HVH2, MKP-2, MKP2, TYP | dual specificity phosphatase 4 (EC:3.1.3.16 3.1.3.48) |
C00004631
|
| 1847 | DUSP5, DUSP, HVH3 | dual specificity phosphatase 5 (EC:3.1.3.16 3.1.3.48) |
C00004631
|
| 1848 | DUSP6, HH19, MKP3, PYST1 | dual specificity phosphatase 6 (EC:3.1.3.16 3.1.3.48) |
C00004631
|
| 84332 | DYDC2 | DPY30 domain containing 2 |
C00004631
|
| 8655 | DYNLL1, DLC1, DLC8, DNCL1, DNCLC1, LC8, LC8a, PIN, hdlc1 | dynein, light chain, LC8-type 1 |
C00004631
|
| 6990 | DYNLT3, RP3, TCTE1L, TCTEX1L | dynein, light chain, Tctex-type 3 |
C00004631
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00004631
|
| 1870 | E2F2, E2F-2 | E2F transcription factor 2 |
C00004631
|
| 1871 | E2F3, E2F-3 | E2F transcription factor 3 |
C00004631
|
| 1875 | E2F5, E2F-5 | E2F transcription factor 5, p130-binding |
C00004631
|
| 144455 | E2F7 | E2F transcription factor 7 |
C00004631
|
| 55840 | EAF2, BM040, TRAITS, U19 | ELL associated factor 2 |
C00004631
|
| 11319 | ECD, GCR2, HSGT1 | ecdysoneless homolog (Drosophila) |
C00004631
|
| 1894 | ECT2, ARHGEF31 | epithelial cell transforming sequence 2 oncogene |
C00004631
|
| 60401 | EDA2R, EDA-A2R, EDAA2R, TNFRSF27, XEDAR | ectodysplasin A2 receptor |
C00004631
|
| 80153 | EDC3, LSM16, YJDC, YJEFN2 | enhancer of mRNA decapping 3 |
C00004631
|
| 1906 | EDN1, ET1, HDLCQ7, PPET1 | endothelin 1 |
C00004631
|
| 1910 | EDNRB, ABCDS, ET-B, ET-BR, ETB, ETBR, ETRB, HSCR, HSCR2, WS4A | endothelin receptor type B |
C00004631
|
| 90141 | EFCAB11, C14orf143 | EF-hand calcium binding domain 11 |
C00004631
|
| 84455 | EFCAB7, RP4-534K7.1 | EF-hand calcium binding domain 7 |
C00004631
|
| 388795 | EFCAB8 | EF-hand calcium binding domain 8 |
C00004631
|
| 2202 | EFEMP1, DHRD, DRAD, FBLN3, FBNL, FIBL-3, MLVT, MTLV, S1-5 | EGF containing fibulin-like extracellular matrix protein 1 |
C00004631
|
| 1942 | EFNA1, B61, ECKLG, EFL1, EPLG1, LERK-1, LERK1, TNFAIP4 | ephrin-A1 |
C00004631
|
| 1945 | EFNA4, EFL4, EPLG4, LERK4 | ephrin-A4 |
C00004631
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00004631
|
| 23167 | EFR3A | EFR3 homolog A (S. cerevisiae) |
C00004631
|
| 63874 | ABHD4, ABH4 | abhydrolase domain containing 4 |
C00004631
|
| 84836 | ABHD14B, CIB | abhydrolase domain containing 14B (EC:3.-.-.-) |
C00004631
|
| 54583 | EGLN1, C1orf12, ECYT3, HIF-PH2, HIFPH2, HPH-2, HPH2, PHD2, SM20, ZMYND6 | egl-9 family hypoxia-inducible factor 1 (EC:1.14.11.29) |
C00004631
|
| 112399 | EGLN3, HIFP4H3, HIFPH3, PHD3 | egl-9 family hypoxia-inducible factor 3 (EC:1.14.11.29) |
C00004631
|
| 64241 | ABCG8, GBD4, STSL | ATP-binding cassette, sub-family G (WHITE), member 8 |
C00004631
|
| 1959 | EGR2, AT591, CMT1D, CMT4E, KROX20 | early growth response 2 |
C00004631
|
| 1960 | EGR3, EGR-3, PILOT | early growth response 3 |
C00004631
|
| 1961 | EGR4, NGFI-C, NGFIC, PAT133 | early growth response 4 |
C00004631
|
| 23301 | EHBP1, HPC12, NACSIN | EH domain binding protein 1 |
C00004631
|
| 1962 | EHHADH, ECHD, L-PBE, LBFP, LBP, PBFE | enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (EC:1.1.1.35 5.3.3.8 4.2.1.17) |
C00004631
|
| 9538 | EI24, EPG4, PIG8, TP53I8 | etoposide induced 2.4 |
C00004631
|
| 126272 | EID2B, EID-3 | EP300 interacting inhibitor of differentiation 2B |
C00004631
|
| 493861 | EID3, NS4EB, NSE4B, NSMCE4B | EP300 interacting inhibitor of differentiation 3 |
C00004631
|
| 10209 | EIF1, A121, EIF-1, EIF1A, ISO1, SUI1 | eukaryotic translation initiation factor 1 |
C00004631
|
| 83939 | EIF2A, EIF-2A, MST089, MSTP004, MSTP089 | eukaryotic translation initiation factor 2A, 65kDa |
C00004631
|
| 5610 | EIF2AK2, EIF2AK1, PKR, PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 (EC:2.7.11.1 2.7.10.2) |
C00004631
|
| 8892 | EIF2B2, EIF-2Bbeta, EIF2B | eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
C00004631
|
| 1965 | EIF2S1, EIF-2, EIF-2A, EIF-2alpha, EIF2, EIF2A | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
C00004631
|
| 8894 | EIF2S2, EIF2, EIF2B, EIF2beta, PPP1R67, eIF-2-beta | eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
C00004631
|
| 8661 | EIF3A, EIF3, EIF3S10, P167, TIF32, eIF3-p170, eIF3-theta, p180, p185 | eukaryotic translation initiation factor 3, subunit A |
C00004631
|
| 1973 | EIF4A1, DDX2A, EIF-4A, EIF4A, eIF-4A-I, eIF4A-I | eukaryotic translation initiation factor 4A1 (EC:3.6.4.13) |
C00004631
|
| 9470 | EIF4E2, 4E-LP, 4EHP, EIF4EL3, IF4e | eukaryotic translation initiation factor 4E family member 2 |
C00004631
|
| 1999 | ELF3, EPR-1, ERT, ESE-1, ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
C00004631
|
| 2000 | ELF4, ELFR, MEF | E74-like factor 4 (ets domain transcription factor) |
C00004631
|
| 2005 | ELK4, SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) |
C00004631
|
| 22936 | ELL2 | elongation factor, RNA polymerase II, 2 |
C00004631
|
| 255520 | ELMOD2, 9830169G11Rik | ELMO/CED-12 domain containing 2 |
C00004631
|
| 64834 | ELOVL1, Ssc1 | ELOVL fatty acid elongase 1 (EC:2.3.1.199) |
C00004631
|
| 83401 | ELOVL3, CIG-30, CIG30 | ELOVL fatty acid elongase 3 (EC:2.3.1.199) |
C00004631
|
| 79071 | ELOVL6, FACE, FAE, LCE | ELOVL fatty acid elongase 6 (EC:2.3.1.199) |
C00004631
|
| 24139 | EML2, ELP70, EMAP-2, EMAP2 | echinoderm microtubule associated protein like 2 |
C00004631
|
| 2012 | EMP1, CL-20, EMP-1, TMP | epithelial membrane protein 1 |
C00004631
|
| 2014 | EMP3, YMP | epithelial membrane protein 3 |
C00004631
|
| 55740 | ENAH, ENA, MENA, NDPP1 | enabled homolog (Drosophila) |
C00004631
|
| 23052 | ENDOD1 | endonuclease domain containing 1 |
C00004631
|
| 2023 | ENO1, ENO1L1, MPB1, NNE, PPH | enolase 1, (alpha) (EC:4.2.1.11) |
C00004631
|
| 55556 | ENOSF1, HSRTSBETA, RTS, TYMSAS | enolase superfamily member 1 |
C00004631
|
| 5167 | ENPP1, ARHR2, M6S1, NPP1, NPPS, PC-1, PCA1, PDNP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 (EC:3.1.4.1 3.6.1.9) |
C00004631
|
| 5168 | ENPP2, ATX, ATX-X, AUTOTAXIN, LysoPLD, NPP2, PD-IALPHA, PDNP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 (EC:3.1.4.39) |
C00004631
|
| 5169 | ENPP3, B10, CD203c, NPP3, PD-IBETA, PDNP3 | ectonucleotide pyrophosphatase/phosphodiesterase 3 (EC:3.1.4.1 3.6.1.9) |
C00004631
|
| 957 | ENTPD5, CD39L4, NTPDase-5, PCPH | ectonucleoside triphosphate diphosphohydrolase 5 (EC:3.6.1.6 3.6.1.42) |
C00004631
|
| 285203 | EOGT, AER61, AOS4, C3orf64, EOGT1 | EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase (EC:2.4.1.255) |
C00004631
|
| 2034 | EPAS1, ECYT4, HIF2A, HLF, MOP2, PASD2, bHLHe73 | endothelial PAS domain protein 1 |
C00004631
|
| 2036 | EPB41L1, 4.1N, MRD11 | erythrocyte membrane protein band 4.1-like 1 |
C00004631
|
| 114915 | EPB41L4A-AS1, C5orf26, NCRNA00219, TIGA1 | EPB41L4A antisense RNA 1 |
C00004631
|
| 57669 | EPB41L5, BE37, YMO1 | erythrocyte membrane protein band 4.1 like 5 |
C00004631
|
| 1969 | EPHA2, ARCC2, CTPA, CTPP1, CTRCT6, ECK | EPH receptor A2 (EC:2.7.10.1) |
C00004631
|
| 2052 | EPHX1, EPHX, EPOX, HYL1, MEH | epoxide hydrolase 1, microsomal (xenobiotic) (EC:3.3.2.9) |
C00004631
|
| 2053 | EPHX2, CEH, SEH | epoxide hydrolase 2, cytoplasmic (EC:3.3.2.10 3.1.3.76) |
C00004631
|
| 55040 | EPN3 | epsin 3 |
C00004631
|
| 2056 | EPO, EP, MVCD2 | erythropoietin |
C00004631
|
| 2060 | EPS15, AF-1P, AF1P, MLLT5 | epidermal growth factor receptor pathway substrate 15 |
C00004631
|
| 2059 | EPS8 | epidermal growth factor receptor pathway substrate 8 |
C00004631
|
| 64787 | EPS8L2, EPS8R2 | EPS8-like 2 |
C00004631
|
| 64240 | ABCG5, STSL | ATP-binding cassette, sub-family G (WHITE), member 5 |
C00004631
|
| 2065 | ERBB3, ErbB-3, HER3, LCCS2, MDA-BF-1, c-erbB-3, c-erbB3, erbB3-S, p180-ErbB3, p45-sErbB3, p85-sErbB3 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 (EC:2.7.10.1) |
C00004631
|
| 2067 | ERCC1, COFS4, RAD10, UV20 | excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
C00004631
|
| 2073 | ERCC5, COFS3, ERCM2, UVDR, XPG, XPGC | excision repair cross-complementing rodent repair deficiency, complementation group 5 |
C00004631
|
| 54821 | ERCC6L, PICH, RAD26L | excision repair cross-complementing rodent repair deficiency, complementation group 6-like (EC:3.6.4.12) |
C00004631
|
| 2069 | EREG, ER | epiregulin |
C00004631
|
| 51290 | ERGIC2, Erv41, PTX1, cd002 | ERGIC and golgi 2 |
C00004631
|
| 121506 | ERP27, C12orf46, PDIA8 | endoplasmic reticulum protein 27 |
C00004631
|
| 10961 | ERP29, C12orf8, ERp28, ERp31, PDI-DB, PDIA9 | endoplasmic reticulum protein 29 |
C00004631
|
| 90952 | ESAM, W117m | endothelial cell adhesion molecule |
C00004631
|
| 11082 | ESM1, endocan | endothelial cell-specific molecule 1 |
C00004631
|
| 83715 | ESPN, DFNB36 | espin |
C00004631
|
| 2109 | ETFB, MADD | electron-transfer-flavoprotein, beta polypeptide |
C00004631
|
| 64850 | ETNPPL, AGXT2L1 | ethanolamine-phosphate phospho-lyase (EC:4.2.3.2) |
C00004631
|
| 2118 | ETV4, E1A-F, E1AF, PEA3, PEAS3 | ets variant 4 |
C00004631
|
| 84141 | EVA1A, FAM176A, TMEM166 | eva-1 homolog A (C. elegans) |
C00004631
|
| 55194 | EVA1B, C1orf78, FAM176B, RP11-268J15.2 | eva-1 homolog B (C. elegans) |
C00004631
|
| 733318 |
C00004631
|
||
| 55770 | EXOC2, SEC5, SEC5L1, Sec5p | exocyst complex component 2 |
C00004631
|
| 60412 | EXOC4, SEC8, SEC8L1, Sec8p | exocyst complex component 4 |
C00004631
|
| 10640 | EXOC5, HSEC10, PRO1912, SEC10, SEC10L1, SEC10P | exocyst complex component 5 |
C00004631
|
| 54536 | EXOC6, EXOC6A, SEC15, SEC15L, SEC15L1, SEC15L3, Sec15p | exocyst complex component 6 |
C00004631
|
| 149371 | EXOC8, EXO84, Exo84p, SEC84 | exocyst complex component 8 |
C00004631
|
| 11340 | EXOSC8, CIP3, EAP2, OIP2, RP11-421P11.3, RRP43, Rrp43p, bA421P11.3, p9 | exosome component 8 (EC:3.1.13.-) |
C00004631
|
| 5393 | EXOSC9, PM/Scl-75, PMSCL1, RRP45, Rrp45p, p5, p6 | exosome component 9 (EC:3.1.13.-) |
C00004631
|
| 2135 | EXTL2, EXTR2 | exostosin-like glycosyltransferase 2 (EC:2.4.1.223) |
C00004631
|
| 2159 | F10, FX, FXA | coagulation factor X (EC:3.4.21.6) |
C00004631
|
| 2147 | F2, PT, RPRGL2, THPH1 | coagulation factor II (thrombin) (EC:3.4.21.5) |
C00004631
|
| 2149 | F2R, CF2R, HTR, PAR-1, PAR1, TR | coagulation factor II (thrombin) receptor |
C00004631
|
| 2150 | F2RL1, GPR11, PAR2 | coagulation factor II (thrombin) receptor-like 1 |
C00004631
|
| 2152 | F3, CD142, TF, TFA | coagulation factor III (thromboplastin, tissue factor) |
C00004631
|
| 2155 | F7, SPCA | coagulation factor VII (serum prothrombin conversion accelerator) (EC:3.4.21.21) |
C00004631
|
| 2168 | FABP1, FABPL, L-FABP | fatty acid binding protein 1, liver |
C00004631
|
| 2172 | FABP6, I-15P, I-BABP, I-BALB, I-BAP, ILBP, ILBP3, ILLBP | fatty acid binding protein 6, ileal |
C00004631
|
| 8772 | FADD, MORT1 | Fas (TNFRSF6)-associated via death domain |
C00004631
|
| 3995 | FADS3, CYB5RP, LLCDL3 | fatty acid desaturase 3 (EC:1.14.19.-) |
C00004631
|
| 2184 | FAH | fumarylacetoacetate hydrolase (fumarylacetoacetase) (EC:3.7.1.2) |
C00004631
|
| 55179 | FAIM, FAIM1 | Fas apoptotic inhibitory molecule |
C00004631
|
| 284611 | FAM102B, SYM-3B | family with sequence similarity 102, member B |
C00004631
|
| 90362 | FAM110B, C8orf72 | family with sequence similarity 110, member B |
C00004631
|
| 92689 | FAM114A1, Noxp20 | family with sequence similarity 114, member A1 |
C00004631
|
| 81558 | FAM117A | family with sequence similarity 117, member A |
C00004631
|
| 150864 | FAM117B, ALS2CR13 | family with sequence similarity 117, member B |
C00004631
|
| 546486 |
C00004631
|
||
| 54954 | FAM120C, CXorf17, ORF34 | family with sequence similarity 120C |
C00004631
|
| 159090 | FAM122B, SPACIA2 | family with sequence similarity 122B |
C00004631
|
| 57579 | FAM135A, KIAA1411 | family with sequence similarity 135, member A |
C00004631
|
| 10144 | FAM13A, ARHGAP48, FAM13A1 | family with sequence similarity 13, member A |
C00004631
|
| 51306 | FAM13B, ARHGAP49, C5orf5, FAM13B1, KHCHP, N61 | family with sequence similarity 13, member B |
C00004631
|
| 220965 | FAM13C, FAM13C1 | family with sequence similarity 13, member C |
C00004631
|
| 285016 | FAM150B, PRO1097, RGPG542 | family with sequence similarity 150, member B |
C00004631
|
| 84140 | FAM161A, RP28 | family with sequence similarity 161, member A |
C00004631
|
| 26355 | FAM162A, C3orf28, E2IG5, HGTD-P | family with sequence similarity 162, member A |
C00004631
|
| 221061 | FAM171A1, C10orf38 | family with sequence similarity 171, member A1 |
C00004631
|
| 345757 | FAM174A, TMEM157, UNQ1912 | family with sequence similarity 174, member A |
C00004631
|
| 84142 | FAM175A, ABRA1, CCDC98 | family with sequence similarity 175, member A |
C00004631
|
| 283635 | FAM177A1, C14orf24 | family with sequence similarity 177, member A1 |
C00004631
|
| 8603 | FAM193A, C4orf8 | family with sequence similarity 193, member A |
C00004631
|
| 54540 | FAM193B, IRIZIO | family with sequence similarity 193, member B |
C00004631
|
| 51313 | FAM198B, AD036, C4orf18 | family with sequence similarity 198, member B |
C00004631
|
| 151647 | FAM19A4, TAFA-4, TAFA4 | family with sequence similarity 19 (chemokine (C-C motif)-like), member A4 |
C00004631
|
| 25817 | FAM19A5, QLLK5208, TAFA-5, TAFA5, UNQ5208 | family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
C00004631
|
| 23272 | FAM208A, C3orf63, RAP140, se89-1 | family with sequence similarity 208, member A |
C00004631
|
| 200232 | FAM209A, C20orf106, dJ1153D9.3 | family with sequence similarity 209, member A |
C00004631
|
| 388799 | FAM209B, C20orf107, dJ1153D9.4 | family with sequence similarity 209, member B |
C00004631
|
| 55924 | FAM212B, C1orf183 | family with sequence similarity 212, member B |
C00004631
|
| 84915 | FAM222A, C12orf34 | family with sequence similarity 222, member A |
C00004631
|
| 375190 | FAM228B | family with sequence similarity 228, member B |
C00004631
|
| 54097 | FAM3B, 2-21, C21orf11, C21orf76, ORF9, PANDER | family with sequence similarity 3, member B |
C00004631
|
| 55603 | FAM46A, C6orf37, XTP11 | family with sequence similarity 46, member A |
C00004631
|
| 115572 | FAM46B, RP11-344H11.8 | family with sequence similarity 46, member B |
C00004631
|
| 54855 | FAM46C | family with sequence similarity 46, member C |
C00004631
|
| 9130 | FAM50A, 9F, DXS9928E, HXC-26, HXC26, XAP5 | family with sequence similarity 50, member A |
C00004631
|
| 51307 | FAM53C, C5orf6 | family with sequence similarity 53, member C |
C00004631
|
| 81610 | FAM83D, C20orf129, CHICA, dJ616B8.3 | family with sequence similarity 83, member D |
C00004631
|
| 25940 | FAM98A | family with sequence similarity 98, member A |
C00004631
|
| 283742 | FAM98B | family with sequence similarity 98, member B |
C00004631
|
| 22909 | FAN1, KIAA1018, KMIN, MTMR15 | FANCD2/FANCI-associated nuclease 1 (EC:3.1.4.1) |
C00004631
|
| 2187 | FANCB, FA2, FAAP90, FAAP95, FAB, FACB | Fanconi anemia, complementation group B |
C00004631
|
| 2189 | FANCG, FAG, XRCC9 | Fanconi anemia, complementation group G |
C00004631
|
| 55215 | FANCI, KIAA1794 | Fanconi anemia, complementation group I |
C00004631
|
| 55120 | FANCL, FAAP43, PHF9, POG | Fanconi anemia, complementation group L |
C00004631
|
| 10160 | FARP1, CDEP, PLEKHC2, PPP1R75 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
C00004631
|
| 9855 | FARP2, FIR, FRG, PLEKHC3 | FERM, RhoGEF and pleckstrin domain protein 2 |
C00004631
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00004631
|
| 2194 | FASN, FAS, OA-519, SDR27X1 | fatty acid synthase (EC:2.3.1.85 2.3.1.38 2.3.1.39 2.3.1.41 3.1.2.14 4.2.1.59 1.1.1.100 1.3.1.39) |
C00004631
|
| 79675 | FASTKD1 | FAST kinase domains 1 |
C00004631
|
| 22868 | FASTKD2, KIAA0971 | FAST kinase domains 2 |
C00004631
|
| 2199 | FBLN2 | fibulin 2 |
C00004631
|
| 25827 | FBXL2, FBL2, FBL3 | F-box and leucine-rich repeat protein 2 |
C00004631
|
| 26223 | FBXL21, FBL3B, FBXL3B, FBXL3P, Fbl21 | F-box and leucine-rich repeat protein 21 (gene/pseudogene) |
C00004631
|
| 26233 | FBXL6, FBL6, FBL6A, PP14630 | F-box and leucine-rich repeat protein 6 |
C00004631
|
| 26267 | FBXO10, FBX10, PRMT11 | F-box protein 10 |
C00004631
|
| 115290 | FBXO17, FBG4, FBX26, FBXO26, Fbx17 | F-box protein 17 |
C00004631
|
| 26232 | FBXO2, FBG1, FBX2, Fbs1, NFB42, OCP1 | F-box protein 2 |
C00004631
|
| 26260 | FBXO25, FBX25 | F-box protein 25 |
C00004631
|
| 23219 | FBXO28, CENP-30, Fbx28 | F-box protein 28 |
C00004631
|
| 26273 | FBXO3, FBA, FBX3 | F-box protein 3 |
C00004631
|
| 84085 | FBXO30, Fbx30 | F-box protein 30 |
C00004631
|
| 79791 | FBXO31, FBX14, FBXO14, Fbx31, pp2386 | F-box protein 31 |
C00004631
|
| 150726 | FBXO41, FBX41 | F-box protein 41 |
C00004631
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00004631
|
| 2207 | FCER1G, FCRG | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
C00004631
|
| 2213 | FCGR2B, CD32, CD32B, FCG2, FCGR2, IGFR2 | Fc fragment of IgG, low affinity IIb, receptor (CD32) |
C00004631
|
| 9873 | FCHSD2, NWK, SH3MD3 | FCH and double SH3 domains 2 |
C00004631
|
| 2222 | FDFT1, DGPT, ERG9, SQS, SS | farnesyl-diphosphate farnesyltransferase 1 (EC:2.5.1.21) |
C00004631
|
| 2224 | FDPS, FPPS, FPS | farnesyl diphosphate synthase (EC:2.5.1.1 2.5.1.10) |
C00004631
|
| 2232 | FDXR, ADXR | ferredoxin reductase (EC:1.18.1.6) |
C00004631
|
| 56929 | FEM1C, EUROIMAGE686608, EUROIMAGE783647, FEM1A | fem-1 homolog c (C. elegans) |
C00004631
|
| 55612 | FERMT1, C20orf42, DTGCU2, KIND1, UNC112A, URP1 | fermitin family member 1 |
C00004631
|
| 10979 | FERMT2, KIND2, MIG2, PLEKHC1, UNC112, UNC112B, mig-2 | fermitin family member 2 |
C00004631
|
| 9637 | FEZ2, HUM3CL | fasciculation and elongation protein zeta 2 (zygin II) |
C00004631
|
| 2243 | FGA, Fib2 | fibrinogen alpha chain |
C00004631
|
| 2244 | FGB | fibrinogen beta chain |
C00004631
|
| 55785 | FGD6, ZFYVE24 | FYVE, RhoGEF and PH domain containing 6 |
C00004631
|
| 8817 | FGF18, FGF-18, ZFGF5 | fibroblast growth factor 18 |
C00004631
|
| 2252 | FGF7, HBGF-7, KGF | fibroblast growth factor 7 |
C00004631
|
| 143282 | FGFBP3, C10orf13, FGF-BP3 | fibroblast growth factor binding protein 3 |
C00004631
|
| 2261 | FGFR3, ACH, CD333, CEK2, HSFGFR3EX, JTK4 | fibroblast growth factor receptor 3 (EC:2.7.10.1) |
C00004631
|
| 2264 | FGFR4, CD334, JTK2, TKF | fibroblast growth factor receptor 4 (EC:2.7.10.1) |
C00004631
|
| 2266 | FGG | fibrinogen gamma chain |
C00004631
|
| 10875 | FGL2, T49, pT49 | fibrinogen-like 2 |
C00004631
|
| 2271 | FH, HLRCC, LRCC, MCL, MCUL1 | fumarate hydratase (EC:4.2.1.2) |
C00004631
|
| 2272 | FHIT, AP3Aase, FRA3B | fragile histidine triad (EC:3.6.1.29) |
C00004631
|
| 9896 | FIG4, ALS11, CMT4J, KIAA0274, SAC3, YVS, dJ249I4.1 | FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
C00004631
|
| 2277 | FIGF, VEGF-D, VEGFD | c-fos induced growth factor (vascular endothelial growth factor D) |
C00004631
|
| 55137 | FIGN | fidgetin |
C00004631
|
| 63979 | FIGNL1 | fidgetin-like 1 |
C00004631
|
| 11259 | FILIP1L, DOC-1, DOC1, GIP130, GIP90 | filamin A interacting protein 1-like |
C00004631
|
| 38488 |
C00004631
|
||
| 60681 | FKBP10, FKBP65, OI11, OI6, PPIASE, hFKBP65 | FK506 binding protein 10, 65 kDa (EC:5.2.1.8) |
C00004631
|
| 23770 | FKBP8, FKBP38, FKBPr38 | FK506 binding protein 8, 38kDa (EC:5.2.1.8) |
C00004631
|
| 80308 | FLAD1, FAD1, FADS, RP11-307C12.7 | flavin adenine dinucleotide synthetase 1 (EC:2.7.7.2) |
C00004631
|
| 201163 | FLCN, BHD, FLCL | folliculin |
C00004631
|
| 2316 | FLNA, ABP-280, ABPX, CSBS, CVD1, FLN, FLN-A, FLN1, FMD, MNS, NHBP, OPD, OPD1, OPD2, XLVD, XMVD | filamin A, alpha |
C00004631
|
| 2317 | FLNB, ABP-278, ABP-280, AOI, FH1, FLN-B, FLN1L, LRS1, SCT, TABP, TAP | filamin B, beta |
C00004631
|
| 23767 | FLRT3, HH21 | fibronectin leucine rich transmembrane protein 3 |
C00004631
|
| 55640 | FLVCR2, C14orf58, CCT, EPV, FLVCRL14q, MFSD7C, PVHH | feline leukemia virus subgroup C cellular receptor family, member 2 |
C00004631
|
| 2326 | FMO1 | flavin containing monooxygenase 1 (EC:1.14.13.8) |
C00004631
|
| 2330 | FMO5 | flavin containing monooxygenase 5 (EC:1.14.13.8) |
C00004631
|
| 2332 | FMR1, FMRP, FRAXA, POF, POF1 | fragile X mental retardation 1 |
C00004631
|
| 2335 | FN1, CIG, ED-B, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | fibronectin 1 |
C00004631
|
| 54874 | FNBP1L, C1orf39, TOCA1 | formin binding protein 1-like |
C00004631
|
| 64778 | FNDC3B, FAD104, PRO4979, YVTM2421 | fibronectin type III domain containing 3B |
C00004631
|
| 57600 | FNIP2, FNIPL, MAPO1 | folliculin interacting protein 2 |
C00004631
|
| 8061 | FOSL1, FRA, FRA1, fra-1 | FOS-like antigen 1 |
C00004631
|
| 3169 | FOXA1, HNF3A, TCF3A | forkhead box A1 |
C00004631
|
| 3171 | FOXA3, FKHH3, HNF3G, TCF3G | forkhead box A3 |
C00004631
|
| 2305 | FOXM1, FKHL16, FOXM1B, HFH-11, HFH11, HNF-3, INS-1, MPHOSPH2, MPP-2, MPP2, PIG29, TGT3, TRIDENT | forkhead box M1 |
C00004631
|
| 1112 | FOXN3, C14orf116, CHES1, PRO1635 | forkhead box N3 |
C00004631
|
| 27086 | FOXP1, 12CC4, QRF1, hFKH1B | forkhead box P1 |
C00004631
|
| 8790 | FPGT, GFPP | fucose-1-phosphate guanylyltransferase (EC:2.7.7.30) |
C00004631
|
| 2358 | FPR2, ALXR, FMLP-R-II, FMLPX, FPR2A, FPRH1, FPRH2, FPRL1, HM63, LXA4R | formyl peptide receptor 2 |
C00004631
|
| 10023 | FRAT1 | frequently rearranged in advanced T-cell lymphomas |
C00004631
|
| 23401 | FRAT2 | frequently rearranged in advanced T-cell lymphomas 2 |
C00004631
|
| 158326 | FREM1, BNAR, C9orf143, C9orf145, C9orf154, MOTA, TILRR, TRIGNO2 | FRAS1 related extracellular matrix 1 |
C00004631
|
| 2444 | FRK, GTK, PTK5, RAK | fyn-related kinase (EC:2.7.10.2) |
C00004631
|
| 257019 | FRMD3, 4.1O, EPB41L4O, EPB41LO, P410 | FERM domain containing 3 |
C00004631
|
| 23150 | FRMD4B, 6030440G05Rik, GRSP1 | FERM domain containing 4B |
C00004631
|
| 84978 | FRMD5 | FERM domain containing 5 |
C00004631
|
| 2487 | FRZB, FRE, FRITZ, FRP-3, FRZB-1, FRZB-PEN, FRZB1, FZRB, OS1, SFRP3, SRFP3, hFIZ | frizzled-related protein |
C00004631
|
| 10272 | FSTL3, FLRG, FSRP | follistatin-like 3 (secreted glycoprotein) |
C00004631
|
| 10841 | FTCD, LCHC1 | formimidoyltransferase cyclodeaminase (EC:2.1.2.5 4.3.1.4) |
C00004631
|
| 170384 | FUT11 | fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
C00004631
|
| 2528 | FUT6, FCT3A, FT1A, Fuc-TVI, FucT-VI | fucosyltransferase 6 (alpha (1,3) fucosyltransferase) (EC:2.4.1.65) |
C00004631
|
| 2530 | FUT8 | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) (EC:2.4.1.68) |
C00004631
|
| 5348 | FXYD1, PLM | FXYD domain containing ion transport regulator 1 |
C00004631
|
| 486 | FXYD2, ATP1G1, HOMG2 | FXYD domain containing ion transport regulator 2 |
C00004631
|
| 53827 | FXYD5, DYSAD, IWU1, KCT1, OIT2, PRO6241, RIC | FXYD domain containing ion transport regulator 5 |
C00004631
|
| 2535 | FZD2, Fz2, fz-2, fzE2, hFz2 | frizzled family receptor 2 |
C00004631
|
| 8324 | FZD7, FzE3 | frizzled family receptor 7 |
C00004631
|
| 50486 | G0S2, RP1-28O10.2 | G0/G1switch 2 |
C00004631
|
| 55632 | G2E3, KIAA1333, PHF7B | G2/M-phase specific E3 ubiquitin protein ligase |
C00004631
|
| 2538 | G6PC, G6PC1, G6PT, GSD1, GSD1a | glucose-6-phosphatase, catalytic subunit (EC:3.1.3.9) |
C00004631
|
| 2539 | G6PD, G6PD1 | glucose-6-phosphate dehydrogenase (EC:1.1.1.49) |
C00004631
|
| 2548 | GAA, LYAG | glucosidase, alpha; acid (EC:3.2.1.20) |
C00004631
|
| 126626 | GABPB2 | GA binding protein transcription factor, beta subunit 2 |
C00004631
|
| 2564 | GABRE | gamma-aminobutyric acid (GABA) A receptor, epsilon |
C00004631
|
| 2571 | GAD1, CPSQ1, GAD, SCP | glutamate decarboxylase 1 (brain, 67kDa) (EC:4.1.1.15) |
C00004631
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00004631
|
| 9514 | GAL3ST1, CST | galactose-3-O-sulfotransferase 1 (EC:2.8.2.11) |
C00004631
|
| 130589 | GALM, IBD1 | galactose mutarotase (aldose 1-epimerase) (EC:5.1.3.3) |
C00004631
|
| 2589 | GALNT1, GALNAC-T1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (EC:2.4.1.41) |
C00004631
|
| 55568 | GALNT10, GALNACT10, PPGALNACT10, PPGANTASE10 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) (EC:2.4.1.41) |
C00004631
|
| 63917 | GALNT11, GALNACT11 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) (EC:2.4.1.41) |
C00004631
|
| 2591 | GALNT3, GalNAc-T3, HFTC, HHS | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (EC:2.4.1.41) |
C00004631
|
| 8811 | GALR2, GAL2-R, GALNR2, GALR-2 | galanin receptor 2 |
C00004631
|
| 8139 | GAN, GAN1, KLHL16 | gigaxonin |
C00004631
|
| 64762 | GAREM, C18orf11, FAM59A, Gm944 | GRB2 associated, regulator of MAPK1 |
C00004631
|
| 2620 | GAS2 | growth arrest-specific 2 |
C00004631
|
| 283431 | GAS2L3, G2L3 | growth arrest-specific 2 like 3 |
C00004631
|
| 2623 | GATA1, ERYF1, GATA-1, GF-1, GF1, NF-E1, NFE1, XLANP, XLTDA, XLTT | GATA binding protein 1 (globin transcription factor 1) |
C00004631
|
| 2627 | GATA6 | GATA binding protein 6 |
C00004631
|
| 2628 | GATM, AGAT, AT, CCDS3 | glycine amidinotransferase (L-arginine:glycine amidinotransferase) (EC:2.1.4.1) |
C00004631
|
| 2632 | GBE1, APBD, GBE, GSD4 | glucan (1,4-alpha-), branching enzyme 1 (EC:2.4.1.18) |
C00004631
|
| 8729 | GBF1, ARF1GEF | golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
C00004631
|
| 25801 | GCA, GCL | grancalcin, EF-hand calcium binding protein |
C00004631
|
| 6936 | GCFC2, C2orf3, DNABF, GCF, TCF9 | GC-rich sequence DNA-binding factor 2 |
C00004631
|
| 2646 | GCKR, FGQTL5, GKRP | glucokinase (hexokinase 4) regulator |
C00004631
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00004631
|
| 2651 | GCNT2, CCAT, CTRCT13, GCNT2C, GCNT5, IGNT, II, NACGT1, NAGCT1, ULG3, bA360O19.2, bA421M1.1 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) (EC:2.4.1.150) |
C00004631
|
| 9245 | GCNT3, C2/4GnT, C24GNT, C2GNT2, C2GNTM, GNTM | glucosaminyl (N-acetyl) transferase 3, mucin type (EC:2.4.1.102 2.4.1.150) |
C00004631
|
| 145781 | GCOM1, GRINL1A, Gcom2, MYZAP, MYZAP-POLR2M, gcom | GRINL1A complex locus 1 |
C00004631
|
| 9615 | GDA, CYPIN, GUANASE, NEDASIN | guanine deaminase (EC:3.5.4.3) |
C00004631
|
| 10220 | GDF11, BMP-11, BMP11 | growth differentiation factor 11 |
C00004631
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00004631
|
| 2664 | GDI1, 1A, GDIL, MRX41, MRX48, OPHN2, RABGD1A, RABGDIA, XAP-4 | GDP dissociation inhibitor 1 |
C00004631
|
| 81544 | GDPD5, GDE2, PP1665 | glycerophosphodiester phosphodiesterase domain containing 5 |
C00004631
|
| 2669 | GEM, KIR | GTP binding protein overexpressed in skeletal muscle |
C00004631
|
| 8487 | GEMIN2, SIP1, SIP1-delta | gem (nuclear organelle) associated protein 2 |
C00004631
|
| 79833 | GEMIN6 | gem (nuclear organelle) associated protein 6 |
C00004631
|
| 85476 | GFM1, COXPD1, EFG, EFG1, EFGM, EGF1, GFM, hEFG1 | G elongation factor, mitochondrial 1 |
C00004631
|
| 54438 | GFOD1, ADG-90, C6orf114 | glucose-fructose oxidoreductase domain containing 1 |
C00004631
|
| 84514 | GHDC, D11LGP1, LGP1 | GH3 domain containing |
C00004631
|
| 84296 | GINS4, SLD5 | GINS complex subunit 4 (Sld5 homolog) |
C00004631
|
| 10755 | GIPC1, C19orf3, GIPC, GLUT1CBP, Hs.6454, IIP-1, NIP, RGS19IP1, SEMCAP, SYNECTIIN, SYNECTIN, TIP-2 | GIPC PDZ domain containing family, member 1 |
C00004631
|
| 54810 | GIPC2, SEMCAP-2, SEMCAP2 | GIPC PDZ domain containing family, member 2 |
C00004631
|
| 2705 | GJB1, CMTX, CMTX1, CX32 | gap junction protein, beta 1, 32kDa |
C00004631
|
| 2717 | GLA, GALA | galactosidase, alpha (EC:3.2.1.22) |
C00004631
|
| 113263 | GLCCI1, FAM117C, GCTR, GIG18, TSSN1 | glucocorticoid induced transcript 1 |
C00004631
|
| 26035 | GLCE, HSEPI | glucuronic acid epimerase (EC:5.1.3.17) |
C00004631
|
| 2731 | GLDC, GCE, GCSP, HYGN1 | glycine dehydrogenase (decarboxylating) (EC:1.4.4.2) |
C00004631
|
| 11010 | GLIPR1, CRISP7, GLIPR, RTVP1 | GLI pathogenesis-related 1 |
C00004631
|
| 11146 | GLMN, FAP, FAP48, FAP68, FKBPAP, GLML, GVM, VMGLOM | glomulin, FKBP associated protein |
C00004631
|
| 2739 | GLO1, GLOD1, GLYI | glyoxalase I (EC:4.4.1.5) |
C00004631
|
| 2741 | GLRA1, HKPX1, STHE | glycine receptor, alpha 1 |
C00004631
|
| 51022 | GLRX2, CGI-133, GRX2 | glutaredoxin 2 |
C00004631
|
| 2744 | GLS, AAD20, GAC, GAM, GLS1, KGA | glutaminase (EC:3.5.1.2) |
C00004631
|
| 27165 | GLS2, GA, GLS, LGA, hLGA | glutaminase 2 (liver, mitochondrial) (EC:3.5.1.2) |
C00004631
|
| 55830 | GLT8D1, MSTP139 | glycosyltransferase 8 domain containing 1 |
C00004631
|
| 388323 | GLTPD2 | glycolipid transfer protein domain containing 2 |
C00004631
|
| 2746 | GLUD1, GDH, GDH1, GLUD | glutamate dehydrogenase 1 (EC:1.4.1.3) |
C00004631
|
| 2747 | GLUD2, GDH2, GLUDP1 | glutamate dehydrogenase 2 (EC:1.4.1.3) |
C00004631
|
| 2762 | GMDS, GMD, SDR3E1 | GDP-mannose 4,6-dehydratase (EC:4.2.1.47) |
C00004631
|
| 9535 | GMFG, GMF-GAMMA | glia maturation factor, gamma |
C00004631
|
| 29925 | GMPPB, MDDGA14, MDDGB14, MDDGC14 | GDP-mannose pyrophosphorylase B (EC:2.7.7.13) |
C00004631
|
| 10672 | GNA13, G13 | guanine nucleotide binding protein (G protein), alpha 13 |
C00004631
|
| 2769 | GNA15, GNA16 | guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
C00004631
|
| 2770 | GNAI1, Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
C00004631
|
| 10399 | GNB2L1, Gnb2-rs1, H12.3, HLC-7, PIG21, RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
C00004631
|
| 2786 | GNG4 | guanine nucleotide binding protein (G protein), gamma 4 |
C00004631
|
| 27232 | GNMT | glycine N-methyltransferase (EC:2.1.1.20) |
C00004631
|
| 79158 | GNPTAB, GNPTA, ICD | N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (EC:2.7.8.17) |
C00004631
|
| 23015 | GOLGA8A, GM88 | golgin A8 family, member A |
C00004631
|
| 785067 |
C00004631
|
||
| 127845 | GOLT1A, CGI-141, GOT1, HMFN1187, RP11-203F10.3, YMR292W | golgi transport 1A |
C00004631
|
| 8733 | GPAA1, GAA1, hGAA1 | glycosylphosphatidylinositol anchor attachment 1 |
C00004631
|
| 57678 | GPAM, GPAT, GPAT1 | glycerol-3-phosphate acyltransferase, mitochondrial (EC:2.3.1.15) |
C00004631
|
| 7918 | GPANK1, ANKRD59, BAT4, D6S54E, G5, GPATCH10 | G patch domain and ankyrin repeats 1 |
C00004631
|
| 55668 | GPATCH2L, C14orf118 | G patch domain containing 2-like |
C00004631
|
| 2817 | GPC1, glypican | glypican 1 |
C00004631
|
| 10082 | GPC6, OMIMD1 | glypican 6 |
C00004631
|
| 23171 | GPD1L, GPD1-L | glycerol-3-phosphate dehydrogenase 1-like (EC:1.1.1.8) |
C00004631
|
| 2852 | GPER1, CEPR, CMKRL2, DRY12, FEG-1, GPCR-Br, GPER, GPR30, LERGU, LERGU2, LyGPR | G protein-coupled estrogen receptor 1 |
C00004631
|
| 170589 | GPHA2, A2, GPA2, ZSIG51 | glycoprotein hormone alpha 2 |
C00004631
|
| 2821 | GPI, AMF, GNPI, NLK, PGI, PHI, SA-36, SA36 | glucose-6-phosphate isomerase (EC:5.3.1.9) |
C00004631
|
| 11321 | GPN1, ATPBD1A, MBDIN, NTPBP, RPAP4, XAB1 | GPN-loop GTPase 1 |
C00004631
|
| 10457 | GPNMB, HGFIN, NMB | glycoprotein (transmembrane) nmb |
C00004631
|
| 166647 | GPR125, PGR21, TEM5L | G protein-coupled receptor 125 |
C00004631
|
| 115330 | GPR146, PGR8 | G protein-coupled receptor 146 |
C00004631
|
| 26996 | GPR160, GPCR1, GPCR150 | G protein-coupled receptor 160 |
C00004631
|
| 2859 | GPR35 | G protein-coupled receptor 35 |
C00004631
|
| 9289 | GPR56, BFPP, TM7LN4, TM7XN1 | G protein-coupled receptor 56 |
C00004631
|
| 10888 | GPR83, GIR, GPR72 | G protein-coupled receptor 83 |
C00004631
|
| 9052 | GPRC5A, GPCR5A, RAI3, RAIG1 | G protein-coupled receptor, family C, group 5, member A |
C00004631
|
| 55890 | GPRC5C, RAIG-3, RAIG3 | G protein-coupled receptor, family C, group 5, member C |
C00004631
|
| 29899 | GPSM2, CMCS, DFNB82, LGN, PINS | G-protein signaling modulator 2 |
C00004631
|
| 84706 | GPT2, ALT2 | glutamic pyruvate transaminase (alanine aminotransferase) 2 (EC:2.6.1.2) |
C00004631
|
| 2876 | GPX1, GPXD, GSHPX1 | glutathione peroxidase 1 (EC:1.11.1.9) |
C00004631
|
| 2877 | GPX2, GI-GPx, GPRP, GPRP-2, GPx-2, GPx-GI, GSHPX-GI, GSHPx-2 | glutathione peroxidase 2 (gastrointestinal) (EC:1.11.1.9) |
C00004631
|
| 2878 | GPX3, GPx-P, GSHPx-3, GSHPx-P | glutathione peroxidase 3 (plasma) (EC:1.11.1.9) |
C00004631
|
| 493869 | GPX8, EPLA847, GPx-8, GSHPx-8, UNQ847 | glutathione peroxidase 8 (putative) (EC:1.11.1.9) |
C00004631
|
| 2887 | GRB10, GRB-IR, Grb-10, IRBP, MEG1, RSS | growth factor receptor-bound protein 10 |
C00004631
|
| 2888 | GRB14 | growth factor receptor-bound protein 14 |
C00004631
|
| 80273 | GRPEL1, HMGE | GrpE-like 1, mitochondrial (E. coli) |
C00004631
|
| 55876 | GSDMB, GSDML, PRO2521 | gasdermin B |
C00004631
|
| 23199 | GSE1, KIAA0182 | Gse1 coiled-coil protein |
C00004631
|
| 2931 | GSK3A | glycogen synthase kinase 3 alpha (EC:2.7.11.1 2.7.11.26) |
C00004631
|
| 2932 | GSK3B | glycogen synthase kinase 3 beta (EC:2.7.11.1 2.7.11.26) |
C00004631
|
| 2934 | GSN, ADF, AGEL | gelsolin |
C00004631
|
| 2936 | GSR | glutathione reductase (EC:1.8.1.7) |
C00004631
|
| 2941 | GSTA4, GSTA4-4, GTA4 | glutathione S-transferase alpha 4 (EC:2.5.1.18) |
C00004631
|
| 373156 | GSTK1, GST, GST_13-13, GST13, GST13-13, GSTK1-1, hGSTK1 | glutathione S-transferase kappa 1 (EC:2.5.1.18) |
C00004631
|
| 2948 | GSTM4, GSTM4-4, GTM4 | glutathione S-transferase mu 4 (EC:2.5.1.18) |
C00004631
|
| 119391 | GSTO2, GSTO_2-2, bA127L20.1 | glutathione S-transferase omega 2 (EC:2.5.1.18 1.8.5.1 1.20.4.2) |
C00004631
|
| 2961 | GTF2E2, FE, TF2E2, TFIIE-B | general transcription factor IIE, polypeptide 2, beta 34kDa |
C00004631
|
| 2965 | GTF2H1, BTF2, P62, TFB1, TFIIH | general transcription factor IIH, polypeptide 1, 62kDa |
C00004631
|
| 2968 | GTF2H4, P52, TFB2, TFIIH | general transcription factor IIH, polypeptide 4, 52kDa |
C00004631
|
| 404672 | GTF2H5, C6orf175, TFB5, TFIIH, TGF2H5, TTD, TTD-A, TTDA, bA120J8.2 | general transcription factor IIH, polypeptide 5 |
C00004631
|
| 29083 | GTPBP8, HSPC135 | GTP-binding protein 8 (putative) |
C00004631
|
| 2979 | GUCA1B, GCAP2, GUCA2, RP48 | guanylate cyclase activator 1B (retina) |
C00004631
|
| 83606 | GUCD1, C22orf13, LLN4 | guanylyl cyclase domain containing 1 |
C00004631
|
| 60558 | GUF1, EF-4 | GUF1 GTPase homolog (S. cerevisiae) |
C00004631
|
| 51454 | GULP1, CED-6, CED6, GULP | GULP, engulfment adaptor PTB domain containing 1 |
C00004631
|
| 91316 | GUSBP11 | glucuronidase, beta pseudogene 11 |
C00004631
|
| 8908 | GYG2, GN-2, GN2 | glycogenin 2 (EC:2.4.1.186) |
C00004631
|
| 2997 | GYS1, GSY, GYS | glycogen synthase 1 (muscle) (EC:2.4.1.11) |
C00004631
|
| 8971 | H1FX, H1X | H1 histone family, member X |
C00004631
|
| 26061 | HACL1, 2-HPCL, HPCL, HPCL2, PHYH2 | 2-hydroxyacyl-CoA lyase 1 |
C00004631
|
| 3034 | HAL, HIS, HSTD | histidine ammonia-lyase (EC:4.3.1.3) |
C00004631
|
| 57817 | HAMP, HEPC, HFE2B, LEAP1, PLTR | hepcidin antimicrobial peptide |
C00004631
|
| 79441 | HAUS3, C4orf15, IT1, dgt3 | HAUS augmin-like complex, subunit 3 |
C00004631
|
| 1839 | HBEGF, DTR, DTS, DTSF, HEGFL | heparin-binding EGF-like growth factor |
C00004631
|
| 3048 | HBG2, TNCY | hemoglobin, gamma G |
C00004631
|
| 26959 | HBP1 | HMG-box transcription factor 1 |
C00004631
|
| 54985 | HCFC1R1, HPIP | host cell factor C1 regulator 1 (XPO1 dependent) |
C00004631
|
| 3059 | HCLS1, CTTNL, HS1 | hematopoietic cell-specific Lyn substrate 1 |
C00004631
|
| 3067 | HDC | histidine decarboxylase (EC:4.1.1.22) |
C00004631
|
| 55127 | HEATR1, BAP28, UTP10 | HEAT repeat containing 1 |
C00004631
|
| 55027 | HEATR3, SYO1 | HEAT repeat containing 3 |
C00004631
|
| 23593 | HEBP2, C6ORF34B, C6orf34, PP23, SOUL | heme binding protein 2 |
C00004631
|
| 143279 | HECTD2 | HECT domain containing E3 ubiquitin protein ligase 2 (EC:6.3.2.-) |
C00004631
|
| 283450 | HECTD4, C12orf51 | HECT domain containing E3 ubiquitin protein ligase 4 (EC:6.3.2.-) |
C00004631
|
| 3070 | HELLS, LSH, PASG, SMARCA6 | helicase, lymphoid-specific |
C00004631
|
| 9931 | HELZ, DHRC, DRHC, HUMORF5 | helicase with zinc finger |
C00004631
|
| 9843 | HEPH, CPL | hephaestin |
C00004631
|
| 8924 | HERC2, D15F37S1, SHEP1, jdf2, p528 | HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
C00004631
|
| 9709 | HERPUD1, HERP, Mif1, SUP | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
C00004631
|
| 3280 | HES1, HES-1, HHL, HRY, bHLHb39 | hes family bHLH transcription factor 1 |
C00004631
|
| 57801 | HES4, bHLHb42 | hes family bHLH transcription factor 4 |
C00004631
|
| 3074 | HEXB, ENC-1AS | hexosaminidase B (beta polypeptide) (EC:3.2.1.52) |
C00004631
|
| 124790 | HEXIM2 | hexamethylene bis-acetamide inducible 2 |
C00004631
|
| 9146 | HGS, HRS | hepatocyte growth factor-regulated tyrosine kinase substrate |
C00004631
|
| 3087 | HHEX, HEX, HMPH, HOX11L-PEN, PRH, PRHX | hematopoietically expressed homeobox |
C00004631
|
| 11147 | HHLA3 | HERV-H LTR-associating 3 |
C00004631
|
| 26275 | HIBCH, HIBYLCOAH | 3-hydroxyisobutyryl-CoA hydrolase (EC:3.1.2.4) |
C00004631
|
| 5825 | ABCD3, ABC43, PMP70, PXMP1, ZWS2 | ATP-binding cassette, sub-family D (ALD), member 3 |
C00004631
|
| 29923 | HILPDA, C7orf68, HIG-2, HIG2 | hypoxia inducible lipid droplet-associated |
C00004631
|
| 135114 | HINT3, HINT4 | histidine triad nucleotide binding protein 3 (EC:3.-.-.-) |
C00004631
|
| 3006 | HIST1H1C, H1.2, H1C, H1F2, H1s-1 | histone cluster 1, H1c |
C00004631
|
| 8334 | HIST1H2AC, H2A/l, H2AFL, dJ221C16.4 | histone cluster 1, H2ac |
C00004631
|
| 3012 | HIST1H2AE, H2A.1, H2A.2, H2A/a, H2AFA | histone cluster 1, H2ae |
C00004631
|
| 3017 | HIST1H2BD, H2B.1B, H2B/b, H2BFB, HIRIP2, dJ221C16.6 | histone cluster 1, H2bd |
C00004631
|
| 8339 | HIST1H2BG, H2B.1A, H2B/a, H2BFA, dJ221C16.8 | histone cluster 1, H2bg |
C00004631
|
| 8358 | HIST1H3B, H3/l, H3FL | histone cluster 1, H3b |
C00004631
|
| 8351 | HIST1H3D, H3/b, H3FB | histone cluster 1, H3d |
C00004631
|
| 8364 | HIST1H4C, H4/g, H4FG, dJ221C16.1 | histone cluster 1, H4c |
C00004631
|
| 8290 | HIST3H3, H3.4, H3/g, H3FT, H3t | histone cluster 3, H3 |
C00004631
|
| 3097 | HIVEP2, HIV-EP2, MBP-2, MIBP1, SHN2, ZAS2, ZNF40B | human immunodeficiency virus type I enhancer binding protein 2 |
C00004631
|
| 3099 | HK2, HKII, HXK2 | hexokinase 2 (EC:2.7.1.1) |
C00004631
|
| 284459 | HKR1, ZNF875 | HKR1, GLI-Kruppel zinc finger family member |
C00004631
|
| 3108 | HLA-DMA, D6S222E, DMA, HLADM, RING6 | major histocompatibility complex, class II, DM alpha |
C00004631
|
| 3131 | HLF | hepatic leukemia factor |
C00004631
|
| 6596 | HLTF, HIP116, HIP116A, HLTF1, RNF80, SMARCA3, SNF2L3, ZBU1 | helicase-like transcription factor |
C00004631
|
| 3145 | HMBS, PBG-D, PBGD, PORC, UPS | hydroxymethylbilane synthase (EC:2.5.1.61) |
C00004631
|
| 3159 | HMGA1, HMG-R, HMGA1A, HMGIY | high mobility group AT-hook 1 |
C00004631
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00004631
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00004631
|
| 3155 | HMGCL, HL | 3-hydroxymethyl-3-methylglutaryl-CoA lyase (EC:4.1.3.4) |
C00004631
|
| 3156 | HMGCR, LDLCQ3 | 3-hydroxy-3-methylglutaryl-CoA reductase (EC:1.1.1.34) |
C00004631
|
| 3157 | HMGCS1, HMGCS | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) (EC:2.3.3.10) |
C00004631
|
| 9324 | HMGN3, PNAS-25, TRIP7 | high mobility group nucleosomal binding domain 3 |
C00004631
|
| 23526 | HMHA1, ARHGAP45, HA-1, HLA-HA1 | histocompatibility (minor) HA-1 |
C00004631
|
| 3161 | HMMR, CD168, IHABP, RHAMM | hyaluronan-mediated motility receptor (RHAMM) |
C00004631
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00004631
|
| 3174 | HNF4G, NR2A2, NR2A3 | hepatocyte nuclear factor 4, gamma |
C00004631
|
| 3176 | HNMT, HMT, HNMT-S1, HNMT-S2 | histamine N-methyltransferase (EC:2.1.1.8) |
C00004631
|
| 3181 | HNRNPA2B1, HNRNPA2, HNRNPB1, HNRPA2, HNRPA2B1, HNRPB1, IBMPFD2, RNPA2, SNRPB1 | heterogeneous nuclear ribonucleoprotein A2/B1 |
C00004631
|
| 3184 | HNRNPD, AUF1, AUF1A, HNRPD, P37, hnRNPD0 | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
C00004631
|
| 9987 | HNRNPDL, HNRNP, HNRPDL, JKTBP, JKTBP2, laAUF1 | heterogeneous nuclear ribonucleoprotein D-like |
C00004631
|
| 3187 | HNRNPH1, HNRPH, HNRPH1, hnRNPH | heterogeneous nuclear ribonucleoprotein H1 (H) |
C00004631
|
| 3189 | HNRNPH3, 2H9, HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) |
C00004631
|
| 380185 |
C00004631
|
||
| 51361 | HOOK1, HK1 | hook microtubule-tethering protein 1 |
C00004631
|
| 29911 | HOOK2, HK2 | hook microtubule-tethering protein 2 |
C00004631
|
| 84376 | HOOK3, HK3 | hook microtubule-tethering protein 3 |
C00004631
|
| 3200 | HOXA3, HOX1, HOX1E | homeobox A3 |
C00004631
|
| 3201 | HOXA4, HOX1, HOX1D | homeobox A4 |
C00004631
|
| 3202 | HOXA5, HOX1, HOX1.3, HOX1C | homeobox A5 |
C00004631
|
| 3231 | HOXD1, HOX4, HOX4G, Hox-4.7 | homeobox D1 |
C00004631
|
| 3240 | HP, BP, HP2ALPHA2, HPA1S | haptoglobin |
C00004631
|
| 3249 | HPN, TMPRSS1 | hepsin (EC:3.4.21.106) |
C00004631
|
| 3250 | HPR, A-259H10.2, HP | haptoglobin-related protein |
C00004631
|
| 3265 | HRAS, C-BAS/HAS, C-H-RAS, C-HA-RAS1, CTLO, H-RASIDX, HAMSV, HRAS1, K-RAS, N-RAS, RASH1 | Harvey rat sarcoma viral oncogene homolog |
C00004631
|
| 64342 | HS1BP3, ETM2, HS1-BP3 | HCLS1 binding protein 3 |
C00004631
|
| 9653 | HS2ST1, dJ604K5.2 | heparan sulfate 2-O-sulfotransferase 1 (EC:2.8.2.-) |
C00004631
|
| 222537 | HS3ST5, 3-OST-5, 3OST5, HS3OST5, NBLA04021 | heparan sulfate (glucosamine) 3-O-sulfotransferase 5 (EC:2.8.2.23) |
C00004631
|
| 3294 | HSD17B2, EDH17B2, HSD17, SDR9C2 | hydroxysteroid (17-beta) dehydrogenase 2 (EC:1.1.1.62 1.1.1.239) |
C00004631
|
| 3295 | HSD17B4, DBP, MFE-2, MPF-2, PRLTS1, SDR8C1 | hydroxysteroid (17-beta) dehydrogenase 4 (EC:4.2.1.107 4.2.1.119 1.1.1.n12) |
C00004631
|
| 8630 | HSD17B6, HSE, RODH, SDR9C6 | hydroxysteroid (17-beta) dehydrogenase 6 (EC:1.1.1.62 1.1.1.239 1.1.1.105) |
C00004631
|
| 3297 | HSF1, HSTF1 | heat shock transcription factor 1 |
C00004631
|
| 3299 | HSF4, CTM, CTRCT5 | heat shock transcription factor 4 |
C00004631
|
| 39078 |
C00004631
|
||
| 7184 | HSP90B1, ECGP, GP96, GRP94, TRA1 | heat shock protein 90kDa beta (Grp94), member 1 |
C00004631
|
| 259217 | HSPA12A | heat shock 70kDa protein 12A |
C00004631
|
| 51182 | HSPA14, HSP70-4, HSP70L1 | heat shock 70kDa protein 14 |
C00004631
|
| 3303 | HSPA1A, HSP70-1, HSP70-1A, HSP70I, HSP72, HSPA1 | heat shock 70kDa protein 1A |
C00004631
|
| 3304 | HSPA1B, HSP70-1B, HSP70-2 | heat shock 70kDa protein 1B |
C00004631
|
| 3308 | HSPA4, APG-2, HS24/P52, HSPH2, RY, hsp70, hsp70RY | heat shock 70kDa protein 4 |
C00004631
|
| 22824 | HSPA4L, APG-1, HSPH3, Osp94 | heat shock 70kDa protein 4-like |
C00004631
|
| 3309 | HSPA5, BIP, GRP78, MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
C00004631
|
| 3315 | HSPB1, CMT2F, HMN2B, HS.76067, HSP27, HSP28, Hsp25, SRP27 | heat shock 27kDa protein 1 |
C00004631
|
| 79663 | HSPBAP1, PASS1 | HSPB (heat shock 27kDa) associated protein 1 |
C00004631
|
| 3329 | HSPD1, CPN60, GROEL, HLD4, HSP-60, HSP60, HSP65, HuCHA60, SPG13 | heat shock 60kDa protein 1 (chaperonin) |
C00004631
|
| 3336 | HSPE1, CPN10, EPF, GROES, HSP10 | heat shock 10kDa protein 1 |
C00004631
|
| 10808 | HSPH1, HSP105, HSP105A, HSP105B, NY-CO-25 | heat shock 105kDa/110kDa protein 1 |
C00004631
|
| 27429 | HTRA2, OMI, PARK13, PRSS25 | HtrA serine peptidase 2 (EC:3.4.21.108) |
C00004631
|
| 219844 | HYLS1, HLS | hydrolethalus syndrome 1 |
C00004631
|
| 3376 | IARS, IARS1, ILERS, ILRS, IRS, PRO0785 | isoleucyl-tRNA synthetase (EC:6.1.1.5) |
C00004631
|
| 3385 | ICAM3, CD50, CDW50, ICAM-R | intercellular adhesion molecule 3 |
C00004631
|
| 3386 | ICAM4, CD242, LW | intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
C00004631
|
| 3398 | ID2, GIG8, ID2A, ID2H, bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
C00004631
|
| 3399 | ID3, HEIR-1, bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
C00004631
|
| 3417 | IDH1, IDCD, IDH, IDP, IDPC, PICD | isocitrate dehydrogenase 1 (NADP+), soluble (EC:1.1.1.42) |
C00004631
|
| 3422 | IDI1, IPP1, IPPI1 | isopentenyl-diphosphate delta isomerase 1 (EC:5.3.3.2) |
C00004631
|
| 3620 | IDO1, IDO, IDO-1, INDO | indoleamine 2,3-dioxygenase 1 (EC:1.13.11.52) |
C00004631
|
| 9592 | IER2, ETR101 | immediate early response 2 |
C00004631
|
| 8870 | IER3, DIF-2, DIF2, GLY96, IEX-1, IEX-1L, IEX1, PRG1 | immediate early response 3 |
C00004631
|
| 51278 | IER5, SBBI48 | immediate early response 5 |
C00004631
|
| 389792 | IER5L, bA247A12.2 | immediate early response 5-like |
C00004631
|
| 122509 | IFI27L1, FAM14B, ISG12C | interferon, alpha-inducible protein 27-like 1 |
C00004631
|
| 10437 | IFI30, GILT, IFI-30, IP30 | interferon, gamma-inducible protein 30 |
C00004631
|
| 2537 | IFI6, 6-16, FAM14C, G1P3, IFI-6-16, IFI616 | interferon, alpha-inducible protein 6 |
C00004631
|
| 64135 | IFIH1, Hlcd, IDDM19, MDA-5, MDA5, RLR-2 | interferon induced with helicase C domain 1 (EC:3.6.4.13) |
C00004631
|
| 3434 | IFIT1, C56, G10P1, IFI-56, IFI-56K, IFI56, IFIT-1, IFNAI1, ISG56, P56, RNM561 | interferon-induced protein with tetratricopeptide repeats 1 |
C00004631
|
| 24138 | IFIT5, P58, RI58 | interferon-induced protein with tetratricopeptide repeats 5 |
C00004631
|
| 3454 | IFNAR1, AVP, IFN-alpha-REC, IFNAR, IFNBR, IFRC | interferon (alpha, beta and omega) receptor 1 |
C00004631
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00004631
|
| 3460 | IFNGR2, AF-1, IFGR2, IFNGT1 | interferon gamma receptor 2 (interferon gamma transducer 1) |
C00004631
|
| 26160 | IFT172, NPHP17, SLB, osm-1, wim | intraflagellar transport 172 homolog (Chlamydomonas) |
C00004631
|
| 57560 | IFT80, ATD2, WDR56 | intraflagellar transport 80 homolog (Chlamydomonas) |
C00004631
|
| 9543 | IGDCC3, HsT18880, PUNC | immunoglobulin superfamily, DCC subclass, member 3 |
C00004631
|
| 57722 | IGDCC4, DDM36, NOPE | immunoglobulin superfamily, DCC subclass, member 4 |
C00004631
|
| 10643 | IGF2BP3, CT98, IMP-3, IMP3, KOC1, VICKZ3 | insulin-like growth factor 2 mRNA binding protein 3 |
C00004631
|
| 3484 | IGFBP1, AFBP, IBP1, IGF-BP25, PP12, hIGFBP-1 | insulin-like growth factor binding protein 1 |
C00004631
|
| 3485 | IGFBP2, IBP2, IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa |
C00004631
|
| 3486 | IGFBP3, BP-53, IBP3 | insulin-like growth factor binding protein 3 |
C00004631
|
| 3487 | IGFBP4, BP-4, HT29-IGFBP, IBP4, IGFBP-4 | insulin-like growth factor binding protein 4 |
C00004631
|
| 3547 | IGSF1, CHTE, IGCD1, IGDC1, INHBP, PGSF2, p120 | immunoglobulin superfamily, member 1 |
C00004631
|
| 121457 | IKBIP, IKIP | IKBKB interacting protein |
C00004631
|
| 8518 | IKBKAP, DYS, ELP1, FD, IKAP, IKI3, TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
C00004631
|
| 3586 | IL10, CSIF, GVHDS, IL-10, IL10A, TGIF | interleukin 10 |
C00004631
|
| 3589 | IL11, AGIF, IL-11 | interleukin 11 |
C00004631
|
| 55540 | IL17RB, CRL4, EVI27, IL17BR, IL17RH1 | interleukin 17 receptor B |
C00004631
|
| 3552 | IL1A, IL-1A, IL1, IL1-ALPHA, IL1F1 | interleukin 1, alpha |
C00004631
|
| 3554 | IL1R1, CD121A, D2S1473, IL-1R-alpha, IL1R, IL1RA, P80 | interleukin 1 receptor, type I |
C00004631
|
| 7850 | IL1R2, CD121b, CDw121b, IL-1R-2, IL-1RT-2, IL-1RT2, IL1R2c, IL1RB | interleukin 1 receptor, type II |
C00004631
|
| 9173 | IL1RL1, DER4, FIT-1, IL33R, ST2, ST2L, ST2V, T1 | interleukin 1 receptor-like 1 |
C00004631
|
| 3558 | IL2, IL-2, TCGF, lymphokine | interleukin 2 |
C00004631
|
| 53833 | IL20RB, DIRS1, FNDC6, IL-20R2 | interleukin 20 receptor beta |
C00004631
|
| 58985 | IL22RA1, CRF2-9, IL22R, IL22R1 | interleukin 22 receptor, alpha 1 |
C00004631
|
| 9235 | IL32, IL-32alpha, IL-32beta, IL-32delta, IL-32gamma, NK4, TAIF, TAIFa, TAIFb, TAIFc, TAIFd | interleukin 32 |
C00004631
|
| 3566 | IL4R, CD124, IL-4RA, IL4RA | interleukin 4 receptor |
C00004631
|
| 3572 | IL6ST, CD130, CDW130, GP130, IL-6RB | interleukin 6 signal transducer (gp130, oncostatin M receptor) |
C00004631
|
| 3574 | IL7, IL-7 | interleukin 7 |
C00004631
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00004631
|
| 196294 | IMMP1L, IMP1, IMP1-LIKE | IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
C00004631
|
| 10989 | IMMT, HMP, MINOS2, P87, P87/89, P89, PIG52 | inner membrane protein, mitochondrial |
C00004631
|
| 55272 | IMP3, BRMS2, C15orf12, MRPS4 | IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
C00004631
|
| 3613 | IMPA2 | inositol(myo)-1(or 4)-monophosphatase 2 (EC:3.1.3.25) |
C00004631
|
| 3625 | INHBB | inhibin, beta B |
C00004631
|
| 83729 | INHBE | inhibin, beta E |
C00004631
|
| 54617 | INO80, INO80A, INOC1, hINO80 | INO80 complex subunit (EC:3.6.4.12) |
C00004631
|
| 83444 | INO80B, HMGA1L4, HMGIYL4, IES2, PAP-1BP, PAPA-1, ZNHIT4, hIes2 | INO80 complex subunit B |
C00004631
|
| 3630 | INS, IDDM2, ILPR, IRDN, MODY10 | insulin |
C00004631
|
| 55656 | INTS8, C8orf52, INT8 | integrator complex subunit 8 |
C00004631
|
| 27152 | INTU, INT, PDZD6, PDZK6 | inturned planar cell polarity protein |
C00004631
|
| 51447 | IP6K2, IHPK2, PIUS | inositol hexakisphosphate kinase 2 (EC:2.7.4.21) |
C00004631
|
| 10526 | IPO8, RANBP8 | importin 8 |
C00004631
|
| 132141 | IQCF1 | IQ motif containing F1 |
C00004631
|
| 8826 | IQGAP1, HUMORFA01, SAR1, p195 | IQ motif containing GTPase activating protein 1 |
C00004631
|
| 10788 | IQGAP2 | IQ motif containing GTPase activating protein 2 |
C00004631
|
| 134728 | IRAK1BP1, AIP70, SIMPL | interleukin-1 receptor-associated kinase 1 binding protein 1 |
C00004631
|
| 64207 | IRF2BPL, C14orf4, EAP1 | interferon regulatory factor 2 binding protein-like |
C00004631
|
| 3661 | IRF3 | interferon regulatory factor 3 |
C00004631
|
| 3667 | IRS1, HIRS-1 | insulin receptor substrate 1 |
C00004631
|
| 79191 | IRX3, IRX-1, IRXB1 | iroquois homeobox 3 |
C00004631
|
| 9636 | ISG15, G1P2, IFI15, IP17, UCRP, hUCRP | ISG15 ubiquitin-like modifier |
C00004631
|
| 145501 | ISM2, TAIL1, THSD3 | isthmin 2 |
C00004631
|
| 51015 | ISOC1 | isochorismatase domain containing 1 |
C00004631
|
| 91464 | ISX, Pix-1, RAXLX | intestine-specific homeobox |
C00004631
|
| 51477 | ISYNA1, INO1, INOS, IPS, IPS_1, IPS-1 | inositol-3-phosphate synthase 1 (EC:5.5.1.4) |
C00004631
|
| 8515 | ITGA10, PRO827 | integrin, alpha 10 |
C00004631
|
| 3673 | ITGA2, BR, CD49B, GPIa, HPA-5, VLA-2, VLAA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
C00004631
|
| 3655 | ITGA6, CD49f, ITGA6B, VLA-6 | integrin, alpha 6 |
C00004631
|
| 3685 | ITGAV, CD51, MSK8, VNRA, VTNR | integrin, alpha V |
C00004631
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00004631
|
| 23421 | ITGB3BP, CENP-R, CENPR, HSU37139, NRIF3, TAP20 | integrin beta 3 binding protein (beta3-endonexin) |
C00004631
|
| 3697 | ITIH1, H1P, IATIH, IGHEP1, ITI-HC1, ITIH, SHAP | inter-alpha-trypsin inhibitor heavy chain 1 |
C00004631
|
| 3699 | ITIH3, H3P | inter-alpha-trypsin inhibitor heavy chain 3 |
C00004631
|
| 3706 | ITPKA, IP3-3KA, IP3KA | inositol-trisphosphate 3-kinase A (EC:2.7.1.127) |
C00004631
|
| 3708 | ITPR1, INSP3R1, IP3R, IP3R1, SCA15, SCA16, SCA29 | inositol 1,4,5-trisphosphate receptor, type 1 |
C00004631
|
| 3709 | ITPR2, IP3R2 | inositol 1,4,5-trisphosphate receptor, type 2 |
C00004631
|
| 3710 | ITPR3, IP3R, IP3R3 | inositol 1,4,5-trisphosphate receptor, type 3 |
C00004631
|
| 85450 | ITPRIP, DANGER, KIAA1754, bA127L20, bA127L20.2 | inositol 1,4,5-trisphosphate receptor interacting protein |
C00004631
|
| 150771 | ITPRIPL1, KIAA1754L | inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
C00004631
|
| 6453 | ITSN1, ITSN, SH3D1A, SH3P17 | intersectin 1 (SH3 domain protein) |
C00004631
|
| 10625 | IVNS1ABP, FLARA3, HSPC068, KLHL39, ND1, NS-1, NS1-BP, NS1BP | influenza virus NS1A binding protein |
C00004631
|
| 182 | JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1 | jagged 1 |
C00004631
|
| 3716 | JAK1, JAK1A, JAK1B, JTK3 | Janus kinase 1 (EC:2.7.10.2) |
C00004631
|
| 3717 | JAK2, JTK10, THCYT3 | Janus kinase 2 (EC:2.7.10.2) |
C00004631
|
| 122953 | JDP2, JUNDM2 | Jun dimerization protein 2 |
C00004631
|
| 133746 | JMY, WHDC1L3 | junction mediating and regulatory protein, p53 cofactor |
C00004631
|
| 368 | ABCC6, ABC34, ARA, EST349056, GACI2, MLP1, MOAT-E, MOATE, MRP6, PXE, PXE1, URG7 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
C00004631
|
| 10057 | ABCC5, ABC33, EST277145, MOAT-C, MOATC, MRP5, SMRP, pABC11 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
C00004631
|
| 23189 | KANK1, ANKRD15, CPSQ2, KANK | KN motif and ankyrin repeat domains 1 |
C00004631
|
| 163782 | KANK4, ANKRD38, RP5-1155K23.5, dJ1078M7.1 | KN motif and ankyrin repeat domains 4 |
C00004631
|
| 8850 | KAT2B, CAF, P, P/CAF, PCAF | K(lysine) acetyltransferase 2B (EC:2.3.1.48) |
C00004631
|
| 84056 | KATNAL1 | katanin p60 subunit A-like 1 (EC:3.6.4.3) |
C00004631
|
| 79768 | KATNBL1, C15orf29 | katanin p80 subunit B-like 1 |
C00004631
|
| 23254 | KAZN, KAZ | kazrin, periplakin interacting protein |
C00004631
|
| 143879 | KBTBD3, BKLHD3 | kelch repeat and BTB (POZ) domain containing 3 |
C00004631
|
| 55709 | KBTBD4, BKLHD4, HSPC252 | kelch repeat and BTB (POZ) domain containing 4 |
C00004631
|
| 89890 | KBTBD6 | kelch repeat and BTB (POZ) domain containing 6 |
C00004631
|
| 84078 | KBTBD7 | kelch repeat and BTB (POZ) domain containing 7 |
C00004631
|
| 3766 | KCNJ10, BIRK-10, KCNJ13-PEN, KIR1.2, KIR4.1, SESAME | potassium inwardly-rectifying channel, subfamily J, member 10 |
C00004631
|
| 3770 | KCNJ14, IRK4, KIR2.4 | potassium inwardly-rectifying channel, subfamily J, member 14 |
C00004631
|
| 253980 | KCTD13, PDIP1, POLDIP1, hBACURD1 | potassium channel tetramerization domain containing 13 |
C00004631
|
| 65987 | KCTD14 | potassium channel tetramerization domain containing 14 |
C00004631
|
| 200845 | KCTD6, KCASH3 | potassium channel tetramerization domain containing 6 |
C00004631
|
| 79070 | KDELC1, EP58, KDEL1 | KDEL (Lys-Asp-Glu-Leu) containing 1 |
C00004631
|
| 221656 | KDM1B, AOF1, C6orf193, LSD2, bA204B7.3, dJ298J15.2 | lysine (K)-specific demethylase 1B |
C00004631
|
| 55818 | KDM3A, JHDM2A, JHMD2A, JMJD1, JMJD1A, TSGA | lysine (K)-specific demethylase 3A |
C00004631
|
| 9817 | KEAP1, INrf2, KLHL19 | kelch-like ECH-associated protein 1 |
C00004631
|
| 23351 | KHNYN, KIAA0323 | KH and NYN domain containing |
C00004631
|
| 9897 | SPG8 | KIAA0196 |
C00004631
|
| 9692 | MRPP3, PRORP | KIAA0391 |
C00004631
|
| 23247 | KIAA0556 |
C00004631
|
|
| 9851 | KIAA0753 |
C00004631
|
|
| 653319 | KIAA0895L | KIAA0895-like |
C00004631
|
| 22889 | BLOM7, RP11-336K24.1 | KIAA0907 |
C00004631
|
| 23240 | TMEM131L | KIAA0922 |
C00004631
|
| 57214 | CCSP1, TMEM2L | KIAA1199 |
C00004631
|
| 57482 | KIAA1211 |
C00004631
|
|
| 57221 | A7322, ARFGEF3, BIG3, C6orf92, PPP1R33, dJ171N11.1 | KIAA1244 |
C00004631
|
| 57650 | CIP2A, p90 | KIAA1524 |
C00004631
|
| 55196 | C12orf35 | KIAA1551 |
C00004631
|
| 57691 | KIAA1586 |
C00004631
|
|
| 85457 | KIAA1737 |
C00004631
|
|
| 85452 | KIAA1751 |
C00004631
|
|
| 3832 | KIF11, EG5, HKSP, KNSL1, MCLMR, TRIP5 | kinesin family member 11 |
C00004631
|
| 9928 | KIF14 | kinesin family member 14 |
C00004631
|
| 56992 | KIF15, HKLP2, KNSL7, NY-BR-62 | kinesin family member 15 |
C00004631
|
| 55614 | KIF16B, C20orf23, KISC20ORF, SNX23 | kinesin family member 16B |
C00004631
|
| 81930 | KIF18A, MS-KIF18A | kinesin family member 18A |
C00004631
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00004631
|
| 9585 | KIF20B, CT90, KRMP1, MPHOSPH1, MPP-1, MPP1 | kinesin family member 20B |
C00004631
|
| 55605 | KIF21A, CFEOM1, FEOM1, FEOM3A | kinesin family member 21A |
C00004631
|
| 9493 | KIF23, CHO1, KNSL5, MKLP-1, MKLP1 | kinesin family member 23 |
C00004631
|
| 3796 | KIF2A, CDCBM3, HK2, KIF2 | kinesin heavy chain member 2A |
C00004631
|
| 11127 | KIF3A, FLA10, KLP-20 | kinesin family member 3A |
C00004631
|
| 24137 | KIF4A, KIF4, KIF4G1 | kinesin family member 4A |
C00004631
|
| 22920 | KIFAP3, FLA3, KAP-1, KAP-3, KAP3, SMAP, Smg-GDS, dJ190I16.1 | kinesin-associated protein 3 |
C00004631
|
| 90990 | KIFC2 | kinesin family member C2 |
C00004631
|
| 89953 | KLC4, KNSL8, bA387M24.3 | kinesin light chain 4 |
C00004631
|
| 51274 | KLF3, BKLF | Kruppel-like factor 3 (basic) |
C00004631
|
| 9314 | KLF4, EZF, GKLF | Kruppel-like factor 4 (gut) |
C00004631
|
| 688 | KLF5, BTEB2, CKLF, IKLF | Kruppel-like factor 5 (intestinal) |
C00004631
|
| 1316 | KLF6, BCD1, CBA1, COPEB, CPBP, GBF, PAC1, ST12, ZF9 | Kruppel-like factor 6 |
C00004631
|
| 23588 | KLHDC2, HCLP-1, HCLP1, LCP | kelch domain containing 2 |
C00004631
|
| 57565 | KLHL14 | kelch-like family member 14 |
C00004631
|
| 11275 | KLHL2, ABP-KELCH, MAV, MAYVEN | kelch-like family member 2 |
C00004631
|
| 9903 | KLHL21 | kelch-like family member 21 |
C00004631
|
| 51088 | KLHL5 | kelch-like family member 5 |
C00004631
|
| 55958 | KLHL9 | kelch-like family member 9 |
C00004631
|
| 3817 | KLK2, KLK2A2, hGK-1, hK2 | kallikrein-related peptidase 2 (EC:3.4.21.35) |
C00004631
|
| 354 | KLK3, APS, KLK2A1, PSA, hK3 | kallikrein-related peptidase 3 (EC:3.4.21.77) |
C00004631
|
| 3827 | KNG1, BDK, BK, KNG | kininogen 1 |
C00004631
|
| 3839 | KPNA3, IPOA4, SRP1, SRP1gamma, SRP4, hSRP1 | karyopherin alpha 3 (importin alpha 4) |
C00004631
|
| 3840 | KPNA4, IPOA3, QIP1, SRP3 | karyopherin alpha 4 (importin alpha 3) |
C00004631
|
| 3845 | KRAS, C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RASK2 | Kirsten rat sarcoma viral oncogene homolog |
C00004631
|
| 51315 | KRCC1, CHBP2 | lysine-rich coiled-coil 1 |
C00004631
|
| 889 | KRIT1, CAM, CCM1 | KRIT1, ankyrin repeat containing |
C00004631
|
| 3848 | KRT1, CK1, EHK, EHK1, EPPK, K1, KRT1A, NEPPK | keratin 1 |
C00004631
|
| 3866 | KRT15, CK15, K15, K1CO | keratin 15 |
C00004631
|
| 3880 | KRT19, CK19, K19, K1CS | keratin 19 |
C00004631
|
| 54474 | KRT20, CD20, CK-20, CK20, K20, KRT21 | keratin 20 |
C00004631
|
| 25984 | KRT23, CK23, HAIK1, K23 | keratin 23 (histone deacetylase inducible) |
C00004631
|
| 112970 | KTI12, RP11-91A18.3, TOT4 | KTI12 homolog, chromatin associated (S. cerevisiae) |
C00004631
|
| 114294 | LACTB, G24, MRPL56 | lactamase, beta |
C00004631
|
| 51110 | LACTB2 | lactamase, beta 2 |
C00004631
|
| 3898 | LAD1, LadA | ladinin 1 |
C00004631
|
| 3904 | LAIR2, CD306 | leukocyte-associated immunoglobulin-like receptor 2 |
C00004631
|
| 51056 | LAP3, LAP, LAPEP, PEPS | leucine aminopeptidase 3 (EC:3.4.11.5 3.4.11.1) |
C00004631
|
| 7805 | LAPTM5, CLAST6 | lysosomal protein transmembrane 5 |
C00004631
|
| 55323 | LARP6, ACHN | La ribonucleoprotein domain family, member 6 |
C00004631
|
| 81606 | LBH | limb bud and heart development |
C00004631
|
| 3930 | LBR, DHCR14B, LMN2R, PHA, TDRD18 | lamin B receptor |
C00004631
|
| 151534 | LBX2-AS1 | LBX2 antisense RNA 1 |
C00004631
|
| 3931 | LCAT | lecithin-cholesterol acyltransferase (EC:2.3.1.43) |
C00004631
|
| 389812 | LCN15, PRO6093, UNQ2541 | lipocalin 15 |
C00004631
|
| 3938 | LCT, LAC, LPH, LPH1 | lactase (EC:3.2.1.108 3.2.1.62) |
C00004631
|
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00004631
|
| 116842 | LEAP2, LEAP-2 | liver expressed antimicrobial peptide 2 |
C00004631
|
| 114823 | LENG8, pp13842 | leukocyte receptor cluster (LRC) member 8 |
C00004631
|
| 137994 | LETM2 | leucine zipper-EF-hand containing transmembrane protein 2 |
C00004631
|
| 3956 | LGALS1, GAL1, GBP | lectin, galactoside-binding, soluble, 1 |
C00004631
|
| 3957 | LGALS2, HL14 | lectin, galactoside-binding, soluble, 2 |
C00004631
|
| 3958 | LGALS3, CBP35, GAL3, GALBP, GALIG, L31, LGALS2, MAC2 | lectin, galactoside-binding, soluble, 3 |
C00004631
|
| 3959 | LGALS3BP, 90K, BTBD17B, MAC-2-BP, TANGO10B | lectin, galactoside-binding, soluble, 3 binding protein |
C00004631
|
| 3964 | LGALS8, Gal-8, PCTA-1, PCTA1, Po66-CBP | lectin, galactoside-binding, soluble, 8 |
C00004631
|
| 55366 | LGR4, BNMD17, GPR48 | leucine-rich repeat containing G protein-coupled receptor 4 |
C00004631
|
| 8549 | LGR5, FEX, GPR49, GPR67, GRP49, HG38 | leucine-rich repeat containing G protein-coupled receptor 5 |
C00004631
|
| 51557 | LGSN, GLULD1, LGS | lengsin, lens protein with glutamine synthetase domain |
C00004631
|
| 3976 | LIF, CDF, DIA, HILDA, MLPLI | leukemia inhibitory factor |
C00004631
|
| 3977 | LIFR, CD118, LIF-R, SJS2, STWS, SWS | leukemia inhibitory factor receptor alpha |
C00004631
|
| 3982 | LIM2, CTRCT19, MP17, MP19 | lens intrinsic membrane protein 2, 19kDa |
C00004631
|
| 51474 | LIMA1, EPLIN, SREBP3 | LIM domain and actin binding 1 |
C00004631
|
| 389421 | LIN28B, CSDD2 | lin-28 homolog B (C. elegans) |
C00004631
|
| 140828 | LINC00261, C20orf56, HCCDR1, NCRNA00261, bA216C10.1, dJ788L20.2 | long intergenic non-protein coding RNA 261 |
C00004631
|
| 654434 | LINC00338, C17orf86, NCRNA00338 | long intergenic non-protein coding RNA 338 |
C00004631
|
| 147525 | LINC00526, C18orf18, HsT959 | long intergenic non-protein coding RNA 526 |
C00004631
|
| 3988 | LIPA, CESD, LAL | lipase A, lysosomal acid, cholesterol esterase (EC:3.1.1.13) |
C00004631
|
| 3990 | LIPC, HDLCQ12, HL, HTGL, LIPH | lipase, hepatic (EC:3.1.1.3) |
C00004631
|
| 9388 | LIPG, EDL, EL, PRO719 | lipase, endothelial (EC:3.1.1.3) |
C00004631
|
| 200879 | LIPH, AH, ARWH2, LAH2, LPDLR, PLA1B, mPA-PLA1 | lipase, member H (EC:2.7.11.30) |
C00004631
|
| 51601 | LIPT1 | lipoyltransferase 1 (EC:2.3.1.181) |
C00004631
|
| 3998 | LMAN1, ERGIC-53, ERGIC53, F5F8D, FMFD1, MCFD1, MR60, gp58 | lectin, mannose-binding, 1 |
C00004631
|
| 55788 | LMBRD1, C6orf209, LMBD1, MAHCF, NESI | LMBR1 domain containing 1 |
C00004631
|
| 29995 | LMCD1 | LIM and cysteine-rich domains 1 |
C00004631
|
| 4000 | LMNA, CDCD1, CDDC, CMD1A, CMT2B1, EMD2, FPL, FPLD, FPLD2, HGPS, IDC, LDP1, LFP, LGMD1B, LMN1, LMNC, LMNL1, PRO1 | lamin A/C |
C00004631
|
| 4001 | LMNB1, ADLD, LMN, LMN2, LMNB | lamin B1 |
C00004631
|
| 4008 | LMO7, FBX20, FBXO20, LOMP | LIM domain 7 |
C00004631
|
| 348801 | LNP1 | leukemia NUP98 fusion partner 1 |
C00004631
|
| 4012 | LNPEP, CAP, IRAP, P-LAP, PLAP | leucyl/cystinyl aminopeptidase (EC:3.4.11.3) |
C00004631
|
| 222484 | LNX2, PDZRN1 | ligand of numb-protein X 2 |
C00004631
|
| 91694 | LONRF1, RNF191 | LON peptidase N-terminal domain and ring finger 1 |
C00004631
|
| 4015 | LOX | lysyl oxidase (EC:1.4.3.13) |
C00004631
|
| 9926 | LPGAT1, FAM34A, FAM34A1, NET8 | lysophosphatidylglycerol acyltransferase 1 |
C00004631
|
| 23266 | LPHN2, CIRL2, CL2, LEC1, LPHH1 | latrophilin 2 |
C00004631
|
| 54886 | LPPR1, PRG-3 | lipid phosphate phosphatase-related protein type 1 |
C00004631
|
| 9404 | LPXN, LDPL | leupaxin |
C00004631
|
| 23143 | LRCH1, CHDC1, NP81 | leucine-rich repeats and calponin homology (CH) domain containing 1 |
C00004631
|
| 57913 |
C00004631
|
||
| 116844 | LRG1, HMFT1766, LRG | leucine-rich alpha-2-glycoprotein 1 |
C00004631
|
| 26018 | LRIG1, LIG-1, LIG1 | leucine-rich repeats and immunoglobulin-like domains 1 |
C00004631
|
| 121227 | LRIG3, LIG3 | leucine-rich repeats and immunoglobulin-like domains 3 |
C00004631
|
| 26020 | LRP10, LRP9, MST087 | low density lipoprotein receptor-related protein 10 |
C00004631
|
| 91355 | LRP5L | low density lipoprotein receptor-related protein 5-like |
C00004631
|
| 10234 | LRRC17, P37NB | leucine rich repeat containing 17 |
C00004631
|
| 374819 | LRRC37A3, LRRC37A | leucine rich repeat containing 37, member A3 |
C00004631
|
| 55144 | LRRC8D, LRRC5 | leucine rich repeat containing 8 family, member D |
C00004631
|
| 127255 | LRRIQ3, LRRC44 | leucine-rich repeats and IQ motif containing 3 |
C00004631
|
| 25804 | LSM4, GRP, YER112W | LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00004631
|
| 286006 | LSMEM1, C7orf53 | leucine-rich single-pass membrane protein 1 |
C00004631
|
| 51599 | LSR, ILDR3, LISCH7 | lipolysis stimulated lipoprotein receptor |
C00004631
|
| 4047 | LSS, OSC | lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (EC:5.4.99.7) |
C00004631
|
| 1241 | LTB4R, BLT1, BLTR, CMKRL1, GPR16, LTB4R1, LTBR1, P2RY7, P2Y7 | leukotriene B4 receptor |
C00004631
|
| 23643 | LY96, ESOP-1, MD-2, MD2, ly-96 | lymphocyte antigen 96 |
C00004631
|
| 4067 | LYN, JTK8, p53Lyn, p56Lyn | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (EC:2.7.10.2) |
C00004631
|
| 127018 | LYPLAL1 | lysophospholipase-like 1 (EC:3.1.2.-) |
C00004631
|
| 256586 | LYSMD2 | LysM, putative peptidoglycan-binding, domain containing 2 |
C00004631
|
| 4069 | LYZ, LZM | lysozyme (EC:3.2.1.17) |
C00004631
|
| 8216 | LZTR1, BTBD29, LZTR-1 | leucine-zipper-like transcription regulator 1 |
C00004631
|
| 28992 | MACROD1, LRP16 | MACRO domain containing 1 |
C00004631
|
| 8379 | MAD1L1, MAD1, PIG9, TP53I9, TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) |
C00004631
|
| 4085 | MAD2L1, HSMAD2, MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) |
C00004631
|
| 23764 | MAFF, U-MAF, hMafF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
C00004631
|
| 4097 | MAFG, hMAF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
C00004631
|
| 9500 | MAGED1, DLXIN-1, NRAGE | melanoma antigen family D, 1 |
C00004631
|
| 9863 | MAGI2, ACVRIP1, AIP-1, AIP1, ARIP1, MAGI-2, SSCAM | membrane associated guanylate kinase, WW and PDZ domain containing 2 |
C00004631
|
| 260425 | MAGI3, MAGI-3, RP4-730K3.1, dJ730K3.2 | membrane associated guanylate kinase, WW and PDZ domain containing 3 |
C00004631
|
| 114569 | MAL2 | mal, T-cell differentiation protein 2 (gene/pseudogene) |
C00004631
|
| 378938 | MALAT1, HCN, LINC00047, MALAT-1, NCRNA00047, NEAT2, PRO2853 | metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
C00004631
|
| 115416 | MALSU1, C7orf30, mtRsfA | mitochondrial assembly of ribosomal large subunit 1 |
C00004631
|
| 10892 | MALT1, IMD12, MLT, MLT1 | mucosa associated lymphoid tissue lymphoma translocation gene 1 (EC:3.4.22.-) |
C00004631
|
| 4121 | MAN1A1, HUMM3, HUMM9, MAN9 | mannosidase, alpha, class 1A, member 1 (EC:3.2.1.113) |
C00004631
|
| 10905 | MAN1A2, MAN1B | mannosidase, alpha, class 1A, member 2 (EC:3.2.1.113) |
C00004631
|
| 4123 | MAN2C1, MAN6A8, MANA, MANA1 | mannosidase, alpha, class 2C, member 1 (EC:3.2.1.24) |
C00004631
|
| 79694 | MANEA, ENDO, hEndo | mannosidase, endo-alpha (EC:3.2.1.130) |
C00004631
|
| 54682 | MANSC1, 9130403P13Rik, LOH12CR3 | MANSC domain containing 1 (EC:3.2.1.14) |
C00004631
|
| 4128 | MAOA, MAO-A | monoamine oxidase A (EC:1.4.3.4) |
C00004631
|
| 4129 | MAOB | monoamine oxidase B (EC:1.4.3.4) |
C00004631
|
| 4131 | MAP1B, FUTSCH, MAP5 | microtubule-associated protein 1B |
C00004631
|
| 81631 | MAP1LC3B, ATG8F, LC3B, MAP1A/1BLC3, MAP1LC3B-a | microtubule-associated protein 1 light chain 3 beta |
C00004631
|
| 5606 | MAP2K3, MAPKK3, MEK3, MKK3, PRKMK3, SAPKK-2, SAPKK2 | mitogen-activated protein kinase kinase 3 (EC:2.7.12.2) |
C00004631
|
| 6416 | MAP2K4, JNKK, JNKK1, MAPKK4, MEK4, MKK4, PRKMK4, SAPKK-1, SAPKK1, SEK1, SERK1, SKK1 | mitogen-activated protein kinase kinase 4 (EC:2.7.12.2) |
C00004631
|
| 5608 | MAP2K6, MAPKK6, MEK6, MKK6, PRKMK6, SAPKK-3, SAPKK3 | mitogen-activated protein kinase kinase 6 (EC:2.7.12.2) |
C00004631
|
| 7786 | MAP3K12, DLK, MEKK12, MUK, ZPK, ZPKP1 | mitogen-activated protein kinase kinase kinase 12 (EC:2.7.11.25) |
C00004631
|
| 9175 | MAP3K13, LZK, MEKK13, MLK | mitogen-activated protein kinase kinase kinase 13 (EC:2.7.11.25) |
C00004631
|
| 9020 | MAP3K14, FTDCR1B, HS, HSNIK, NIK | mitogen-activated protein kinase kinase kinase 14 (EC:2.7.11.25) |
C00004631
|
| 4216 | MAP3K4, MAPKKK4, MEKK4, MTK1, PRO0412 | mitogen-activated protein kinase kinase kinase 4 (EC:2.7.11.25) |
C00004631
|
| 790559 |
C00004631
|
||
| 1326 | MAP3K8, COT, EST, ESTF, MEKK8, TPL2, Tpl-2, c-COT | mitogen-activated protein kinase kinase kinase 8 (EC:2.7.11.25) |
C00004631
|
| 4293 | MAP3K9, MEKK9, MLK1, PRKE1 | mitogen-activated protein kinase kinase kinase 9 (EC:2.7.11.25) |
C00004631
|
| 11184 | MAP4K1, HPK1 | mitogen-activated protein kinase kinase kinase kinase 1 (EC:2.7.11.1) |
C00004631
|
| 9448 | MAP4K4, FLH21957, HGK, MEKKK4, NIK | mitogen-activated protein kinase kinase kinase kinase 4 (EC:2.7.11.1) |
C00004631
|
| 55700 | MAP7D1, PARCC1, RPRC1 | MAP7 domain containing 1 |
C00004631
|
| 5603 | MAPK13, MAPK_13, MAPK-13, PRKM13, SAPK4, p38delta | mitogen-activated protein kinase 13 (EC:2.7.11.24) |
C00004631
|
| 1432 | MAPK14, CSBP, CSBP1, CSBP2, CSPB1, EXIP, Mxi2, PRKM14, PRKM15, RK, SAPK2A, p38, p38ALPHA | mitogen-activated protein kinase 14 (EC:2.7.11.24) |
C00004631
|
| 10257 | ABCC4, EST170205, MOAT-B, MOATB, MRP4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
C00004631
|
| 1244 | ABCC2, ABC30, CMOAT, DJS, MRP2, cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
C00004631
|
| 64757 | MARC1, MOSC1 | mitochondrial amidoxime reducing component 1 |
C00004631
|
| 115123 | MARCH3, MARCH-III, RNF173 | membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase (EC:6.3.2.19) |
C00004631
|
| 92935 | MARS2, MetRS, mtMetRS | methionyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.10) |
C00004631
|
| 84930 | MASTL, GREATWALL, GW, GWL, MAST-L, RP11-85G18.2, THC2, hGWL | microtubule associated serine/threonine kinase-like (EC:2.7.11.1) |
C00004631
|
| 4143 | MAT1A, MAT, MATA1, SAMS, SAMS1 | methionine adenosyltransferase I, alpha (EC:2.5.1.6) |
C00004631
|
| 8930 | MBD4, MED1 | methyl-CpG binding domain protein 4 |
C00004631
|
| 114785 | MBD6 | methyl-CpG binding domain protein 6 |
C00004631
|
| 51562 | MBIP | MAP3K12 binding inhibitory protein 1 |
C00004631
|
| 153364 | MBLAC2 | metallo-beta-lactamase domain containing 2 |
C00004631
|
| 10150 | MBNL2, MBLL, MBLL39, PRO2032 | muscleblind-like splicing regulator 2 |
C00004631
|
| 55796 | MBNL3, CHCR, MBLX, MBLX39, MBXL | muscleblind-like splicing regulator 3 |
C00004631
|
| 4170 | MCL1, BCL2L3, EAT, MCL1-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT | myeloid cell leukemia sequence 1 (BCL2-related) |
C00004631
|
| 55388 | MCM10, CNA43, DNA43 | minichromosome maintenance complex component 10 |
C00004631
|
| 4173 | MCM4, CDC21, CDC54, NKCD, NKGCD, P1-CDC21, hCdc21 | minichromosome maintenance complex component 4 (EC:3.6.4.12) |
C00004631
|
| 57192 | MCOLN1, MG-2, ML4, MLIV, MST080, TRP-ML1, TRPM-L1, TRPML1 | mucolipin 1 |
C00004631
|
| 4192 | MDK, ARAP, MK, NEGF2 | midkine (neurite growth-promoting factor 2) |
C00004631
|
| 56890 | MDM1 | Mdm1 nuclear protein homolog (mouse) |
C00004631
|
| 4193 | MDM2, ACTFS, HDMX, hdm2 | MDM2 oncogene, E3 ubiquitin protein ligase |
C00004631
|
| 4200 | ME2, ODS1 | malic enzyme 2, NAD(+)-dependent, mitochondrial (EC:1.1.1.38) |
C00004631
|
| 84246 | MED10, NUT2, TRG20 | mediator complex subunit 10 |
C00004631
|
| 9412 | MED21, SRB7, SURB7, hSrb7 | mediator complex subunit 21 |
C00004631
|
| 9441 | MED26, CRSP7, CRSP70 | mediator complex subunit 26 |
C00004631
|
| 90390 | MED30, MED30S, THRAP6, TRAP25 | mediator complex subunit 30 |
C00004631
|
| 9443 | MED7, ARC34, CRSP33, CRSP9 | mediator complex subunit 7 |
C00004631
|
| 1953 | MEGF6, EGFL3 | multiple EGF-like-domains 6 |
C00004631
|
| 1954 | MEGF8, C19orf49, CRPT2, EGFL4, SBP1 | multiple EGF-like-domains 8 |
C00004631
|
| 4211 | MEIS1 | Meis homeobox 1 |
C00004631
|
| 4212 | MEIS2, HsT18361, MRG1 | Meis homeobox 2 |
C00004631
|
| 4221 | MEN1, MEAI, SCG2 | multiple endocrine neoplasia I |
C00004631
|
| 4224 | MEP1A, PPHA | meprin A, alpha (PABA peptide hydrolase) (EC:3.4.24.18) |
C00004631
|
| 284207 | METRNL | meteorin, glial cell differentiation regulator-like |
C00004631
|
| 399818 | METTL10, C10orf138, Em:AC068896.3 | methyltransferase like 10 |
C00004631
|
| 751071 | METTL12, U99HG | methyltransferase like 12 (EC:2.1.1.-) |
C00004631
|
| 92342 | METTL18, AsTP2, C1orf156, HPM1 | methyltransferase like 18 |
C00004631
|
| 151194 | METTL21A, FAM119A, HCA557b, HSPA-KMT | methyltransferase like 21A |
C00004631
|
| 29081 | METTL5, HSPC133 | methyltransferase like 5 |
C00004631
|
| 25840 | METTL7A, AAM-B | methyltransferase like 7A |
C00004631
|
| 196410 | METTL7B, ALDI | methyltransferase like 7B |
C00004631
|
| 9258 | MFHAS1, LRRC65, MASL1 | malignant fibrous histiocytoma amplified sequence 1 |
C00004631
|
| 55669 | MFN1, hfzo1, hfzo2 | mitofusin 1 |
C00004631
|
| 10227 | MFSD10, TETRAN, TETTRAN | major facilitator superfamily domain containing 10 |
C00004631
|
| 79157 | MFSD11, ET | major facilitator superfamily domain containing 11 |
C00004631
|
| 256471 | MFSD8, CLN7 | major facilitator superfamily domain containing 8 |
C00004631
|
| 23269 | MGA, MAD5, MXD5 | MGA, MAX dimerization protein |
C00004631
|
| 4258 | MGST2, GST2, MGST-II | microsomal glutathione S-transferase 2 (EC:2.5.1.18) |
C00004631
|
| 117153 | MIA2 | melanoma inhibitory activity 2 |
C00004631
|
| 142678 | MIB2, ZZANK1, ZZZ5 | mindbomb E3 ubiquitin protein ligase 2 |
C00004631
|
| 64780 | MICAL1, MICAL, MICAL-1, NICAL | microtubule associated monooxygenase, calponin and LIM domain containing 1 |
C00004631
|
| 57553 | MICAL3, MICAL-3 | microtubule associated monooxygenase, calponin and LIM domain containing 3 |
C00004631
|
| 85377 | MICALL1, MICAL-L1, MIRAB13 | MICAL-like 1 |
C00004631
|
| 79778 | MICALL2, JRAB, MICAL-L2 | MICAL-like 2 |
C00004631
|
| 4277 | MICB, PERB11.2 | MHC class I polypeptide-related sequence B |
C00004631
|
| 90007 | MIDN | midnolin |
C00004631
|
| 4282 | MIF, GIF, GLIF, MMIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) (EC:5.3.3.12 5.3.2.1) |
C00004631
|
| 54468 | MIOS | missing oocyte, meiosis regulator, homolog (Drosophila) |
C00004631
|
| 145282 | MIPOL1 | mirror-image polydactyly 1 |
C00004631
|
| 84981 | MIR22HG, C17orf91 | MIR22 host gene (non-protein coding) |
C00004631
|
| 100422952 | MIR4271 | microRNA 4271 |
C00004631
|
| 55320 | MIS18BP1, C14orf106, HSA242977, KNL2, M18BP1 | MIS18 binding protein 1 |
C00004631
|
| 129531 | MITD1 | MIT, microtubule interacting and transport, domain containing 1 |
C00004631
|
| 4286 | MITF, CMM8, MI, WS2, WS2A, bHLHe32 | microphthalmia-associated transcription factor |
C00004631
|
| 83881 | MIXL1, MILD1, MIX, MIXL | Mix paired-like homeobox |
C00004631
|
| 4288 | MKI67, KIA, MIB-1 | marker of proliferation Ki-67 |
C00004631
|
| 2872 | MKNK2, GPRK7, MNK2 | MAP kinase interacting serine/threonine kinase 2 (EC:2.7.11.1) |
C00004631
|
| 79682 | CENPU, CENP50, CENPU50, KLIP1, MLF1IP, PBIP1 | centromere protein U |
C00004631
|
| 4300 | MLLT3, AF9, YEATS3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
C00004631
|
| 653483 | MLLT4-AS1, C6orf124, HGC6.4, dJ431P23.3 | MLLT4 antisense RNA 1 (head to head) |
C00004631
|
| 79083 | MLPH, SLAC2-A | melanophilin |
C00004631
|
| 326625 | MMAB, ATR, cblB, cob | methylmalonic aciduria (cobalamin deficiency) cblB type (EC:2.5.1.17) |
C00004631
|
| 5244 | ABCB4, ABC21, GBD1, ICP3, MDR2, MDR2/3, MDR3, PFIC-3, PGY3 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 (EC:3.6.3.44) |
C00004631
|
| 4319 | MMP10, SL-2, STMY2 | matrix metallopeptidase 10 (stromelysin 2) (EC:3.4.24.22) |
C00004631
|
| 4320 | MMP11, SL-3, ST3, STMY3 | matrix metallopeptidase 11 (stromelysin 3) (EC:3.4.24.-) |
C00004631
|
| 4321 | MMP12, HME, ME, MME, MMP-12 | matrix metallopeptidase 12 (macrophage elastase) (EC:3.4.24.65) |
C00004631
|
| 4322 | MMP13, CLG3, MANDP1 | matrix metallopeptidase 13 (collagenase 3) (EC:3.4.24.-) |
C00004631
|
| 4314 | MMP3, CHDS6, MMP-3, SL-1, STMY, STMY1, STR1 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) (EC:3.4.24.17) |
C00004631
|
| 4316 | MMP7, MMP-7, MPSL1, PUMP-1 | matrix metallopeptidase 7 (matrilysin, uterine) (EC:3.4.24.23) |
C00004631
|
| 10347 | ABCA7, ABCA-SSN, ABCX | ATP-binding cassette, sub-family A (ABC1), member 7 |
C00004631
|
| 4330 | MN1, MGCR, MGCR1, MGCR1-PEN, dJ353E16.2 | meningioma (disrupted in balanced translocation) 1 |
C00004631
|
| 55329 | MNS1, SPATA40 | meiosis-specific nuclear structural 1 |
C00004631
|
| 422647 |
C00004631
|
||
| 55034 | MOCOS, HMCS, MCS, MOS | molybdenum cofactor sulfurase (EC:2.8.1.9) |
C00004631
|
| 4338 | MOCS2, MCBPE, MOCO1, MOCODB, MPTS | molybdenum cofactor synthesis 2 (EC:2.8.1.12) |
C00004631
|
| 116255 | MOGAT1, DGAT2L, DGAT2L1, MGAT1 | monoacylglycerol O-acyltransferase 1 (EC:2.3.1.22) |
C00004631
|
| 80168 | MOGAT2, DGAT2L5, MGAT2 | monoacylglycerol O-acyltransferase 2 (EC:2.3.1.22) |
C00004631
|
| 79710 | MORC4, ZCW4, ZCWCC2, dJ75H8.2 | MORC family CW-type zinc finger 4 |
C00004631
|
| 729967 | MORN2, BLOCK27, MOPT | MORN repeat containing 2 |
C00004631
|
| 51660 | MPC1, BRP44L, MPYCD, dJ68L15.3 | mitochondrial pyruvate carrier 1 |
C00004631
|
| 25874 | MPC2, BRP44 | mitochondrial pyruvate carrier 2 |
C00004631
|
| 8777 | MPDZ, HYC2, MUPP1 | multiple PDZ domain protein |
C00004631
|
| 10198 | MPHOSPH9, MPP-9, MPP9 | M-phase phosphoprotein 9 |
C00004631
|
| 64398 | MPP5, PALS1 | membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
C00004631
|
| 51678 | MPP6, PALS2, VAM-1, VAM1, p55T | membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
C00004631
|
| 143098 | MPP7 | membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
C00004631
|
| 4359 | MPZ, CHM, CMT1, CMT1B, CMT2I, CMT2J, CMT4E, CMTDI3, CMTDID, DSS, HMSNIB, MPP, P0 | myelin protein zero |
C00004631
|
| 3140 | MR1, HLALS | major histocompatibility complex, class I-related |
C00004631
|
| 4361 | MRE11A, ATLD, HNGS1, MRE11, MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
C00004631
|
| 117195 | MRGPRX3, GPCR, MRGX3, SNSR1, SNSR2 | MAS-related GPR, member X3 |
C00004631
|
| 84245 | MRI1, MRDI, MTNA, Ypr118w | methylthioribose-1-phosphate isomerase 1 (EC:5.3.1.23) |
C00004631
|
| 727957 | MROH1, HEATR7A | maestro heat-like repeat family member 1 |
C00004631
|
| 140699 | MROH8, C20orf131, C20orf132, dJ621N11.3, dJ621N11.4 | maestro heat-like repeat family member 8 |
C00004631
|
| 28998 | MRPL13, L13, L13A, L13mt, RPL13, RPML13 | mitochondrial ribosomal protein L13 |
C00004631
|
| 55052 | MRPL20, L20mt, MRP-L20 | mitochondrial ribosomal protein L20 |
C00004631
|
| 6150 | MRPL23, L23MRP, RPL23, RPL23L | mitochondrial ribosomal protein L23 |
C00004631
|
| 54148 | MRPL39, C21orf92, L39mt, MRP-L5, MRPL5, PRED22, PRED66, RPML5 | mitochondrial ribosomal protein L39 |
C00004631
|
| 57129 | MRPL47, L47mt, MRP-L47, NCM1 | mitochondrial ribosomal protein L47 |
C00004631
|
| 740 | MRPL49, C11orf4, L49mt, MRP-L49, NOF, NOF1 | mitochondrial ribosomal protein L49 |
C00004631
|
| 54534 | MRPL50, MRP-L50 | mitochondrial ribosomal protein L50 |
C00004631
|
| 56945 | MRPS22, C3orf5, COXPD5, GIBT, MRP-S22, RPMS22 | mitochondrial ribosomal protein S22 |
C00004631
|
| 10240 | MRPS31, IMOGN38, MRP-S31, S31mt | mitochondrial ribosomal protein S31 |
C00004631
|
| 64968 | MRPS6, C21orf101, MRP-S6, RPMS6, S6mt | mitochondrial ribosomal protein S6 |
C00004631
|
| 345222 | MSANTD1, C4orf44 | Myb/SANT-like DNA-binding domain containing 1 |
C00004631
|
| 91283 | MSANTD3, C9orf30 | Myb/SANT-like DNA-binding domain containing 3 |
C00004631
|
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00004631
|
| 4439 | MSH5, G7, MUTSH5, NG23 | mutS homolog 5 |
C00004631
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00004631
|
| 4485 | MST1, D3F15S2, DNF15S2, HGFL, MSP, NF15S2 | macrophage stimulating 1 (hepatocyte growth factor-like) |
C00004631
|
| 11223 | MST1L, BRF-1, D1F15S1A, MSPL-7, MSPL7, MST1P9, MSTP7, MSTP9 | macrophage stimulating 1-like |
C00004631
|
| 51765 | MST4, MASK | serine/threonine protein kinase MST4 (EC:2.7.11.1) |
C00004631
|
| 4487 | MSX1, ECTD3, HOX7, HYD1, STHAG1 | msh homeobox 1 |
C00004631
|
| 4495 | MT1G, MT1, MT1K | metallothionein 1G |
C00004631
|
| 4499 | MT1M, MT-1M, MT-IM, MT1, MT1K | metallothionein 1M |
C00004631
|
| 4501 | MT1X, MT-1l, MT1 | metallothionein 1X |
C00004631
|
| 9112 | MTA1 | metastasis associated 1 |
C00004631
|
| 51001 | MTERFD1, mTERF3 | MTERF domain containing 1 |
C00004631
|
| 80298 | MTERFD3, mTERF2, mTERFL | MTERF domain containing 3 |
C00004631
|
| 9650 | MTFR1, CHPPR, FAM54A2 | mitochondrial fission regulator 1 |
C00004631
|
| 113115 | MTFR2, DUFD1, FAM54A | mitochondrial fission regulator 2 |
C00004631
|
| 441024 | MTHFD2L | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (EC:3.5.4.9 1.5.1.15) |
C00004631
|
| 4528 | MTIF2 | mitochondrial translational initiation factor 2 |
C00004631
|
| 219402 | MTIF3, IF3mt | mitochondrial translational initiation factor 3 |
C00004631
|
| 9110 | MTMR4, FYVE-DSP2, ZFYVE11 | myotubularin related protein 4 (EC:3.1.3.48) |
C00004631
|
| 4548 | MTR, HMAG, MS, cblG | 5-methyltetrahydrofolate-homocysteine methyltransferase (EC:2.1.1.13) |
C00004631
|
| 9788 | MTSS1, MIM, MIMA, MIMB | metastasis suppressor 1 |
C00004631
|
| 4547 | MTTP, ABL, MTP | microsomal triglyceride transfer protein |
C00004631
|
| 57509 | MTUS1, ATBP, ATIP, ICIS, MP44, MTSG1 | microtubule associated tumor suppressor 1 |
C00004631
|
| 56667 | MUC13, DRCC1, MUC-13 | mucin 13, cell surface associated |
C00004631
|
| 143662 | MUC15, MUC-15, PAS3, PASIII | mucin 15, cell surface associated |
C00004631
|
| 4583 | MUC2, MLP, MUC-2, SMUC | mucin 2, oligomeric mucus/gel-forming |
C00004631
|
| 4597 | MVD, FP17780, MPD | mevalonate (diphospho) decarboxylase (EC:4.1.1.33) |
C00004631
|
| 9961 | MVP, LRP, VAULT1 | major vault protein |
C00004631
|
| 4084 | MXD1, BHLHC58, MAD, MAD1 | MAX dimerization protein 1 |
C00004631
|
| 4601 | MXI1, MAD2, MXD2, MXI, bHLHc11 | MAX interactor 1, dimerization protein |
C00004631
|
| 4602 | MYB, Cmyb, c-myb, c-myb_CDS, efg | v-myb avian myeloblastosis viral oncogene homolog |
C00004631
|
| 4603 | MYBL1, A-MYB, AMYB | v-myb avian myeloblastosis viral oncogene homolog-like 1 |
C00004631
|
| 4605 | MYBL2, B-MYB, BMYB | v-myb avian myeloblastosis viral oncogene homolog-like 2 |
C00004631
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00004631
|
| 4615 | MYD88, MYD88D | myeloid differentiation primary response 88 |
C00004631
|
| 4621 | MYH3, HEMHC, MYHC-EMB, MYHSE1, SMHCE | myosin, heavy chain 3, skeletal muscle, embryonic |
C00004631
|
| 10627 | MYL12A, MLCB, MRCL3, MRLC3, MYL2B | myosin, light chain 12A, regulatory, non-sarcomeric |
C00004631
|
| 4636 | MYL5 | myosin, light chain 5, regulatory |
C00004631
|
| 10398 | MYL9, LC20, MLC-2C, MLC2, MRLC1, MYRL2 | myosin, light chain 9, regulatory |
C00004631
|
| 29116 | MYLIP, IDOL, MIR | myosin regulatory light chain interacting protein (EC:6.3.2.-) |
C00004631
|
| 4638 | MYLK, AAT7, KRP, MLCK, MLCK1, MLCK108, MLCK210, MSTP083, MYLK1, smMLCK | myosin light chain kinase (EC:2.7.11.18) |
C00004631
|
| 4640 | MYO1A, BBMI, DFNA48, MIHC, MYHL | myosin IA |
C00004631
|
| 4430 | MYO1B, myr1 | myosin IB |
C00004631
|
| 4644 | MYO5A, GS1, MYH12, MYO5, MYR12 | myosin VA (heavy chain 12, myoxin) |
C00004631
|
| 4646 | MYO6, DFNA22, DFNB37 | myosin VI |
C00004631
|
| 4647 | MYO7A, DFNA11, DFNB2, MYOVIIA, MYU7A, NSRD2, USH1B | myosin VIIA |
C00004631
|
| 26509 | MYOF, FER1L3 | myoferlin |
C00004631
|
| 90634 | N4BP2L1, CG018 | NEDD4 binding protein 2-like 1 |
C00004631
|
| 10443 | N4BP2L2, 92M18.3, CG005, PFAAP5 | NEDD4 binding protein 2-like 2 |
C00004631
|
| 29104 | N6AMT1, C21orf127, HEMK2, MTQ2, N6AMT | N-6 adenine-specific DNA methyltransferase 1 (putative) (EC:2.1.1.72) |
C00004631
|
| 221143 | N6AMT2, ESP13 | N-6 adenine-specific DNA methyltransferase 2 (putative) |
C00004631
|
| 80155 | NAA15, Ga19, NARG1, NAT1P, NATH, TBDN, TBDN100 | N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
C00004631
|
| 10003 | NAALAD2, GCPIII, GPCIII, NAADALASE2, NAALADASE2 | N-acetylated alpha-linked acidic dipeptidase 2 (EC:3.4.17.21) |
C00004631
|
| 64859 | NABP1, OBFC2A, SOSS-B2, SSB2 | nucleic acid binding protein 1 |
C00004631
|
| 138151 | NACC2, BEND9, BTBD14, BTBD14A, BTBD31, RBB | NACC family member 2, BEN and BTB (POZ) domain containing |
C00004631
|
| 133686 | NADK2, C5orf33, MNADK, NADKD1 | NAD kinase 2, mitochondrial (EC:2.7.1.23) |
C00004631
|
| 4669 | NAGLU, MPS-IIIB, MPS3B, NAG, UFHSD | N-acetylglucosaminidase, alpha (EC:3.2.1.50) |
C00004631
|
| 162417 | NAGS, AGAS, ARGA | N-acetylglutamate synthase (EC:2.3.1.1) |
C00004631
|
| 266812 | NAP1L5, DRLM | nucleosome assembly protein 1-like 5 |
C00004631
|
| 63908 | NAPB, SNAP-BETA, SNAPB | N-ethylmaleimide-sensitive factor attachment protein, beta |
C00004631
|
| 222236 | NAPEPLD, FMP30, NAPE-PLD | N-acyl phosphatidylethanolamine phospholipase D (EC:3.1.4.54) |
C00004631
|
| 79664 | NARG2, BRCC1 | NMDA receptor regulated 2 |
C00004631
|
| 57106 | NAT14, KLP1 | N-acetyltransferase 14 (GCN5-related, putative) |
C00004631
|
| 24142 | NAT6, FUS-2, FUS2 | N-acetyltransferase 6 (GCN5-related) (EC:2.3.1.-) |
C00004631
|
| 9027 | NAT8, ATase2, CML1, GLA, Hcml1, TSC501, TSC510 | N-acetyltransferase 8 (GCN5-related, putative) |
C00004631
|
| 26151 | NAT9, EBSP, hNATL | N-acetyltransferase 9 (GCN5-related, putative) |
C00004631
|
| 89797 | NAV2, HELAD1, POMFIL2, RAINB1, STEERIN2, UNC53H2 | neuron navigator 2 (EC:3.6.4.12) |
C00004631
|
| 26960 | NBEA, BCL8B, LYST2 | neurobeachin |
C00004631
|
| 4683 | NBN, AT-V1, AT-V2, ATV, NBS, NBS1, P95 | nibrin |
C00004631
|
| 64151 | NCAPG, CAPG, CHCG, NY-MEL-3 | non-SMC condensin I complex, subunit G |
C00004631
|
| 54892 | NCAPG2, CAP-G2, CAPG2, LUZP5, MTB, hCAP-G2 | non-SMC condensin II complex, subunit G2 |
C00004631
|
| 57552 | NCEH1, AADACL1, NCEH | neutral cholesterol ester hydrolase 1 (EC:3.1.1.-) |
C00004631
|
| 653361 | NCF1, NCF1A, NOXO2, SH3PXD1A, p47phox | neutrophil cytosolic factor 1 |
C00004631
|
| 4688 | NCF2, NCF-2, NOXA2, P67-PHOX, P67PHOX | neutrophil cytosolic factor 2 |
C00004631
|
| 10787 | NCKAP1, HEM2, NAP1, NAP125, p125Nap1 | NCK-associated protein 1 |
C00004631
|
| 51517 | NCKIPSD, AF3P21, DIP, DIP1, ORF1, SPIN90, VIP54, WASLBP, WISH | NCK interacting protein with SH3 domain |
C00004631
|
| 8648 | NCOA1, F-SRC-1, KAT13A, RIP160, SRC1, bHLHe42, bHLHe74 | nuclear receptor coactivator 1 (EC:2.3.1.48) |
C00004631
|
| 10499 | NCOA2, GRIP1, KAT13C, NCoA-2, SRC2, TIF2, bHLHe75 | nuclear receptor coactivator 2 |
C00004631
|
| 8031 | NCOA4, ARA70, ELE1, PTC3, RFG | nuclear receptor coactivator 4 |
C00004631
|
| 10403 | NDC80, HEC, HEC1, HsHec1, KNTC2, TID3, hsNDC80 | NDC80 kinetochore complex component |
C00004631
|
| 54602 | NDFIP2, N4WBP5A | Nedd4 family interacting protein 2 |
C00004631
|
| 10397 | NDRG1, CAP43, CMT4D, DRG-1, DRG1, GC4, HMSNL, NDR1, NMSL, PROXY1, RIT42, RTP, TARG1, TDD5 | N-myc downstream regulated 1 |
C00004631
|
| 57447 | NDRG2, SYLD | NDRG family member 2 |
C00004631
|
| 4696 | NDUFA3, B9, CI-B9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
C00004631
|
| 4697 | NDUFA4, CI-9k, CI-MLRQ, MLRQ | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa (EC:1.6.5.3 1.6.99.3) |
C00004631
|
| 4719 | NDUFS1, CI-75Kd, CI-75k, PRO1304 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) (EC:1.6.5.3 1.6.99.3) |
C00004631
|
| 4703 | NEB, NEB177D, NEM2 | nebulin |
C00004631
|
| 23327 | NEDD4L, NEDD4-2, NEDD4.2, RSP5, hNEDD4-2 | neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00004631
|
| 4738 | NEDD8, NEDD-8 | neural precursor cell expressed, developmentally down-regulated 8 |
C00004631
|
| 4739 | NEDD9, CAS-L, CAS2, CASL, CASS2, HEF1 | neural precursor cell expressed, developmentally down-regulated 9 |
C00004631
|
| 4741 | NEFM, NEF3, NF-M, NFM | neurofilament, medium polypeptide |
C00004631
|
| 4750 | NEK1, NY-REN-55, SRPS2, SRPS2A | NIMA-related kinase 1 (EC:2.7.11.1) |
C00004631
|
| 4751 | NEK2, HsPK21, NEK2A, NLK1 | NIMA-related kinase 2 (EC:2.7.11.1) |
C00004631
|
| 6787 | NEK4, NRK2, STK2, pp12301 | NIMA-related kinase 4 (EC:2.7.11.1) |
C00004631
|
| 7936 | NELFE, D6S45, NELF-E, RD, RDBP, RDP | negative elongation factor complex member E |
C00004631
|
| 10276 | NET1, ARHGEF8, NET1A | neuroepithelial cell transforming 1 |
C00004631
|
| 81831 | NETO2, BTCL2, NEOT2 | neuropilin (NRP) and tolloid (TLL)-like 2 |
C00004631
|
| 91624 | NEXN, CMH20, NELIN | nexilin (F actin binding protein) |
C00004631
|
| 374987 | NEXN-AS1, C1orf118 | NEXN antisense RNA 1 |
C00004631
|
| 10725 | NFAT5, NF-AT5, NFATL1, NFATZ, OREBP, TONEBP | nuclear factor of activated T-cells 5, tonicity-responsive |
C00004631
|
| 4778 | NFE2, NF-E2, p45 | nuclear factor, erythroid 2 |
C00004631
|
| 9603 | NFE2L3, NRF3 | nuclear factor, erythroid 2-like 3 |
C00004631
|
| 4783 | NFIL3, E4BP4, IL3BP1, NF-IL3A, NFIL3A | nuclear factor, interleukin 3 regulated |
C00004631
|
| 23461 | ABCA5, ABC13, EST90625 | ATP-binding cassette, sub-family A (ABC1), member 5 |
C00004631
|
| 4791 | NFKB2, H2TF1, LYT-10, LYT10, NF-kB2, p105, p52 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
C00004631
|
| 4800 | NFYA, CBF-A, CBF-B, HAP2, NF-YA | nuclear transcription factor Y, alpha |
C00004631
|
| 25791 | NGEF, ARHGEF27, EPHEXIN | neuronal guanine nucleotide exchange factor |
C00004631
|
| 55768 | NGLY1, CDG1V, PNG1, PNGase | N-glycanase 1 (EC:3.5.1.52) |
C00004631
|
| 60491 | NIF3L1, ALS2CR1, CALS-7 | NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
C00004631
|
| 51199 | NIN, SCKL7 | ninein (GSK3B interacting protein) |
C00004631
|
| 4815 | NINJ2 | ninjurin 2 |
C00004631
|
| 25934 | NIPSNAP3A, NIPSNAP4, TASSC | nipsnap homolog 3A (C. elegans) |
C00004631
|
| 56954 | NIT2 | nitrilase family, member 2 (EC:3.5.1.3) |
C00004631
|
| 79570 | NKAIN1, FAM77C | Na+/K+ transporting ATPase interacting 1 |
C00004631
|
| 85409 | NKD2, Naked2 | naked cuticle homolog 2 (Drosophila) |
C00004631
|
| 28512 | NKIRAS1, KBRAS1, kappaB-Ras1 | NFKB inhibitor interacting Ras-like 1 |
C00004631
|
| 28511 | NKIRAS2, KBRAS2, kappaB-Ras2 | NFKB inhibitor interacting Ras-like 2 |
C00004631
|
| 4824 | NKX3-1, BAPX2, NKX3, NKX3.1, NKX3A | NK3 homeobox 1 |
C00004631
|
| 54475 | NLE1, Nle | notchless homolog 1 (Drosophila) |
C00004631
|
| 51701 | NLK | nemo-like kinase (EC:2.7.11.24) |
C00004631
|
| 4828 | NMB | neuromedin B |
C00004631
|
| 4833 | NME4, NDPK-D, NM23H4, nm23-H4 | NME/NM23 nucleoside diphosphate kinase 4 (EC:2.7.4.6) |
C00004631
|
| 29922 | NME7, MN23H7, NDK_7, NDK7, nm23-H7 | NME/NM23 family member 7 (EC:2.7.4.6) |
C00004631
|
| 9111 | NMI | N-myc (and STAT) interactor |
C00004631
|
| 9397 | NMT2 | N-myristoyltransferase 2 (EC:2.3.1.97) |
C00004631
|
| 161424 | NOP9, C14orf21 | NOP9 nucleolar protein |
C00004631
|
| 404036 |
C00004631
|
||
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00004631
|
| 4883 | NPR3, ANP-C, ANPR-C, ANPRC, C5orf23, GUCY2B, NPR-C, NPRC | natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
C00004631
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00004631
|
| 4835 | NQO2, DHQV, DIA6, NMOR2, QR2 | NAD(P)H dehydrogenase, quinone 2 (EC:1.10.99.2) |
C00004631
|
| 8431 | NR0B2, SHP, SHP1 | nuclear receptor subfamily 0, group B, member 2 |
C00004631
|
| 9971 | NR1H4, BAR, FXR, HRR-1, HRR1, RIP14 | nuclear receptor subfamily 1, group H, member 4 |
C00004631
|
| 7026 | NR2F2, ARP1, COUPTFB, COUPTFII, NF-E3, NR2F1, SVP40, TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 |
C00004631
|
| 2908 | NR3C1, GCCR, GCR, GR, GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
C00004631
|
| 3164 | NR4A1, GFRP1, HMR, N10, NAK-1, NGFIB, NP10, NUR77, TR3 | nuclear receptor subfamily 4, group A, member 1 |
C00004631
|
| 4893 | NRAS, ALPS4, N-ras, NRAS1, NS6 | neuroblastoma RAS viral (v-ras) oncogene homolog |
C00004631
|
| 9315 | NREP, C5orf13, D4S114, P311, PRO1873, PTZ17, SEZ17 | neuronal regeneration related protein |
C00004631
|
| 8829 | NRP1, BDCA4, CD304, NP1, NRP, VEGF165R | neuropilin 1 |
C00004631
|
| 26012 | NSMF, HH9, NELF | NMDA receptor synaptonuclear signaling and neuronal migration factor |
C00004631
|
| 221078 | NSUN6, 4933414E04Rik, NOPD1 | NOP2/Sun domain family, member 6 |
C00004631
|
| 4907 | NT5E, CALJA, CD73, E5NT, NT, NT5, NTE, eN, eNT | 5'-nucleotidase, ecto (CD73) (EC:3.1.3.5) |
C00004631
|
| 123803 | NTAN1, PNAA, PNAD | N-terminal asparagine amidase |
C00004631
|
| 59277 | NTN4, PRO3091 | netrin 4 |
C00004631
|
| 9891 | NUAK1, ARK5 | NUAK family, SNF1-like kinase, 1 (EC:2.7.11.1) |
C00004631
|
| 80224 | NUBPL, C14orf127, IND1, huInd1 | nucleotide binding protein-like |
C00004631
|
| 4924 | NUCB1, CALNUC, NUC | nucleobindin 1 |
C00004631
|
| 134492 | NUDCD2 | NudC domain containing 2 |
C00004631
|
| 4521 | NUDT1, MTH1 | nudix (nucleoside diphosphate linked moiety X)-type motif 1 (EC:3.6.1.55 3.6.1.56) |
C00004631
|
| 256281 | NUDT14, UGPP, UGPPase | nudix (nucleoside diphosphate linked moiety X)-type motif 14 (EC:3.6.1.45) |
C00004631
|
| 53343 | NUDT9, NUDT10 | nudix (nucleoside diphosphate linked moiety X)-type motif 9 (EC:3.6.1.13) |
C00004631
|
| 83540 | NUF2, CDCA1, CT106, NUF2R | NUF2, NDC80 kinetochore complex component |
C00004631
|
| 23225 | NUP210, GP210, POM210 | nucleoporin 210kDa |
C00004631
|
| 129401 | NUP35, MP44, NP44, NUP53 | nucleoporin 35kDa |
C00004631
|
| 54830 | NUP62CL | nucleoporin 62kDa C-terminal like |
C00004631
|
| 79902 | NUP85, Nup75 | nucleoporin 85kDa |
C00004631
|
| 26471 | NUPR1, COM1, P8 | nuclear protein, transcriptional regulator, 1 |
C00004631
|
| 51203 | NUSAP1, ANKT, BM037, LNP, NUSAP, PRO0310p1, Q0310, SAPL | nucleolar and spindle associated protein 1 |
C00004631
|
| 220323 | OAF, NS5ATP13TP2 | OAF homolog (Drosophila) |
C00004631
|
| 4938 | OAS1, IFI-4, OIAS, OIASI | 2'-5'-oligoadenylate synthetase 1, 40/46kDa (EC:2.7.7.84) |
C00004631
|
| 4940 | OAS3, p100 | 2'-5'-oligoadenylate synthetase 3, 100kDa (EC:2.7.7.84) |
C00004631
|
| 79991 | OBFC1, AAF-44, AAF44, RP11-541N10.2, RPA-32, STN1, bA541N10.2 | oligonucleotide/oligosaccharide-binding fold containing 1 |
C00004631
|
| 100506658 | OCLN, BLCPMG | occludin (EC:2.1.1.67) |
C00004631
|
| 54959 | ODAM, APIN | odontogenic, ameloblast asssociated |
C00004631
|
| 4953 | ODC1, ODC | ornithine decarboxylase 1 (EC:4.1.1.17) |
C00004631
|
| 79627 | OGFRL1, dJ331H24.1 | opioid growth factor receptor-like 1 |
C00004631
|
| 4968 | OGG1, HMMH, HOGG1, MUTM, OGH1 | 8-oxoguanine DNA glycosylase (EC:4.2.99.18) |
C00004631
|
| 11339 | OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18beta, hMIS18beta | Opa interacting protein 5 |
C00004631
|
| 283298 | OLFML1, UNQ564 | olfactomedin-like 1 |
C00004631
|
| 56944 | OLFML3, HNOEL-iso, OLF44 | olfactomedin-like 3 |
C00004631
|
| 4973 | OLR1, CLEC8A, LOX1, LOXIN, SCARE1, SLOX1 | oxidized low density lipoprotein (lectin-like) receptor 1 |
C00004631
|
| 115209 | OMA1, 2010001O09Rik, DAB1, MPRP-1, YKR087C, ZMPOMA1, peptidase | OMA1 zinc metallopeptidase |
C00004631
|
| 4974 | OMG, OMGP | oligodendrocyte myelin glycoprotein |
C00004631
|
| 9480 | ONECUT2, OC-2, OC2 | one cut homeobox 2 |
C00004631
|
| 4998 | ORC1, HSORC1, ORC1L, PARC1 | origin recognition complex, subunit 1 |
C00004631
|
| 4999 | ORC2, ORC2L | origin recognition complex, subunit 2 |
C00004631
|
| 716175 |
C00004631
|
||
| 5004 | ORM1, AGP-A, AGP1, ORM | orosomucoid 1 |
C00004631
|
| 5005 | ORM2, AGP-B, AGP-B', AGP2 | orosomucoid 2 |
C00004631
|
| 29095 | ORMDL2, MST095, MSTP095, adoplin-2 | ORM1-like 2 (S. cerevisiae) |
C00004631
|
| 114884 | OSBPL10, ORP10, OSBP9 | oxysterol binding protein-like 10 |
C00004631
|
| 114885 | OSBPL11, ORP-11, ORP11, OSBP12, TCCCIA00292 | oxysterol binding protein-like 11 |
C00004631
|
| 26031 | OSBPL3, ORP-3, ORP3, OSBP3 | oxysterol binding protein-like 3 |
C00004631
|
| 114882 | OSBPL8, MST120, MSTP120, ORP8, OSBP10 | oxysterol binding protein-like 8 |
C00004631
|
| 64172 | OSGEPL1, Qri7 | O-sialoglycoprotein endopeptidase-like 1 (EC:3.4.24.57) |
C00004631
|
| 734 | OSGIN2, C8orf1, hT41 | oxidative stress induced growth inhibitor family member 2 |
C00004631
|
| 116039 | OSR2 | odd-skipped related transciption factor 2 |
C00004631
|
| 26578 | OSTF1, OSF, SH3P2, bA235O14.1 | osteoclast stimulating factor 1 |
C00004631
|
| 28962 | OSTM1, GIPN, GL, OPTB5 | osteopetrosis associated transmembrane protein 1 |
C00004631
|
| 78990 | OTUB2, C14orf137, OTB2, OTU2 | OTU domain, ubiquitin aldehyde binding 2 (EC:3.4.19.12) |
C00004631
|
| 51633 | OTUD6B, DUBA-5, DUBA5 | OTU domain containing 6B (EC:3.4.19.12) |
C00004631
|
| 92106 | OXNAD1 | oxidoreductase NAD-binding domain containing 1 |
C00004631
|
| 55074 | OXR1, TLDC3 | oxidation resistance 1 |
C00004631
|
| 5021 | OXTR, OT-R | oxytocin receptor |
C00004631
|
| 5029 | P2RY2, HP2U, P2RU1, P2U, P2U1, P2UR, P2Y2, P2Y2R | purinergic receptor P2Y, G-protein coupled, 2 |
C00004631
|
| 100196098 |
C00004631
|
||
| 5033 | P4HA1, P4HA | prolyl 4-hydroxylase, alpha polypeptide I (EC:1.14.11.2) |
C00004631
|
| 5034 | P4HB, DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB, PO4DB, PO4HB, PROHB | prolyl 4-hydroxylase, beta polypeptide (EC:5.3.4.1) |
C00004631
|
| 5036 | PA2G4, EBP1, HG4-1, p38-2G4 | proliferation-associated 2G4, 38kDa |
C00004631
|
| 80336 | PABPC1L, C20orf119, PABPC1L1, dJ1069P2.3, ePAB | poly(A) binding protein, cytoplasmic 1-like |
C00004631
|
| 5047 | PAEP, GD, GdA, GdF, GdS, PAEG, PEP, PP14 | progestagen-associated endometrial protein |
C00004631
|
| 5048 | PAFAH1B1, LIS1, LIS2, MDCR, MDS, PAFAH | platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) (EC:3.1.1.47) |
C00004631
|
| 5053 | PAH, PH, PKU, PKU1 | phenylalanine hydroxylase (EC:1.14.16.1) |
C00004631
|
| 23022 | PALLD, CGI151, MYN, PNCA1, SIH002 | palladin, cytoskeletal associated protein |
C00004631
|
| 54873 | PALMD, C1orf11, PALML | palmdelphin |
C00004631
|
| 9924 | PAN2, USP52 | PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (EC:3.1.13.4) |
C00004631
|
| 53354 | PANK1, PANK | pantothenate kinase 1 (EC:2.7.1.33) |
C00004631
|
| 80025 | PANK2, C20orf48, HARP, HSS, NBIA1, PKAN | pantothenate kinase 2 (EC:2.7.1.33) |
C00004631
|
| 152559 | PAQR3, RKTG | progestin and adipoQ receptor family member III |
C00004631
|
| 54852 | PAQR5, MPRG | progestin and adipoQ receptor family member V |
C00004631
|
| 85315 | PAQR8, C6orf33, LMPB1, MPRB | progestin and adipoQ receptor family member VIII |
C00004631
|
| 344838 | PAQR9 | progestin and adipoQ receptor family member IX |
C00004631
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00004631
|
| 64761 | PARP12, ARTD12, MST109, MSTP109, ZC3H1, ZC3HDC1 | poly (ADP-ribose) polymerase family, member 12 (EC:2.4.2.30) |
C00004631
|
| 55010 | PARPBP, AROM, C12orf48, PARI | PARP1 binding protein |
C00004631
|
| 25859 | PART1, NCRNA00206 | prostate androgen-regulated transcript 1 (non-protein coding) |
C00004631
|
| 23178 | PASK, PASKIN, STK37 | PAS domain containing serine/threonine kinase (EC:2.7.11.1) |
C00004631
|
| 7849 | PAX8 | paired box 8 |
C00004631
|
| 51260 | PBDC1, CXorf26 | polysaccharide biosynthesis domain containing 1 |
C00004631
|
| 55872 | PBK, CT84, Nori-3, SPK, TOPK | PDZ binding kinase (EC:2.7.12.2) |
C00004631
|
| 55193 | PBRM1, BAF180, PB1 | polybromo 1 |
C00004631
|
| 5090 | PBX3 | pre-B-cell leukemia homeobox 3 |
C00004631
|
| 5095 | PCCA | propionyl CoA carboxylase, alpha polypeptide (EC:6.4.1.3) |
C00004631
|
| 56133 | PCDHB2, PCDH-BETA2 | protocadherin beta 2 |
C00004631
|
| 5105 | PCK1, PEPCK-C, PEPCK1, PEPCKC | phosphoenolpyruvate carboxykinase 1 (soluble) (EC:4.1.1.32) |
C00004631
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00004631
|
| 399909 | PCNXL3 | pecanex-like 3 (Drosophila) |
C00004631
|
| 64430 | PCNXL4, C14orf135, FBP2 | pecanex-like 4 (Drosophila) |
C00004631
|
| 26577 | PCOLCE2, PCPE2 | procollagen C-endopeptidase enhancer 2 |
C00004631
|
| 5125 | PCSK5, PC5, PC6, PC6A, SPC6, subtilase | proprotein convertase subtilisin/kexin type 5 (EC:3.4.21.-) |
C00004631
|
| 255738 | PCSK9, FH3, HCHOLA3, LDLCQ1, NARC-1, NARC1, PC9 | proprotein convertase subtilisin/kexin type 9 |
C00004631
|
| 201626 | PDE12, 2'-PDE | phosphodiesterase 12 |
C00004631
|
| 5143 | PDE4C, DPDE1 | phosphodiesterase 4C, cAMP-specific (EC:3.1.4.17) |
C00004631
|
| 5155 | PDGFB, IBGC5, PDGF-2, PDGF2, SIS, SSV, c-sis | platelet-derived growth factor beta polypeptide |
C00004631
|
| 56034 | PDGFC, FALLOTEIN, SCDGF | platelet derived growth factor C |
C00004631
|
| 5157 | PDGFRL, PDGRL, PRLTS | platelet-derived growth factor receptor-like |
C00004631
|
| 2923 | PDIA3, ER60, ERp57, ERp60, ERp61, GRP57, GRP58, HsT17083, P58, PI-PLC | protein disulfide isomerase family A, member 3 (EC:5.3.4.1) |
C00004631
|
| 149420 | PDIK1L, CLIK1L, STK35L2 | PDLIM1 interacting kinase 1 like (EC:2.7.11.1) |
C00004631
|
| 5163 | PDK1 | pyruvate dehydrogenase kinase, isozyme 1 (EC:2.7.11.2) |
C00004631
|
| 5164 | PDK2, PDHK2, PDKII | pyruvate dehydrogenase kinase, isozyme 2 (EC:2.7.11.2) |
C00004631
|
| 5165 | PDK3, CMTX6 | pyruvate dehydrogenase kinase, isozyme 3 (EC:2.7.11.2) |
C00004631
|
| 5166 | PDK4 | pyruvate dehydrogenase kinase, isozyme 4 (EC:2.7.11.2) |
C00004631
|
| 9260 | PDLIM7, LMP1, LMP3 | PDZ and LIM domain 7 (enigma) |
C00004631
|
| 54704 | PDP1, PDH, PDP, PDPC, PPM2C | pyruvate dehyrogenase phosphatase catalytic subunit 1 (EC:3.1.3.43) |
C00004631
|
| 57546 | PDP2, PPM2C2 | pyruvate dehyrogenase phosphatase catalytic subunit 2 (EC:3.1.3.43) |
C00004631
|
| 23047 | PDS5B, APRIN, AS3, CG008 | PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
C00004631
|
| 57107 | PDSS2, C6orf210, COQ10D3, DLP1, bA59I9.3, hDLP1 | prenyl (decaprenyl) diphosphate synthase, subunit 2 (EC:2.5.1.91) |
C00004631
|
| 8682 | PEA15, HMAT1, HUMMAT1H, MAT1, MAT1H, PEA-15, PED | phosphoprotein enriched in astrocytes 15 |
C00004631
|
| 375033 | PEAR1, JEDI, MEGF12 | platelet endothelial aggregation receptor 1 |
C00004631
|
| 5037 | PEBP1, HCNP, HCNPpp, PBP, PEBP, PEBP-1, RKIP | phosphatidylethanolamine binding protein 1 |
C00004631
|
| 5175 | PECAM1, CD31, CD31/EndoCAM, GPIIA', PECA1, PECAM-1, endoCAM | platelet/endothelial cell adhesion molecule 1 |
C00004631
|
| 100196721 |
C00004631
|
||
| 55825 | PECR, DCRRP, HPDHASE, HSA250303, PVIARL, SDR29C1, TERP | peroxisomal trans-2-enoyl-CoA reductase (EC:1.3.1.38) |
C00004631
|
| 23089 | PEG10, EDR, HB-1, MEF3L, Mar2, Mart2, RGAG3 | paternally expressed 10 |
C00004631
|
| 246330 | PELI3 | pellino E3 ubiquitin protein ligase family member 3 |
C00004631
|
| 53918 | PELO, PRO1770 | pelota homolog (Drosophila) |
C00004631
|
| 64065 | PERP, KCP1, KRTCAP1, PIGPC1, RP3-496H19.1, THW, dJ496H19.1 | PERP, TP53 apoptosis effector |
C00004631
|
| 5194 | PEX13, NALD, PBD11A, PBD11B, ZWS | peroxisomal biogenesis factor 13 |
C00004631
|
| 8504 | PEX3, PBD10A, TRG18 | peroxisomal biogenesis factor 3 |
C00004631
|
| 5190 | PEX6, PAF-2, PAF2, PBD4A, PDB4B, PXAAA1 | peroxisomal biogenesis factor 6 |
C00004631
|
| 5209 | PFKFB3, IPFK2, PFK2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (EC:2.7.1.105 3.1.3.46) |
C00004631
|
| 5210 | PFKFB4 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (EC:2.7.1.105 3.1.3.46) |
C00004631
|
| 5214 | PFKP, PFK-C, PFKF | phosphofructokinase, platelet (EC:2.7.1.11) |
C00004631
|
| 5223 | PGAM1, PGAM-B, PGAMA | phosphoglycerate mutase 1 (brain) (EC:3.1.3.13 5.4.2.4 5.4.2.11) |
C00004631
|
| 323107 |
C00004631
|
||
| 80055 | PGAP1, Bst1, ISPD3024 | post-GPI attachment to proteins 1 |
C00004631
|
| 84547 | PGBD1, HUCEP-4, SCAND4, dJ874C20.4 | piggyBac transposable element derived 1 |
C00004631
|
| 267004 | PGBD3 | piggyBac transposable element derived 3 |
C00004631
|
| 5225 | PGC, PEPC, PGII | progastricsin (pepsinogen C) (EC:3.4.23.3) |
C00004631
|
| 5226 | PGD, 6PGD | phosphogluconate dehydrogenase (EC:1.1.1.44) |
C00004631
|
| 5230 | PGK1, MIG10, PGKA | phosphoglycerate kinase 1 (EC:2.7.2.3) |
C00004631
|
| 5236 | PGM1, CDG1T, GSD14 | phosphoglucomutase 1 (EC:5.4.2.2) |
C00004631
|
| 55276 | PGM2 | phosphoglucomutase 2 (EC:5.4.2.2 5.4.2.7) |
C00004631
|
| 283209 | PGM2L1, BM32A | phosphoglucomutase 2-like 1 (EC:2.7.1.106) |
C00004631
|
| 9489 | PGS1 | phosphatidylglycerophosphate synthase 1 (EC:2.7.8.5) |
C00004631
|
| 9749 | PHACTR2, C6orf56 | phosphatase and actin regulator 2 |
C00004631
|
| 55274 | PHF10, BAF45A, XAP135 | PHD finger protein 10 |
C00004631
|
| 9678 | PHF14 | PHD finger protein 14 |
C00004631
|
| 23133 | PHF8, JHDM1F, MRXSSD, ZNF422 | PHD finger protein 8 (EC:1.14.11.27) |
C00004631
|
| 26227 | PHGDH, 3-PGDH, 3PGDH, PDG, PGAD, PGD, PGDH, SERA | phosphoglycerate dehydrogenase (EC:1.1.1.95) |
C00004631
|
| 5256 | PHKA2, GSD9A, PHK, PYK, PYKL, XLG, XLG2 | phosphorylase kinase, alpha 2 (liver) (EC:2.7.11.19) |
C00004631
|
| 5257 | PHKB | phosphorylase kinase, beta (EC:2.7.11.19) |
C00004631
|
| 22822 | PHLDA1, DT1P1B11, PHRIP, TDAG51 | pleckstrin homology-like domain, family A, member 1 |
C00004631
|
| 7262 | PHLDA2, BRW1C, BWR1C, HLDA2, IPL, TSSC3 | pleckstrin homology-like domain, family A, member 2 |
C00004631
|
| 23612 | PHLDA3, TIH1 | pleckstrin homology-like domain, family A, member 3 |
C00004631
|
| 90102 | PHLDB2, LL5b, LL5beta | pleckstrin homology-like domain, family B, member 2 |
C00004631
|
| 493911 | PHOSPHO2 | phosphatase, orphan 2 (EC:3.1.3.74) |
C00004631
|
| 10745 | PHTF1, PHTF | putative homeodomain transcription factor 1 |
C00004631
|
| 57157 | PHTF2 | putative homeodomain transcription factor 2 |
C00004631
|
| 254295 | PHYHD1 | phytanoyl-CoA dioxygenase domain containing 1 |
C00004631
|
| 84457 | PHYHIPL | phytanoyl-CoA 2-hydroxylase interacting protein-like |
C00004631
|
| 5297 | PI4KA, PI4K-ALPHA, PIK4CA, pi4K230 | phosphatidylinositol 4-kinase, catalytic, alpha (EC:2.7.1.67) |
C00004631
|
| 63895 | PIEZO2, C18orf30, C18orf58, FAM38B, FAM38B2, HsT748, HsT771 | piezo-type mechanosensitive ion channel component 2 |
C00004631
|
| 80119 | PIF1, C15orf20, PIF | PIF1 5'-to-3' DNA helicase (EC:3.6.4.12) |
C00004631
|
| 23556 | PIGN, MCAHS, MCAHS1, MCD4, MDC4, PIG-N | phosphatidylinositol glycan anchor biosynthesis, class N |
C00004631
|
| 80235 | PIGZ, GPI-MT-IV, PIG-Z, SMP3 | phosphatidylinositol glycan anchor biosynthesis, class Z |
C00004631
|
| 118788 | PIK3AP1, BCAP, RP11-34E5.3 | phosphoinositide-3-kinase adaptor protein 1 |
C00004631
|
| 5290 | PIK3CA, CLOVE, CWS5, MCAP, MCM, MCMTC, PI3K, p110-alpha | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (EC:2.7.11.1 2.7.1.153) |
C00004631
|
| 5294 | PIK3CG, PI3CG, PI3K, PI3Kgamma, PIK3, p110gamma, p120-PI3K | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma (EC:2.7.11.1 2.7.1.153) |
C00004631
|
| 29990 | PILRB, FDFACT1, FDFACT2 | paired immunoglobin-like type 2 receptor beta |
C00004631
|
| 5300 | PIN1, DOD, UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 (EC:5.2.1.8) |
C00004631
|
| 8395 | PIP5K1B, MSS4, STM7 | phosphatidylinositol-4-phosphate 5-kinase, type I, beta (EC:2.7.1.68) |
C00004631
|
| 51268 | PIPOX, LPIPOX | pipecolic acid oxidase (EC:1.5.3.1 1.5.3.7) |
C00004631
|
| 8544 | PIR | pirin (iron-binding nuclear protein) (EC:1.13.11.24) |
C00004631
|
| 57095 | PITHD1, C1orf128, HT014, RP5-886K2.4, TXNL1CL | PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
C00004631
|
| 26207 | PITPNC1, M-RDGB-beta, MRDGBbeta, RDGB-BETA, RDGBB, RDGBB1 | phosphatidylinositol transfer protein, cytoplasmic 1 |
C00004631
|
| 9600 | PITPNM1, DRES9, NIR2, PITPNM, RDGB, RDGB1, RDGBA, RDGBA1, Rd9 | phosphatidylinositol transfer protein, membrane-associated 1 |
C00004631
|
| 5308 | PITX2, ARP1, Brx1, IDG2, IGDS, IGDS2, IHG2, IRID2, Otlx2, PTX2, RGS, RIEG, RIEG1, RS | paired-like homeodomain 2 |
C00004631
|
| 91461 | PKDCC, SGK493, Vlk | protein kinase domain containing, cytoplasmic |
C00004631
|
| 5315 | PKM, CTHBP, OIP3, PK3, PKM2, TCB, THBP1 | pyruvate kinase, muscle (EC:2.7.1.40) |
C00004631
|
| 9088 | PKMYT1, MYT1 | protein kinase, membrane associated tyrosine/threonine 1 (EC:2.7.11.1) |
C00004631
|
| 5585 | PKN1, DBK, PAK-1, PAK1, PKN, PKN-ALPHA, PRK1, PRKCL1 | protein kinase N1 (EC:2.7.11.13) |
C00004631
|
| 5318 | PKP2, ARVD9 | plakophilin 2 |
C00004631
|
| 8502 | PKP4, p0071 | plakophilin 4 |
C00004631
|
| 84647 | PLA2G12B, GXIIB, GXIIIsPLA2, PLA2G13 | phospholipase A2, group XIIB (EC:3.1.1.4) |
C00004631
|
| 11145 | PLA2G16, AdPLA, H-REV107-1, HRASLS3, HREV107, HREV107-1, HREV107-3, HRSL3 | phospholipase A2, group XVI (EC:3.1.1.4 3.1.1.32) |
C00004631
|
| 5320 | PLA2G2A, MOM1, PLA2, PLA2B, PLA2L, PLA2S, PLAS1, sPLA2 | phospholipase A2, group IIA (platelets, synovial fluid) (EC:3.1.1.4) |
C00004631
|
| 153770 | PLAC8L1 | PLAC8-like 1 |
C00004631
|
| 5324 | PLAG1, PSA, SGPA, ZNF912 | pleiomorphic adenoma gene 1 |
C00004631
|
| 5325 | PLAGL1, LOT1, ZAC, ZAC1 | pleiomorphic adenoma gene-like 1 |
C00004631
|
| 5327 | PLAT, T-PA, TPA | plasminogen activator, tissue (EC:3.4.21.68) |
C00004631
|
| 5328 | PLAU, ATF, BDPLT5, QPD, UPA, URK, u-PA | plasminogen activator, urokinase (EC:3.4.21.73) |
C00004631
|
| 23236 | PLCB1, EIEE12, PI-PLC, PLC-154, PLC-I, PLC154, PLCB1A, PLCB1B | phospholipase C, beta 1 (phosphoinositide-specific) (EC:3.1.4.11) |
C00004631
|
| 5337 | PLD1 | phospholipase D1, phosphatidylcholine-specific (EC:3.1.4.4) |
C00004631
|
| 23646 | PLD3, HU-K4, HUK4 | phospholipase D family, member 3 (EC:3.1.4.4) |
C00004631
|
| 59339 | PLEKHA2, TAPP2 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
C00004631
|
| 54477 | PLEKHA5, PEPP-2, PEPP2 | pleckstrin homology domain containing, family A member 5 |
C00004631
|
| 22874 | PLEKHA6, PEPP-3, PEPP3 | pleckstrin homology domain containing, family A member 6 |
C00004631
|
| 64857 | PLEKHG2, ARHGEF42, CLG | pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
C00004631
|
| 25894 | PLEKHG4, ARHGEF44, PRTPHN1, SCA4 | pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
C00004631
|
| 57475 | PLEKHH1 | pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
C00004631
|
| 9842 | PLEKHM1, AP162, B2, OPTB6 | pleckstrin homology domain containing, family M (with RUN domain) member 1 |
C00004631
|
| 51177 | PLEKHO1, CKIP-1, OC120 | pleckstrin homology domain containing, family O member 1 |
C00004631
|
| 45915 |
C00004631
|
||
| 26154 | ABCA12, ARCI4A, ARCI4B, ICR2B, LI2 | ATP-binding cassette, sub-family A (ABC1), member 12 |
C00004631
|
| 10769 | PLK2, SNK, hPlk2, hSNK | polo-like kinase 2 (EC:2.7.11.21) |
C00004631
|
| 1263 | PLK3, CNK, FNK, PRK | polo-like kinase 3 (EC:2.7.11.21) |
C00004631
|
| 5352 | PLOD2, LH2, TLH | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (EC:1.14.11.4) |
C00004631
|
| 5354 | PLP1, GPM6C, HLD1, MMPL, PLP, PLP/DM20, PMD, SPG2 | proteolipid protein 1 |
C00004631
|
| 5357 | PLS1 | plastin 1 |
C00004631
|
| 5359 | PLSCR1, MMTRA1B | phospholipid scramblase 1 |
C00004631
|
| 57088 | PLSCR4, TRA1 | phospholipid scramblase 4 |
C00004631
|
| 5360 | PLTP, BPIFE, HDLCQ9 | phospholipid transfer protein |
C00004631
|
| 55558 | PLXNA3, 6.3, HSSEXGENE, PLXN3, PLXN4, XAP-6 | plexin A3 |
C00004631
|
| 23654 | PLXNB2, MM1, Nbla00445, PLEXB2, dJ402G11.3 | plexin B2 |
C00004631
|
| 10154 | PLXNC1, CD232, PLXN-C1, VESPR | plexin C1 |
C00004631
|
| 5366 | PMAIP1, APR, NOXA | phorbol-12-myristate-13-acetate-induced protein 1 |
C00004631
|
| 5376 | PMP22, CMT1A, CMT1E, DSS, GAS-3, HMSNIA, HNPP, Sp110 | peripheral myelin protein 22 |
C00004631
|
| 11284 | PNKP, EIEE10, MCSZ, PNK | polynucleotide kinase 3'-phosphatase (EC:2.7.1.78 3.1.3.32) |
C00004631
|
| 9240 | PNMA1, MA1 | paraneoplastic Ma antigen 1 |
C00004631
|
| 84968 | PNMA6A, MA6, PNMA6 | paraneoplastic Ma antigen family member 6A |
C00004631
|
| 4860 | PNP, NP, PRO1837, PUNP | purine nucleoside phosphorylase (EC:2.4.2.1) |
C00004631
|
| 80339 | PNPLA3, ADPN, C22orf20, iPLA(2)epsilon | patatin-like phospholipase domain containing 3 (EC:3.1.1.3) |
C00004631
|
| 5422 | POLA1, POLA, p180 | polymerase (DNA directed), alpha 1, catalytic subunit (EC:2.7.7.7) |
C00004631
|
| 5425 | POLD2 | polymerase (DNA directed), delta 2, accessory subunit (EC:2.7.7.7) |
C00004631
|
| 57804 | POLD4, POLDS, p12 | polymerase (DNA-directed), delta 4, accessory subunit |
C00004631
|
| 26073 | POLDIP2, PDIP38, POLD4 | polymerase (DNA-directed), delta interacting protein 2 |
C00004631
|
| 5426 | POLE, CRCS12, FILS, POLE1 | polymerase (DNA directed), epsilon, catalytic subunit (EC:2.7.7.7) |
C00004631
|
| 5427 | POLE2, DPE2 | polymerase (DNA directed), epsilon 2, accessory subunit (EC:2.7.7.7) |
C00004631
|
| 5429 | POLH, RAD30, RAD30A, XP-V, XPV | polymerase (DNA directed), eta (EC:2.7.7.7) |
C00004631
|
| 10721 | POLQ, POLH, PRO0327 | polymerase (DNA directed), theta (EC:2.7.7.7) |
C00004631
|
| 51082 | POLR1D, AC19, POLR1C, RPA16, RPA9, RPAC2, RPC16, RPO1-3, TCS2 | polymerase (RNA) I polypeptide D, 16kDa |
C00004631
|
| 55703 | POLR3B, C128, HLD8, RPC2 | polymerase (RNA) III (DNA directed) polypeptide B (EC:2.7.7.6) |
C00004631
|
| 661 | POLR3D, BN51T, RPC4, RPC53, TSBN51 | polymerase (RNA) III (DNA directed) polypeptide D, 44kDa (EC:2.7.7.6) |
C00004631
|
| 10622 | POLR3G, RPC32, RPC7 | polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
C00004631
|
| 5444 | PON1, ESA, MVCD5, PON | paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81) |
C00004631
|
| 64091 | POPDC2, POP2 | popeye domain containing 2 |
C00004631
|
| 5447 | POR, CPR, CYPOR, P450R | P450 (cytochrome) oxidoreductase (EC:1.6.2.4) |
C00004631
|
| 5450 | POU2AF1, BOB1, OBF-1, OBF1, OCAB | POU class 2 associating factor 1 |
C00004631
|
| 27068 | PPA2, SID6-306 | pyrophosphatase (inorganic) 2 (EC:3.6.1.1) |
C00004631
|
| 5465 | PPARA, NR1C1, PPAR, PPARalpha, hPPAR | peroxisome proliferator-activated receptor alpha |
C00004631
|
| 5467 | PPARD, FAAR, NR1C2, NUC1, NUCI, NUCII, PPARB | peroxisome proliferator-activated receptor delta |
C00004631
|
| 5468 | PPARG, CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | peroxisome proliferator-activated receptor gamma |
C00004631
|
| 60490 | PPCDC | phosphopantothenoylcysteine decarboxylase (EC:4.1.1.36) |
C00004631
|
| 8495 | PPFIBP2, Cclp1 | PTPRF interacting protein, binding protein 2 (liprin beta 2) |
C00004631
|
| 5478 | PPIA, CYPA, CYPH | peptidylprolyl isomerase A (cyclophilin A) (EC:5.2.1.8) |
C00004631
|
| 5481 | PPID, CYP-40, CYPD | peptidylprolyl isomerase D (EC:5.2.1.8) |
C00004631
|
| 85313 | PPIL4, HDCME13P | peptidylprolyl isomerase (cyclophilin)-like 4 (EC:5.2.1.8) |
C00004631
|
| 8493 | PPM1D, PP2C-DELTA, WIP1 | protein phosphatase, Mg2+/Mn2+ dependent, 1D (EC:3.1.3.16) |
C00004631
|
| 57460 | PPM1H, ARHCL1, NERPP-2C, URCC2 | protein phosphatase, Mg2+/Mn2+ dependent, 1H (EC:3.1.3.16) |
C00004631
|
| 152926 | PPM1K, BDP, MSUDMV, PP2Ckappa, PP2Cm, PTMP, UG0882E07 | protein phosphatase, Mg2+/Mn2+ dependent, 1K (EC:3.1.3.16) |
C00004631
|
| 132160 | PPM1M, PP2C-eta, PP2CE, PP2Ceta | protein phosphatase, Mg2+/Mn2+ dependent, 1M (EC:3.1.3.16) |
C00004631
|
| 10848 | PPP1R13L, IASPP, NKIP1, RAI, RAI4 | protein phosphatase 1, regulatory subunit 13 like |
C00004631
|
| 23645 | PPP1R15A, GADD34 | protein phosphatase 1, regulatory subunit 15A |
C00004631
|
| 84988 | PPP1R16A, MYPT3 | protein phosphatase 1, regulatory subunit 16A |
C00004631
|
| 170954 | PPP1R18, HKMT1098, KIAA1949 | protein phosphatase 1, regulatory subunit 18 |
C00004631
|
| 5502 | PPP1R1A, I1, IPP1 | protein phosphatase 1, regulatory (inhibitor) subunit 1A (EC:3.1.3.16) |
C00004631
|
| 129285 | PPP1R21, CCDC128, KLRAQ1 | protein phosphatase 1, regulatory subunit 21 |
C00004631
|
| 5507 | PPP1R3C, PPP1R5 | protein phosphatase 1, regulatory subunit 3C |
C00004631
|
| 5509 | PPP1R3D, PPP1R6 | protein phosphatase 1, regulatory subunit 3D (EC:3.1.3.16) |
C00004631
|
| 55607 | PPP1R9A, NRB1, NRBI, Neurabin-I | protein phosphatase 1, regulatory subunit 9A |
C00004631
|
| 5519 | PPP2R1B, PP2A-Abeta, PR65B | protein phosphatase 2, regulatory subunit A, beta (EC:3.1.3.16) |
C00004631
|
| 55012 | PPP2R3C, C14orf10, G4-1, G5pr | protein phosphatase 2, regulatory subunit B'', gamma |
C00004631
|
| 5524 | PPP2R4, PP2A, PR53, PTPA | protein phosphatase 2A activator, regulatory subunit 4 (EC:5.2.1.8) |
C00004631
|
| 5530 | PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B | protein phosphatase 3, catalytic subunit, alpha isozyme (EC:3.1.3.16) |
C00004631
|
| 57718 | PPP4R4, KIAA1622, PP4R4 | protein phosphatase 4, regulatory subunit 4 |
C00004631
|
| 9701 | PPP6R2, KIAA0685, PP6R2, SAP190, SAPS2 | protein phosphatase 6, regulatory subunit 2 |
C00004631
|
| 23398 | PPWD1 | peptidylprolyl isomerase domain and WD repeat containing 1 (EC:5.2.1.8) |
C00004631
|
| 130814 | PQLC3, C2orf22 | PQ loop repeat containing 3 |
C00004631
|
| 306506 |
C00004631
|
||
| 118471 | PRAP1, PRO1195, RP11-122K13.6, UPA | proline-rich acidic protein 1 |
C00004631
|
| 56980 | PRDM10, PFM7 | PR domain containing 10 |
C00004631
|
| 5052 | PRDX1, MSP23, NKEF-A, NKEFA, PAG, PAGA, PAGB, PRX1, PRXI, TDPX2 | peroxiredoxin 1 (EC:1.11.1.15) |
C00004631
|
| 7001 | PRDX2, NKEF-B, NKEFB, PRP, PRX2, PRXII, PTX1, TDPX1, TPX1, TSA | peroxiredoxin 2 (EC:1.11.1.15) |
C00004631
|
| 10549 | PRDX4, AOE37-2, PRX-4 | peroxiredoxin 4 (EC:1.11.1.15) |
C00004631
|
| 153768 | PRELID2 | PRELI domain containing 2 |
C00004631
|
| 5564 | PRKAB1, AMPK, HAMPKb | protein kinase, AMP-activated, beta 1 non-catalytic subunit |
C00004631
|
| 5565 | PRKAB2 | protein kinase, AMP-activated, beta 2 non-catalytic subunit |
C00004631
|
| 5580 | PRKCD, MAY1, PKCD, nPKC-delta | protein kinase C, delta (EC:2.7.10.2 2.7.11.13) |
C00004631
|
| 5584 | PRKCI, DXS1179E, PKCI, nPKC-iota | protein kinase C, iota (EC:2.7.11.13) |
C00004631
|
| 5588 | PRKCQ, PRKCT, nPKC-theta | protein kinase C, theta (EC:2.7.11.13) |
C00004631
|
| 5587 | PRKD1, PKC-MU, PKCM, PKD, PRKCM | protein kinase D1 (EC:2.7.11.13) |
C00004631
|
| 23683 | PRKD3, EPK2, PKC-NU, PKD3, PRKCN, nPKC-NU | protein kinase D3 (EC:2.7.11.13) |
C00004631
|
| 79706 | PRKRIP1, C114, KRBOX3 | PRKR interacting protein 1 (IL11 inducible) |
C00004631
|
| 90826 | PRMT10 | protein arginine methyltransferase 10 (putative) |
C00004631
|
| 10419 | PRMT5, HRMT1L5, IBP72, JBP1, SKB1, SKB1Hs | protein arginine methyltransferase 5 (EC:2.1.1.125) |
C00004631
|
| 5624 | PROC, APC, PC, PROC1, THPH3, THPH4 | protein C (inactivator of coagulation factors Va and VIIIa) (EC:3.4.21.69) |
C00004631
|
| 10544 | PROCR, CCCA, CCD41, EPCR | protein C receptor, endothelial |
C00004631
|
| 58510 | PRODH2, HSPOX1 | proline dehydrogenase (oxidase) 2 (EC:1.5.99.8) |
C00004631
|
| 8842 | PROM1, AC133, CD133, CORD12, MCDR2, PROML1, RP41, STGD4 | prominin 1 |
C00004631
|
| 344405 | PRORSD1P, NCRNA00117, PRDXDD1P, Prdxdd1, Ybakd1 | prolyl-tRNA synthetase associated domain containing 1, pseudogene |
C00004631
|
| 5627 | PROS1, PROS, PS21, PS22, PS23, PS24, PS25, PSA, THPH5, THPH6 | protein S (alpha) |
C00004631
|
| 11212 | PROSC | proline synthetase co-transcribed homolog (bacterial) |
C00004631
|
| 5629 | PROX1 | prospero homeobox 1 |
C00004631
|
| 8899 | PRPF4B, PR4H, PRP4, PRP4H, PRP4K, dJ1013A10.1 | pre-mRNA processing factor 4B (EC:2.7.11.1) |
C00004631
|
| 222171 | PRR15 | proline rich 15 |
C00004631
|
| 255783 | PRR24 | proline rich 24 |
C00004631
|
| 80758 | PRR7 | proline rich 7 (synaptic) |
C00004631
|
| 84726 | PRRC2B, BAT2L, BAT2L1, KIAA0515, LQFBS-1 | proline-rich coiled-coil 2B |
C00004631
|
| 80863 | PRRT1, C6orf31, DSPD1, IFITMD7, NG5 | proline-rich transmembrane protein 1 |
C00004631
|
| 124221 | PRSS30P, Disp, TMPRSS8, TMPRSS8P | protease, serine, 30, pseudogene |
C00004631
|
| 58497 | PRUNE, DRES-17, DRES17, HTCD37 | prune exopolyphosphatase (EC:3.6.1.1) |
C00004631
|
| 29968 | PSAT1, EPIP, PSA, PSAT | phosphoserine aminotransferase 1 (EC:2.6.1.52) |
C00004631
|
| 23362 | PSD3, EFA6R, HCA67 | pleckstrin and Sec7 domain containing 3 |
C00004631
|
| 23550 | PSD4, EFA6B, TIC | pleckstrin and Sec7 domain containing 4 |
C00004631
|
| 11168 | PSIP1, DFS70, LEDGF, PAIP, PSIP2, p52, p75 | PC4 and SFRS1 interacting protein 1 |
C00004631
|
| 29893 | PSMC3IP, GT198, HOP2, HUMGT198A, ODG3, TBPIP | PSMC3 interacting protein |
C00004631
|
| 5708 | PSMD2, P97, RPN1, S2, TRAP2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
C00004631
|
| 5711 | PSMD5, S5B | proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
C00004631
|
| 23198 | PSME4, PA200 | proteasome (prosome, macropain) activator subunit 4 |
C00004631
|
| 8624 | PSMG1, C21LRP, DSCR2, LRPC21, PAC-1, PAC1 | proteasome (prosome, macropain) assembly chaperone 1 |
C00004631
|
| 114796 | PSMG3-AS1 | PSMG3 antisense RNA 1 (head to head) |
C00004631
|
| 389362 | PSMG4, C6orf86, PAC4, bA506K6.2 | proteasome (prosome, macropain) assembly chaperone 4 |
C00004631
|
| 5723 | PSPH, PSP, PSPHD | phosphoserine phosphatase (EC:3.1.3.3) |
C00004631
|
| 84722 | PSRC1, DDA3, FP3214 | proline/serine-rich coiled-coil 1 |
C00004631
|
| 9051 | PSTPIP1, CD2BP1, CD2BP1L, CD2BP1S, H-PIP, PAPAS, PSTPIP | proline-serine-threonine phosphatase interacting protein 1 |
C00004631
|
| 5725 | PTBP1, HNRNP-I, HNRNPI, HNRPI, PTB, PTB-1, PTB-T, PTB2, PTB3, PTB4, pPTB | polypyrimidine tract binding protein 1 |
C00004631
|
| 58155 | PTBP2, PTB, PTBLP, brPTB, nPTB, nPTB5, nPTB6, nPTB7, nPTB8 | polypyrimidine tract binding protein 2 |
C00004631
|
| 5727 | PTCH1, BCNS, HPE7, NBCCS, PTC, PTC1, PTCH, PTCH11 | patched 1 |
C00004631
|
| 442213 | PTCHD4, C6orf138, dJ402H5.2 | patched domain containing 4 |
C00004631
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00004631
|
| 10728 | PTGES3, P23, TEBP, cPGES | prostaglandin E synthase 3 (cytosolic) (EC:5.3.99.3) |
C00004631
|
| 5738 | PTGFRN, CD315, CD9P-1, EWI-F, FPRP, SMAP-6 | prostaglandin F2 receptor inhibitor |
C00004631
|
| 22949 | PTGR1, LTB4DH, PGR1, ZADH3 | prostaglandin reductase 1 (EC:1.3.1.48 1.3.1.74) |
C00004631
|
| 145482 | PTGR2, PGR2, ZADH1 | prostaglandin reductase 2 (EC:1.3.1.48) |
C00004631
|
| 5742 | PTGS1, COX1, COX3, PCOX1, PES-1, PGG/HS, PGHS-1, PGHS1, PHS1, PTGHS | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00004631
|
| 5744 | PTHLH, BDE2, HHM, PLP, PTHR, PTHRP | parathyroid hormone-like hormone |
C00004631
|
| 19 | ABCA1, ABC-1, ABC1, CERP, HDLDT1, TGD | ATP-binding cassette, sub-family A (ABC1), member 1 |
C00004631
|
| 11156 | PTP4A3, PRL-3, PRL-R, PRL3 | protein tyrosine phosphatase type IVA, member 3 (EC:3.1.3.48) |
C00004631
|
| 201562 | PTPLB, HACD2 | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b (EC:4.2.1.134) |
C00004631
|
| 114971 | PTPMT1, DUSP23, MOSP, PLIP | protein tyrosine phosphatase, mitochondrial 1 (EC:3.1.3.16 3.1.3.48 3.1.3.27) |
C00004631
|
| 5793 | PTPRG, HPTPG, PTPG, R-PTP-GAMMA, RPTPG | protein tyrosine phosphatase, receptor type, G (EC:3.1.3.48) |
C00004631
|
| 5794 | PTPRH, SAP1 | protein tyrosine phosphatase, receptor type, H (EC:3.1.3.48) |
C00004631
|
| 5796 | PTPRK, R-PTP-kappa | protein tyrosine phosphatase, receptor type, K (EC:3.1.3.48) |
C00004631
|
| 5799 | PTPRN2, IA-2beta, IAR, ICAAR, PTPRP, R-PTP-N2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 (EC:3.1.3.48) |
C00004631
|
| 10076 | PTPRU, FMI, PCP-2, PTP, PTP-J, PTP-PI, PTP-RO, PTPPSI, PTPRO, PTPU2, R-PTP-PSI, R-PTP-U, hPTP-J | protein tyrosine phosphatase, receptor type, U (EC:3.1.3.48) |
C00004631
|
| 284119 | PTRF, CAVIN, CAVIN1, CGL4, cavin-1 | polymerase I and transcript release factor |
C00004631
|
| 9232 | PTTG1, EAP1, HPTTG, PTTG, TUTR1 | pituitary tumor-transforming 1 |
C00004631
|
| 150962 | PUS10, CCDC139, DOBI | pseudouridylate synthase 10 |
C00004631
|
| 81607 | PVRL4, EDSS1, LNIR, PRR4, nectin-4 | poliovirus receptor-related 4 |
C00004631
|
| 11137 | PWP1, IEF-SSP-9502 | PWP1 homolog (S. cerevisiae) |
C00004631
|
| 361820 |
C00004631
|
||
| 100689047 |
C00004631
|
||
| 29920 | PYCR2, P5CR2 | pyrroline-5-carboxylate reductase family, member 2 (EC:1.5.1.2) |
C00004631
|
| 5834 | PYGB, GPBB | phosphorylase, glycogen; brain (EC:2.4.1.1) |
C00004631
|
| 5860 | QDPR, DHPR, PKU2, SDR33C1 | quinoid dihydropteridine reductase (EC:1.5.1.34) |
C00004631
|
| 25797 | QPCT, GCT, QC, sQC | glutaminyl-peptide cyclotransferase (EC:2.3.2.5) |
C00004631
|
| 84074 | QRICH2 | glutamine rich 2 |
C00004631
|
| 5768 | QSOX1, Q6, QSCN6 | quiescin Q6 sulfhydryl oxidase 1 (EC:1.8.3.2) |
C00004631
|
| 84440 | RAB11FIP4, RAB11-FIP4 | RAB11 family interacting protein 4 (class II) |
C00004631
|
| 81876 | RAB1B | RAB1B, member RAS oncogene family |
C00004631
|
| 55647 | RAB20 | RAB20, member RAS oncogene family |
C00004631
|
| 51715 | RAB23, HSPC137 | RAB23, member RAS oncogene family |
C00004631
|
| 9364 | RAB28, CORD18 | RAB28, member RAS oncogene family |
C00004631
|
| 11031 | RAB31, Rab22B | RAB31, member RAS oncogene family |
C00004631
|
| 5865 | RAB3B | RAB3B, member RAS oncogene family |
C00004631
|
| 10966 | RAB40B, RAR, SEC4L | RAB40B, member RAS oncogene family |
C00004631
|
| 115273 | RAB42, RP4-669K10.6 | RAB42, member RAS oncogene family |
C00004631
|
| 51762 | RAB8B | RAB8B, member RAS oncogene family |
C00004631
|
| 10567 | RABAC1, PRA1, PRAF1, YIP3 | Rab acceptor 1 (prenylated) |
C00004631
|
| 10244 | RABEPK, RAB9P40, bA65N13.1, p40 | Rab9 effector protein with kelch motifs |
C00004631
|
| 9910 | RABGAP1L, HHL, TBC1D18 | RAB GTPase activating protein 1-like |
C00004631
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00004631
|
| 25788 | RAD54B, RDH54 | RAD54 homolog B (S. cerevisiae) |
C00004631
|
| 8438 | RAD54L, HR54, RAD54A, hHR54, hRAD54 | RAD54-like (S. cerevisiae) |
C00004631
|
| 26064 | RAI14, NORPEG, RAI13 | retinoic acid induced 14 |
C00004631
|
| 5898 | RALA, RAL | v-ral simian leukemia viral oncogene homolog A (ras related) |
C00004631
|
| 5899 | RALB | v-ral simian leukemia viral oncogene homolog B |
C00004631
|
| 5900 | RALGDS, RGDS, RGF, RalGEF | ral guanine nucleotide dissociation stimulator |
C00004631
|
| 10267 | RAMP1 | receptor (G protein-coupled) activity modifying protein 1 |
C00004631
|
| 57610 | RANBP10 | RAN binding protein 10 |
C00004631
|
| 26953 | RANBP6 | RAN binding protein 6 |
C00004631
|
| 9693 | RAPGEF2, CNrasGEF, NRAPGEP, PDZ-GEF1, PDZGEF1, RA-GEF, RA-GEF-1, Rap-GEP, nRap_GEP | Rap guanine nucleotide exchange factor (GEF) 2 |
C00004631
|
| 65059 | RAPH1, ALS2CR18, ALS2CR9, LPD, PREL-2, PREL2, RMO1, RalGDS/AF-6 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
C00004631
|
| 5913 | RAPSN, RAPSYN, RNF205 | receptor-associated protein of the synapse |
C00004631
|
| 5915 | RARB, HAP, NR1B2, RRB2 | retinoic acid receptor, beta |
C00004631
|
| 5919 | RARRES2, HP10433, TIG2 | retinoic acid receptor responder (tazarotene induced) 2 |
C00004631
|
| 5921 | RASA1, CM-AVM, CMAVM, GAP, PKWS, RASA, RASGAP, p120GAP, p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 |
C00004631
|
| 51655 | RASD1, AGS1, DEXRAS1, MGC:26290 | RAS, dexamethasone-induced 1 |
C00004631
|
| 11186 | RASSF1, 123F2, NORE2A, RASSF1A, RDA32, REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 |
C00004631
|
| 55225 | RAVER2 | ribonucleoprotein, PTB-binding 2 |
C00004631
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00004631
|
| 5930 | RBBP6, MY038, P2P-R, PACT, RBQ-1, SNAMA | retinoblastoma binding protein 6 |
C00004631
|
| 5932 | RBBP8, COM1, CTIP, JWDS, RIM, SAE2, SCKL2 | retinoblastoma binding protein 8 |
C00004631
|
| 10616 | RBCK1, C20orf18, HOIL-1, HOIL1, RBCK2, RNF54, UBCE7IP3, XAP3, XAP4, ZRANB4 | RanBP-type and C3HC4-type zinc finger containing 1 |
C00004631
|
| 64080 | RBKS, RBSK | ribokinase (EC:2.7.1.15) |
C00004631
|
| 10137 | RBM12, HRIHFB2091, SWAN | RNA binding motif protein 12 |
C00004631
|
| 9904 | RBM19 | RNA binding motif protein 19 |
C00004631
|
| 55147 | RBM23, CAPERbeta, RNPC4 | RNA binding motif protein 23 |
C00004631
|
| 221662 | RBM24, RNPC6, dJ259A10.1 | RNA binding motif protein 24 |
C00004631
|
| 5935 | RBM3, IS1-RNPL, RNPL | RNA binding motif (RNP1, RRM) protein 3 |
C00004631
|
| 55544 | RBM38, HSRNASEB, RNPC1, SEB4B, SEB4D, dJ800J21.2 | RNA binding motif protein 38 |
C00004631
|
| 5936 | RBM4, LARK, RBM4A, ZCCHC21, ZCRB3A | RNA binding motif protein 4 |
C00004631
|
| 129831 | RBM45, DRB1 | RNA binding motif protein 45 |
C00004631
|
| 83759 | RBM4B, RBM30, RBM4L, ZCCHC15, ZCCHC21B, ZCRB3B | RNA binding motif protein 4B |
C00004631
|
| 5937 | RBMS1, C2orf12, HCC-4, MSSP, MSSP-1, MSSP-2, MSSP-3, SCR2, YC1 | RNA binding motif, single stranded interacting protein 1 |
C00004631
|
| 5939 | RBMS2, SCR3 | RNA binding motif, single stranded interacting protein 2 |
C00004631
|
| 5947 | RBP1, CRABP-I, CRBP, CRBP1, CRBPI, RBPC | retinol binding protein 1, cellular |
C00004631
|
| 5950 | RBP4, RDCCAS | retinol binding protein 4, plasma |
C00004631
|
| 55213 | RCBTB1, CLLD7, CLLL7, GLP, RP11-185C18.1 | regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
C00004631
|
| 145226 | RDH12, LCA13, LCA3, RP53, SDR7C2 | retinol dehydrogenase 12 (all-trans/9-cis/11-cis) (EC:1.1.1.-) |
C00004631
|
| 8434 | RECK, ST15 | reversion-inducing-cysteine-rich protein with kazal motifs |
C00004631
|
| 5965 | RECQL, RECQL1, RecQ1 | RecQ protein-like (DNA helicase Q1-like) (EC:3.6.4.12) |
C00004631
|
| 9401 | RECQL4, RECQ4 | RecQ protein-like 4 (EC:3.6.4.12) |
C00004631
|
| 221035 | REEP3, C10orf74 | receptor accessory protein 3 |
C00004631
|
| 92840 | REEP6, C19orf32, DP1L1, TB2L1 | receptor accessory protein 6 |
C00004631
|
| 10157 | AASS, LKR/SDH, LKRSDH, LORSDH | aminoadipate-semialdehyde synthase (EC:1.5.1.8 1.5.1.9) |
C00004631
|
| 5971 | RELB, I-REL, IREL, REL-B | v-rel avian reticuloendotheliosis viral oncogene homolog B |
C00004631
|
| 54884 | RETSAT | retinol saturase (all-trans-retinol 13,14-reductase) (EC:1.3.99.23) |
C00004631
|
| 5980 | REV3L, POLZ, REV3 | REV3-like, polymerase (DNA directed), zeta, catalytic subunit (EC:2.7.7.7) |
C00004631
|
| 5983 | RFC3, RFC38 | replication factor C (activator 1) 3, 38kDa |
C00004631
|
| 5984 | RFC4, A1, RFC37 | replication factor C (activator 1) 4, 37kDa |
C00004631
|
| 117584 | RFFL, CARP-2, CARP2, FRING, RIFIFYLIN, RNF189, RNF34L | ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
C00004631
|
| 5986 | RFNG | RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (EC:2.4.1.222) |
C00004631
|
| 10737 | RFPL3S, NCRNA00005, RFPL3-AS, RFPL3-AS1, RFPL3ANT, RFPL3AS | RFPL3 antisense |
C00004631
|
| 55159 | RFWD3, RNF201 | ring finger and WD repeat domain 3 |
C00004631
|
| 64864 | RFX7, RFXDC2 | regulatory factor X, 7 |
C00004631
|
| 5994 | RFXAP | regulatory factor X-associated protein |
C00004631
|
| 28984 | RGCC, C13orf15, RGC-32, RGC32, bA157L14.2 | regulator of cell cycle |
C00004631
|
| 23179 | RGL1, RGL | ral guanine nucleotide dissociation stimulator-like 1 |
C00004631
|
| 285704 | RGMB, DRAGON | repulsive guidance molecule family member b |
C00004631
|
| 9104 | RGN, GNL, RC, SMP30 | regucalcin (EC:3.1.1.17) |
C00004631
|
| 5996 | RGS1, 1R20, BL34, IER1, IR20 | regulator of G-protein signaling 1 |
C00004631
|
| 6001 | RGS10 | regulator of G-protein signaling 10 |
C00004631
|
| 10287 | RGS19, GAIP, RGSGAIP | regulator of G-protein signaling 19 |
C00004631
|
| 5997 | RGS2, G0S8 | regulator of G-protein signaling 2, 24kDa |
C00004631
|
| 8601 | RGS20, RGSZ1, ZGAP1, g(z)GAP, gz-GAP | regulator of G-protein signaling 20 |
C00004631
|
| 8490 | RGS5, MST092, MST106, MST129, MSTP032, MSTP092, MSTP106, MSTP129 | regulator of G-protein signaling 5 |
C00004631
|
| 6000 | RGS7 | regulator of G-protein signaling 7 |
C00004631
|
| 57414 | RHBDD2, NPD007, RHBDL7 | rhomboid domain containing 2 |
C00004631
|
| 25807 | RHBDD3, C22orf3, HS984G1A, PTAG | rhomboid domain containing 3 |
C00004631
|
| 79651 | RHBDF2, RHBDL5, RHBDL6, TOC | rhomboid 5 homolog 2 (Drosophila) |
C00004631
|
| 54933 | RHBDL2, RRP2 | rhomboid, veinlet-like 2 (Drosophila) (EC:3.4.21.105) |
C00004631
|
| 57127 | RHBG, SLC42A2 | Rh family, B glycoprotein (gene/pseudogene) |
C00004631
|
| 9886 | RHOBTB1 | Rho-related BTB domain containing 1 |
C00004631
|
| 22836 | RHOBTB3 | Rho-related BTB domain containing 3 |
C00004631
|
| 389 | RHOC, ARH9, ARHC, H9, RHOH9 | ras homolog family member C |
C00004631
|
| 54509 | RHOF, ARHF, RIF | ras homolog family member F (in filopodia) |
C00004631
|
| 55288 | RHOT1, ARHT1, MIRO-1, MIRO1 | ras homolog family member T1 |
C00004631
|
| 58480 | RHOU, ARHU, CDC42L1, DJ646B12.2, WRCH1, fJ646B12.2, hG28K | ras homolog family member U |
C00004631
|
| 196383 | RILPL2, RLP2 | Rab interacting lysosomal protein-like 2 |
C00004631
|
| 284716 | RIMKLA, FAM80A, NAAGS, NAAGS-II | ribosomal modification protein rimK-like family member A |
C00004631
|
| 54453 | RIN2, MACS, RASSF4 | Ras and Rab interactor 2 |
C00004631
|
| 60561 | RINT1, RINT-1 | RAD50 interactor 1 |
C00004631
|
| 8767 | RIPK2, CARD3, CARDIAK, CCK, GIG30, RICK, RIP2 | receptor-interacting serine-threonine kinase 2 (EC:2.7.11.1 2.7.10.2) |
C00004631
|
| 53820 | RIPPLY3, DSCR6 | ripply transcriptional repressor 3 |
C00004631
|
| 6014 | RIT2, RIBA, RIN, ROC2 | Ras-like without CAAX 2 |
C00004631
|
| 6018 | RLF, ZN-15L, ZNF292L | rearranged L-myc fusion |
C00004631
|
| 55005 | RMND1, C6orf96, COXPD11, RMD1, bA351K16, bA351K16.3 | required for meiotic nuclear division 1 homolog (S. cerevisiae) |
C00004631
|
| 6038 | RNASE4, RNS4 | ribonuclease, RNase A family, 4 (EC:3.1.27.-) |
C00004631
|
| 10535 | RNASEH2A, AGS4, JUNB, RNASEHI, RNHIA, RNHL | ribonuclease H2, subunit A (EC:3.1.26.4) |
C00004631
|
| 567505 |
C00004631
|
||
| 27289 | RND1, ARHS, RHO6, RHOS | Rho family GTPase 1 |
C00004631
|
| 9604 | RNF14, ARA54, HFB30, TRIAD2 | ring finger protein 14 |
C00004631
|
| 50862 | RNF141, ZFP26, ZNF230 | ring finger protein 141 |
C00004631
|
| 81847 | RNF146, RP3-351K20.1, dJ351K20.1 | ring finger protein 146 |
C00004631
|
| 127544 | RNF19B, IBRDC3, NKLAM | ring finger protein 19B |
C00004631
|
| 57674 | RNF213, ALO17, C17orf27, KIAA1618, MYMY2, MYSTR, NET57 | ring finger protein 213 |
C00004631
|
| 11237 | RNF24, G1L | ring finger protein 24 |
C00004631
|
| 54894 | RNF43, RNF124, URCC | ring finger protein 43 |
C00004631
|
| 51136 | RNFT1 | ring finger protein, transmembrane 1 |
C00004631
|
| 6091 | ROBO1, DUTT1, SAX3 | roundabout, axon guidance receptor, homolog 1 (Drosophila) |
C00004631
|
| 6093 | ROCK1, P160ROCK, ROCK-I | Rho-associated, coiled-coil containing protein kinase 1 (EC:2.7.11.1) |
C00004631
|
| 9475 | ROCK2, ROCK-II | Rho-associated, coiled-coil containing protein kinase 2 (EC:2.7.11.1) |
C00004631
|
| 6095 | RORA, NR1F1, ROR1, ROR2, ROR3, RZR-ALPHA, RZRA | RAR-related orphan receptor A |
C00004631
|
| 441212 | RP9P | retinitis pigmentosa 9 pseudogene |
C00004631
|
| 26015 | RPAP1 | RNA polymerase II associated protein 1 |
C00004631
|
| 6120 | RPE, RPE2-1 | ribulose-5-phosphate-3-epimerase (EC:5.1.3.1) |
C00004631
|
| 23322 | RPGRIP1L, CORS3, FTM, JBTS7, MKS5, NPHP8 | RPGRIP1-like |
C00004631
|
| 6138 | RPL15, EC45, L15, RPL10, RPLY10, RPYL10 | ribosomal protein L15 |
C00004631
|
| 6157 | RPL27A, L27A | ribosomal protein L27a |
C00004631
|
| 6158 | RPL28, L28 | ribosomal protein L28 (EC:3.6.5.3) |
C00004631
|
| 132241 | RPL32P3 | ribosomal protein L32 pseudogene 3 |
C00004631
|
| 10557 | RPP38 | ribonuclease P/MRP 38kDa subunit (EC:3.1.26.5) |
C00004631
|
| 10799 | RPP40, RNASEP1, bA428J1.3 | ribonuclease P/MRP 40kDa subunit (EC:3.1.26.5) |
C00004631
|
| 85495 | RPPH1, H1RNA, RPPH1-1 | ribonuclease P RNA component H1 |
C00004631
|
| 6228 | RPS23, S23 | ribosomal protein S23 |
C00004631
|
| 6195 | RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1) |
C00004631
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00004631
|
| 9252 | RPS6KA5, MSK1, MSPK1, RLPK | ribosomal protein S6 kinase, 90kDa, polypeptide 5 (EC:2.7.11.1) |
C00004631
|
| 3921 | RPSA, 37LRP, 67LR, LAMBR, LAMR1, LBP, LBP/p40, LRP, LRP/LR, NEM/1CHD4, SA, lamR, p40 | ribosomal protein SA |
C00004631
|
| 9125 | RQCD1, CNOT9, CT129, RCD-1, RCD1 | RCD1 required for cell differentiation1 homolog (S. pombe) |
C00004631
|
| 6236 | RRAD, RAD, RAD1, REM3 | Ras-related associated with diabetes |
C00004631
|
| 6237 | RRAS | related RAS viral (r-ras) oncogene homolog |
C00004631
|
| 6238 | RRBP1, ES/130, ES130, RRp, hES | ribosome binding protein 1 |
C00004631
|
| 6240 | RRM1, R1, RIR1, RR1 | ribonucleotide reductase M1 (EC:1.17.4.1) |
C00004631
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00004631
|
| 50484 | RRM2B, MTDPS8A, MTDPS8B, P53R2 | ribonucleotide reductase M2 B (TP53 inducible) (EC:1.17.4.1) |
C00004631
|
| 100131998 | RRN3P3 | RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene 3 |
C00004631
|
| 51093 | RRNAD1, C1orf66, CGI-41 | ribosomal RNA adenine dimethylase domain containing 1 |
C00004631
|
| 27341 | RRP7A, BK126B4.3, CGI-96 | ribosomal RNA processing 7 homolog A (S. cerevisiae) |
C00004631
|
| 23212 | RRS1 | RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
C00004631
|
| 55316 | RSAD1 | radical S-adenosyl methionine domain containing 1 |
C00004631
|
| 91543 | RSAD2, 2510004L01Rik, cig33, cig5, vig1 | radical S-adenosyl methionine domain containing 2 |
C00004631
|
| 26156 | RSL1D1, CSIG, PBK1, UTP30 | ribosomal L1 domain containing 1 |
C00004631
|
| 51319 | RSRC1, SFRS21, SRrp53 | arginine/serine-rich coiled-coil 1 |
C00004631
|
| 8634 | RTCA, RPC, RTC1, RTCD1 | RNA 3'-terminal phosphate cyclase (EC:6.5.1.4) |
C00004631
|
| 84816 | RTN4IP1, NIMP | reticulon 4 interacting protein 1 |
C00004631
|
| 80230 | RUFY1, RABIP4, ZFYVE12 | RUN and FYVE domain containing 1 |
C00004631
|
| 154661 | RUNDC3B, RPIB9, RPIP9 | RUN domain containing 3B |
C00004631
|
| 860 | RUNX2, AML3, CBF-alpha-1, CBFA1, CCD, CCD1, CLCD, OSF-2, OSF2, PEA2aA, PEBP2aA | runt-related transcription factor 2 |
C00004631
|
| 112611 | RWDD2A, RWDD2, dJ747H23.2 | RWD domain containing 2A |
C00004631
|
| 10069 | RWDD2B, C21orf6 | RWD domain containing 2B |
C00004631
|
| 6258 | RXRG, NR2B3, RXRC | retinoid X receptor, gamma |
C00004631
|
| 6259 | RYK, D3S3195, JTK5, JTK5A, RYK1 | receptor-like tyrosine kinase (EC:2.7.10.1) |
C00004631
|
| 6271 | S100A1, S100, S100-alpha, S100A | S100 calcium binding protein A1 |
C00004631
|
| 6282 | S100A11, MLN70, S100C | S100 calcium binding protein A11 |
C00004631
|
| 6284 | S100A13 | S100 calcium binding protein A13 |
C00004631
|
| 140576 | S100A16, DT1P1A7, S100F | S100 calcium binding protein A16 |
C00004631
|
| 6273 | S100A2, CAN19, S100L | S100 calcium binding protein A2 |
C00004631
|
| 6274 | S100A3, S100E | S100 calcium binding protein A3 |
C00004631
|
| 6277 | S100A6, 2A9, 5B10, CABP, CACY, PRA | S100 calcium binding protein A6 |
C00004631
|
| 6285 | S100B, NEF, S100, S100-B, S100beta | S100 calcium binding protein B |
C00004631
|
| 795 | S100G, CABP, CABP1, CABP9K, CALB3 | S100 calcium binding protein G |
C00004631
|
| 6286 | S100P, MIG9 | S100 calcium binding protein P |
C00004631
|
| 26278 | SACS, ARSACS, DNAJC29 | spastic ataxia of Charlevoix-Saguenay (sacsin) |
C00004631
|
| 6294 | SAFB, HAP, HET, SAF-B1, SAFB1 | scaffold attachment factor B |
C00004631
|
| 9667 | SAFB2 | scaffold attachment factor B2 |
C00004631
|
| 6299 | SALL1, HSAL1, Sal-1, TBS, ZNF794 | spalt-like transcription factor 1 |
C00004631
|
| 23034 | SAMD4A, SAMD4, SMAUG, SMAUG1, SMG, SMGA | sterile alpha motif domain containing 4A |
C00004631
|
| 54809 | SAMD9, C7orf5, DRIF1, NFTC, OEF1, OEF2 | sterile alpha motif domain containing 9 |
C00004631
|
| 25939 | SAMHD1, CHBL2, DCIP, HDDC1, MOP-5, SBBI88 | SAM domain and HD domain 1 |
C00004631
|
| 10284 | SAP18, 2HOR0202, SAP18P | Sin3A-associated protein, 18kDa |
C00004631
|
| 79685 | SAP30L, NS4ATP2 | SAP30-like |
C00004631
|
| 89958 | SAPCD2, C9orf140, p42.3 | suppressor APC domain containing 2 |
C00004631
|
| 9092 | SART1, Ara1, HOMS1, SART1259, SNRNP110, Snu66 | squamous cell carcinoma antigen recognized by T cells |
C00004631
|
| 23328 | SASH1, SH3D6A, dJ323M4, dJ323M4.1 | SAM and SH3 domain containing 1 |
C00004631
|
| 163786 | SASS6, SAS-6, SAS6 | spindle assembly 6 homolog (C. elegans) |
C00004631
|
| 6303 | SAT1, DC21, KFSD, KFSDX, SAT, SSAT, SSAT-1 | spermidine/spermine N1-acetyltransferase 1 (EC:2.3.1.57) |
C00004631
|
| 23314 | SATB2 | SATB homeobox 2 |
C00004631
|
| 66234 |
C00004631
|
||
| 114821 | SCAND3, Buster4, ZBED9, ZFP38-L, ZNF305P2, ZNF452, dJ1186N24.3 | SCAN domain containing 3 |
C00004631
|
| 49855 | SCAPER, ZNF291, Zfp291 | S-phase cyclin A-associated protein in the ER |
C00004631
|
| 677766 | SCARNA2, HBII-382, mgU2-25/61 | small Cajal body-specific RNA 2 |
C00004631
|
| 6319 | SCD, FADS5, MSTP008, SCD1, SCDOS | stearoyl-CoA desaturase (delta-9-desaturase) (EC:1.14.19.1) |
C00004631
|
| 152579 | SCFD2, STXBP1L1 | sec1 family domain containing 2 |
C00004631
|
| 6447 | SCG5, 7B2, P7B2, SGNE1, SgV | secretogranin V (7B2 protein) |
C00004631
|
| 132320 | SCLT1, CAP-1A, CAP1A, hCAP-1A | sodium channel and clathrin linker 1 |
C00004631
|
| 22955 | SCMH1, Scml3 | sex comb on midleg homolog 1 (Drosophila) |
C00004631
|
| 10389 | SCML2 | sex comb on midleg-like 2 (Drosophila) |
C00004631
|
| 6323 | SCN1A, EIEE6, FEB3, FEB3A, FHM3, GEFSP2, HBSCI, NAC1, Nav1.1, SCN1, SMEI | sodium channel, voltage-gated, type I, alpha subunit |
C00004631
|
| 6335 | SCN9A, ETHA, FEB3B, GEFSP7, NE-NA, NENA, Nav1.7, PN1, SFNP | sodium channel, voltage-gated, type IX, alpha subunit |
C00004631
|
| 6337 | SCNN1A, BESC2, ENaCa, ENaCalpha, SCNEA, SCNN1 | sodium channel, non-voltage-gated 1 alpha subunit |
C00004631
|
| 6383 | SDC2, CD362, HSPG, HSPG1, SYND2 | syndecan 2 |
C00004631
|
| 6385 | SDC4, SYND4 | syndecan 4 |
C00004631
|
| 23753 | SDF2L1 | stromal cell-derived factor 2-like 1 |
C00004631
|
| 6391 | SDHC, CYB560, CYBL, PGL3, QPS1, SDH3 | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
C00004631
|
| 8436 | SDPR, CAVIN2, PS-p68, SDR, cavin-2 | serum deprivation response |
C00004631
|
| 90701 | SEC11C, SEC11L3, SPC21, SPCS4C | SEC11 homolog C (S. cerevisiae) (EC:3.4.21.89) |
C00004631
|
| 26984 | SEC22A, SEC22L2 | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
C00004631
|
| 9871 | SEC24D | SEC24 family member D |
C00004631
|
| 195827 | AAED1, C9orf21 | AhpC/TSA antioxidant enzyme domain containing 1 |
C00004631
|
| 8991 | SELENBP1, LPSB, SBP56, SP56, hSBP | selenium binding protein 1 |
C00004631
|
| 83642 | SELO | selenoprotein O |
C00004631
|
| 6403 | SELP, CD62, CD62P, GMP140, GRMP, LECAM3, PADGEM, PSEL | selectin P (granule membrane protein 140kDa, antigen CD62) |
C00004631
|
| 65260 | SELRC1, C1orf163 | Sel1 repeat containing 1 |
C00004631
|
| 10512 | SEMA3C, SEMAE, SemE | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
C00004631
|
| 6405 | SEMA3F, SEMA-IV, SEMA4, SEMAK | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
C00004631
|
| 56920 | SEMA3G, sem2 | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
C00004631
|
| 57715 | SEMA4G | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
C00004631
|
| 57337 | SENP7 | SUMO1/sentrin specific peptidase 7 (EC:3.4.22.68) |
C00004631
|
| 51091 | SEPSECS, LP, PCH2D, SLA, SLA/LP | Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (EC:2.9.1.2) |
C00004631
|
| 84947 | SERAC1 | serine active site containing 1 |
C00004631
|
| 347735 | SERINC2, PRO0899, TDE2, TDE2L | serine incorporator 2 |
C00004631
|
| 51156 | SERPINA10, PZI, ZPI | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
C00004631
|
| 5267 | SERPINA4, KAL, KLST, KST, PI-4, PI4, kallistatin | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 |
C00004631
|
| 866 | SERPINA6, CBG | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
C00004631
|
| 6906 | SERPINA7, TBG | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
C00004631
|
| 1992 | SERPINB1, EI, ELANH2, LEI, M/NEI, MNEI, PI-2, PI2 | serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
C00004631
|
| 5055 | SERPINB2, HsT1201, PAI, PAI-2, PAI2, PLANH2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
C00004631
|
| 6317 | SERPINB3, HsT1196, SCC, SCCA-1, SCCA-PD, SCCA1, T4-A | serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
C00004631
|
| 5272 | SERPINB9, CAP-3, CAP3, PI-9, PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
C00004631
|
| 462 | SERPINC1, AT3, AT3D, ATIII, THPH7 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
C00004631
|
| 3053 | SERPIND1, D22S673, HC2, HCF2, HCII, HLS2, LS2, THPH10 | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
C00004631
|
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00004631
|
| 5270 | SERPINE2, GDN, GDNPF, PI-7, PI7, PN-1, PN1, PNI | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
C00004631
|
| 5176 | SERPINF1, EPC-1, OI12, OI6, PEDF | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
C00004631
|
| 5345 | SERPINF2, A2AP, AAP, ALPHA-2-PI, API, PLI | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
C00004631
|
| 5274 | SERPINI1, PI12, neuroserpin | serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
C00004631
|
| 29950 | SERTAD1, SEI1, TRIP-Br1 | SERTA domain containing 1 |
C00004631
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00004631
|
| 83667 | SESN2, HI95, SES2, SEST2 | sestrin 2 |
C00004631
|
| 143686 | SESN3, SEST3 | sestrin 3 |
C00004631
|
| 55209 | SETD5 | SET domain containing 5 |
C00004631
|
| 79918 | SETD6 | SET domain containing 6 |
C00004631
|
| 83852 | SETDB2, C13orf4, CLLD8, CLLL8, KMT1F | SET domain, bifurcated 2 (EC:2.1.1.43) |
C00004631
|
| 6419 | SETMAR, HsMar1, METNASE, Mar1 | SET domain and mariner transposase fusion gene (EC:2.1.1.43) |
C00004631
|
| 8175 | SF3A2, PRP11, PRPF11, SAP62, SF3a66 | splicing factor 3a, subunit 2, 66kDa |
C00004631
|
| 10262 | SF3B4, AFD1, Hsh49, SAP49, SF3b49 | splicing factor 3b, subunit 4, 49kDa |
C00004631
|
| 6422 | SFRP1, FRP, FRP-1, FRP1, FrzA, SARP2 | secreted frizzled-related protein 1 |
C00004631
|
| 100380358 |
C00004631
|
||
| 100305297 |
C00004631
|
||
| 727087 |
C00004631
|
||
| 375035 | SFT2D2, UNQ512, dJ747L4.C1.2 | SFT2 domain containing 2 |
C00004631
|
| 6446 | SGK1, SGK | serum/glucocorticoid regulated kinase 1 (EC:2.7.11.1) |
C00004631
|
| 23678 | SGK3, CISK, SGK2, SGKL | serum/glucocorticoid regulated kinase family, member 3 (EC:2.7.11.1) |
C00004631
|
| 151246 | SGOL2, SGO2, TRIPIN | shugoshin-like 2 (S. pombe) |
C00004631
|
| 10019 | SH2B3, IDDM20, LNK | SH2B adaptor protein 3 |
C00004631
|
| 400745 | SH2D5 | SH2 domain containing 5 |
C00004631
|
| 6450 | SH3BGR, 21-GARP | SH3 domain binding glutamic acid-rich protein |
C00004631
|
| 6451 | SH3BGRL, SH3BGR | SH3 domain binding glutamic acid-rich protein like |
C00004631
|
| 83699 | SH3BGRL2 | SH3 domain binding glutamic acid-rich protein like 2 |
C00004631
|
| 83442 | SH3BGRL3, SH3BP-1, TIP-B1 | SH3 domain binding glutamic acid-rich protein like 3 |
C00004631
|
| 6452 | SH3BP2, 3BP-2, 3BP2, CRBM, CRPM | SH3-domain binding protein 2 |
C00004631
|
| 152503 | SH3D19, EBP, EVE1, Kryn, SH3P19 | SH3 domain containing 19 |
C00004631
|
| 618322 |
C00004631
|
||
| 9644 | SH3PXD2A, FISH, SH3MD1, TKS5 | SH3 and PX domains 2A |
C00004631
|
| 57630 | SH3RF1, POSH, RNF142, SH3MD2 | SH3 domain containing ring finger 1 |
C00004631
|
| 26751 | SH3YL1, RAY | SH3 and SYLF domain containing 1 |
C00004631
|
| 6462 | SHBG, ABP, SBP, TEBG | sex hormone-binding globulin |
C00004631
|
| 257218 | SHPRH, bA545I5.2 | SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
C00004631
|
| 6476 | SI | sucrase-isomaltase (alpha-glucosidase) (EC:3.2.1.10 3.2.1.48) |
C00004631
|
| 6477 | SIAH1, SIAH1A | siah E3 ubiquitin protein ligase 1 (EC:6.3.2.-) |
C00004631
|
| 6478 | SIAH2, hSiah2 | siah E3 ubiquitin protein ligase 2 (EC:6.3.2.-) |
C00004631
|
| 25942 | SIN3A | SIN3 transcription regulator family member A |
C00004631
|
| 23411 | SIRT1, SIR2L1 | sirtuin 1 |
C00004631
|
| 51547 | SIRT7, SIR2L7 | sirtuin 7 |
C00004631
|
| 220134 | SKA1, C18orf24 | spindle and kinetochore associated complex subunit 1 |
C00004631
|
| 8935 | SKAP2, PRAP, RA70, SAPS, SCAP2, SKAP-HOM, SKAP55R | src kinase associated phosphoprotein 2 |
C00004631
|
| 387640 | SKIDA1, C10orf140, DLN-1 | SKI/DACH domain containing 1 |
C00004631
|
| 23517 | SKIV2L2, Dob1, KIAA0052, Mtr4, fSAP118 | superkiller viralicidic activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13) |
C00004631
|
| 6502 | SKP2, FBL1, FBXL1, FLB1, p45 | S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
C00004631
|
| 4891 | SLC11A2, DCT1, DMT1, NRAMP2 | solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
C00004631
|
| 6558 | SLC12A2, BSC, BSC2, NKCC1 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
C00004631
|
| 6560 | SLC12A4, KCC1 | solute carrier family 12 (potassium/chloride transporter), member 4 |
C00004631
|
| 64849 | SLC13A3, NADC3, SDCT2 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
C00004631
|
| 284111 | SLC13A5, NACT | solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
C00004631
|
| 6566 | SLC16A1, HHF7, MCT, MCT1 | solute carrier family 16 (monocarboxylate transporter), member 1 |
C00004631
|
| 9123 | SLC16A3, MCT_3, MCT_4, MCT-3, MCT-4, MCT3, MCT4 | solute carrier family 16 (monocarboxylate transporter), member 3 |
C00004631
|
| 9122 | SLC16A4, MCT4, MCT5 | solute carrier family 16, member 4 |
C00004631
|
| 9120 | SLC16A6, MCT6, MCT7 | solute carrier family 16, member 6 |
C00004631
|
| 10246 | SLC17A2, NPT3 | solute carrier family 17, member 2 |
C00004631
|
| 63910 | SLC17A9, C20orf59, VNUT | solute carrier family 17 (vesicular nucleotide transporter), member 9 |
C00004631
|
| 116843 | SLC18B1, C6orf192, dJ55C23.6 | solute carrier family 18, subfamily B, member 1 |
C00004631
|
| 80704 | SLC19A3, BBGD, THMD2, THTR2 | solute carrier family 19 (thiamine transporter), member 3 |
C00004631
|
| 6506 | SLC1A2, EAAT2, GLT-1 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
C00004631
|
| 6574 | SLC20A1, GLVR1, Glvr-1, PIT1, PiT-1 | solute carrier family 20 (phosphate transporter), member 1 |
C00004631
|
| 55356 | SLC22A15, FLIPT1, PRO34686 | solute carrier family 22, member 15 |
C00004631
|
| 6581 | SLC22A3, EMT, EMTH, OCT3 | solute carrier family 22 (organic cation transporter), member 3 |
C00004631
|
| 6584 | SLC22A5, CDSP, OCTN2, OCTN2VT | solute carrier family 22 (organic cation/carnitine transporter), member 5 |
C00004631
|
| 114571 | SLC22A9, HOAT4, OAT4, OAT7, UST3H, ust3 | solute carrier family 22 (organic anion transporter), member 9 |
C00004631
|
| 9963 | SLC23A1, SLC23A2, SVCT1, YSPL3 | solute carrier family 23 (ascorbic acid transporter), member 1 |
C00004631
|
| 9962 | SLC23A2, NBTL1, SLC23A1, SVCT2, YSPL2 | solute carrier family 23 (ascorbic acid transporter), member 2 |
C00004631
|
| 6576 | SLC25A1, CTP, D2L2AD, SEA, SLC20A3 | solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
C00004631
|
| 8604 | SLC25A12, AGC1, ARALAR | solute carrier family 25 (aspartate/glutamate carrier), member 12 |
C00004631
|
| 8034 | SLC25A16, D10S105E, GDA, GDC, HGT.1, ML7, hML7 | solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
C00004631
|
| 788 | SLC25A20, CAC, CACT | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
C00004631
|
| 89874 | SLC25A21, ODC, ODC1 | solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
C00004631
|
| 29957 | SLC25A24, APC1, SCAMC-1 | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
C00004631
|
| 253512 | SLC25A30, KMCP1 | solute carrier family 25, member 30 |
C00004631
|
| 51629 | SLC25A39, CGI69 | solute carrier family 25, member 39 |
C00004631
|
| 203427 | SLC25A43 | solute carrier family 25, member 43 |
C00004631
|
| 283130 | SLC25A45 | solute carrier family 25, member 45 |
C00004631
|
| 91137 | SLC25A46 | solute carrier family 25, member 46 |
C00004631
|
| 1836 | SLC26A2, D5S1708, DTD, DTDST, EDM4, MST153, MSTP157 | solute carrier family 26 (anion exchanger), member 2 |
C00004631
|
| 1811 | SLC26A3, CLD, DRA | solute carrier family 26 (anion exchanger), member 3 |
C00004631
|
| 11001 | SLC27A2, ACSVL1, FACVL1, FATP2, HsT17226, VLACS, VLCS, hFACVL1 | solute carrier family 27 (fatty acid transporter), member 2 (EC:6.2.1.3) |
C00004631
|
| 2030 | SLC29A1, ENT1 | solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
C00004631
|
| 55315 | SLC29A3, ENT3, HCLAP, HJCD, PHID | solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
C00004631
|
| 81031 | SLC2A10, ATS, GLUT10 | solute carrier family 2 (facilitated glucose transporter), member 10 |
C00004631
|
| 6514 | SLC2A2, GLUT2 | solute carrier family 2 (facilitated glucose transporter), member 2 |
C00004631
|
| 6515 | SLC2A3, GLUT3 | solute carrier family 2 (facilitated glucose transporter), member 3 |
C00004631
|
| 29988 | SLC2A8, GLUT8, GLUTX1 | solute carrier family 2 (facilitated glucose transporter), member 8 |
C00004631
|
| 56606 | SLC2A9, GLUT9, GLUTX, UAQTL2, URATv1 | solute carrier family 2 (facilitated glucose transporter), member 9 |
C00004631
|
| 7779 | SLC30A1, ZNT1, ZRC1 | solute carrier family 30 (zinc transporter), member 1 |
C00004631
|
| 55532 | SLC30A10, HMDPC, ZNT10, ZNT8, ZRC1, ZnT-10 | solute carrier family 30, member 10 |
C00004631
|
| 9197 | SLC33A1, ACATN, AT-1, AT1, CCHLND, SPG42 | solute carrier family 33 (acetyl-CoA transporter), member 1 |
C00004631
|
| 10559 | SLC35A1, CDG2F, CMPST, CST, hCST | solute carrier family 35 (CMP-sialic acid transporter), member A1 |
C00004631
|
| 7355 | SLC35A2, CDG2M, CDGX, UDP-Gal-Tr, UGALT, UGAT, UGT, UGT1, UGT2, UGTL | solute carrier family 35 (UDP-galactose transporter), member A2 |
C00004631
|
| 23169 | SLC35D1, UGTREL7 | solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
C00004631
|
| 80255 | SLC35F5 | solute carrier family 35, member F5 |
C00004631
|
| 80723 | SLC35G2, TMEM22 | solute carrier family 35, member G2 |
C00004631
|
| 219855 | SLC37A2, pp11662 | solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
C00004631
|
| 81539 | SLC38A1, ATA1, NAT2, SAT1, SNAT1 | solute carrier family 38, member 1 |
C00004631
|
| 151258 | SLC38A11, AVT2 | solute carrier family 38, member 11 |
C00004631
|
| 54407 | SLC38A2, ATA2, PRO1068, SAT2, SNAT2 | solute carrier family 38, member 2 |
C00004631
|
| 10991 | SLC38A3, G17, NAT1, SN1 | solute carrier family 38, member 3 |
C00004631
|
| 55089 | SLC38A4, ATA3, NAT3, PAAT | solute carrier family 38, member 4 |
C00004631
|
| 92745 | SLC38A5, JM24, SN2, SNAT5, pp7194 | solute carrier family 38, member 5 |
C00004631
|
| 145389 | SLC38A6, NAT-1 | solute carrier family 38, member 6 |
C00004631
|
| 153129 | SLC38A9 | solute carrier family 38, member 9 |
C00004631
|
| 57181 | SLC39A10, LZT-Hs2 | solute carrier family 39 (zinc transporter), member 10 |
C00004631
|
| 201266 | SLC39A11, C17orf26, ZIP11 | solute carrier family 39, member 11 |
C00004631
|
| 23516 | SLC39A14, LZT-Hs4, NET34, ZIP14, cig19 | solute carrier family 39 (zinc transporter), member 14 |
C00004631
|
| 55630 | SLC39A4, AEZ, AWMS2, ZIP4 | solute carrier family 39 (zinc transporter), member 4 |
C00004631
|
| 283375 | SLC39A5, LZT-Hs7, ZIP5 | solute carrier family 39 (zinc transporter), member 5 |
C00004631
|
| 6520 | SLC3A2, 4F2, 4F2HC, 4T2HC, CD98, CD98HC, MDU1, NACAE | solute carrier family 3 (amino acid transporter heavy chain), member 2 |
C00004631
|
| 30061 | SLC40A1, FPN1, HFE4, IREG1, MST079, MTP1, SLC11A3 | solute carrier family 40 (iron-regulated transporter), member 1 |
C00004631
|
| 254428 | SLC41A1, MgtE | solute carrier family 41 (magnesium transporter), member 1 |
C00004631
|
| 84102 | SLC41A2, SLC41A1-L1 | solute carrier family 41 (magnesium transporter), member 2 |
C00004631
|
| 126969 | SLC44A3, CTL3 | solute carrier family 44, member 3 |
C00004631
|
| 204962 | SLC44A5, CTL5 | solute carrier family 44, member 5 |
C00004631
|
| 83959 | SLC4A11, BTR1, CDPD1, CHED2, NABC1, dJ794I6.2 | solute carrier family 4, sodium borate transporter, member 11 |
C00004631
|
| 9497 | SLC4A7, NBC2, NBC3, NBCN1, SBC2, SLC4A6 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
C00004631
|
| 200931 | SLC51A, OSTA, OSTalpha | solute carrier family 51, alpha subunit |
C00004631
|
| 123264 | SLC51B, OSTB, OSTBETA | solute carrier family 51, beta subunit |
C00004631
|
| 6526 | SLC5A3, SMIT, SMIT1, SMIT2 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
C00004631
|
| 6528 | SLC5A5, NIS, TDH1 | solute carrier family 5 (sodium/iodide cotransporter), member 5 |
C00004631
|
| 200010 | SLC5A9, SGLT4 | solute carrier family 5 (sodium/sugar cotransporter), member 9 |
C00004631
|
| 6538 | SLC6A11, GAT-3, GAT3, GAT4 | solute carrier family 6 (neurotransmitter transporter), member 11 |
C00004631
|
| 6539 | SLC6A12, BGT-1, BGT1, GAT2 | solute carrier family 6 (neurotransmitter transporter), member 12 |
C00004631
|
| 6532 | SLC6A4, 5-HTT, 5-HTTLPR, 5HTT, HTT, OCD1, SERT, SERT1, hSERT | solute carrier family 6 (neurotransmitter transporter), member 4 |
C00004631
|
| 6535 | SLC6A8, CCDS1, CRT, CRTR, CT1 | solute carrier family 6 (neurotransmitter transporter), member 8 |
C00004631
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00004631
|
| 8140 | SLC7A5, 4F2LC, CD98, D16S469E, E16, LAT1, MPE16, hLAT1 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
C00004631
|
| 84138 | SLC7A6OS | solute carrier family 7, member 6 opposite strand |
C00004631
|
| 11136 | SLC7A9, BAT1, CSNU3 | solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
C00004631
|
| 9368 | SLC9A3R1, EBP50, NHERF, NHERF-1, NHERF1, NPHLOP2 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
C00004631
|
| 10479 | SLC9A6, MRSA, NHE6 | solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 |
C00004631
|
| 23315 | SLC9A8, NHE-8, NHE8 | solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
C00004631
|
| 28231 | SLCO4A1, OATP-E, OATP1, OATP4A1, OATPE, OATPRP1, POAT, SLC21A12 | solute carrier organic anion transporter family, member 4A1 |
C00004631
|
| 353189 | SLCO4C1, OATP-H, OATP-M1, OATP4C1, OATPX, PRO2176, SLC21A20 | solute carrier organic anion transporter family, member 4C1 |
C00004631
|
| 128710 | SLX4IP, C20orf94, bA204H22.1, bA254M13.1, dJ1099D15.3 | SLX4 interacting protein |
C00004631
|
| 4090 | SMAD5, DWFC, JV5-1, MADH5 | SMAD family member 5 |
C00004631
|
| 4092 | SMAD7, CRCS3, MADH7, MADH8 | SMAD family member 7 |
C00004631
|
| 60682 | SMAP1, SMAP-1 | small ArfGAP 1 |
C00004631
|
| 10592 | SMC2, CAP-E, CAPE, SMC-2, SMC2L1 | structural maintenance of chromosomes 2 |
C00004631
|
| 9126 | SMC3, BAM, BMH, CDLS3, CSPG6, HCAP, SMC3L1 | structural maintenance of chromosomes 3 |
C00004631
|
| 79677 | SMC6, SMC-6, SMC6L1, hSMC6 | structural maintenance of chromosomes 6 |
C00004631
|
| 23049 | SMG1, 61E3.4, ATX, LIP | SMG1 phosphatidylinositol 3-kinase-related kinase (EC:2.7.11.1) |
C00004631
|
| 56006 | SMG9, C19orf61, F17127_1 | SMG9 nonsense mediated mRNA decay factor |
C00004631
|
| 114926 | SMIM19, C8orf40 | small integral membrane protein 19 |
C00004631
|
| 389203 | SMIM20, C4orf52 | small integral membrane protein 20 |
C00004631
|
| 85027 | SMIM3, C5orf62, MST150, NID67 | small integral membrane protein 3 |
C00004631
|
| 54498 | SMOX, C20orf16, PAO, PAO-1, PAOH1, SMO | spermine oxidase (EC:1.5.3.16) |
C00004631
|
| 10924 | SMPDL3A, ASM3A, ASML3a, yR36GH4.1 | sphingomyelin phosphodiesterase, acid-like 3A |
C00004631
|
| 6525 | SMTN | smoothelin |
C00004631
|
| 57154 | SMURF1 | SMAD specific E3 ubiquitin protein ligase 1 |
C00004631
|
| 64750 | SMURF2 | SMAD specific E3 ubiquitin protein ligase 2 |
C00004631
|
| 64754 | SMYD3, KMT3E, ZMYND1, ZNFN3A1, bA74P14.1 | SET and MYND domain containing 3 (EC:2.1.1.43) |
C00004631
|
| 6615 | SNAI1, SLUGH2, SNA, SNAH, SNAIL, SNAIL1, dJ710H13.1 | snail family zinc finger 1 |
C00004631
|
| 6621 | SNAPC4, PTFalpha, SNAP190 | small nuclear RNA activating complex, polypeptide 4, 190kDa |
C00004631
|
| 6622 | SNCA, NACP, PARK1, PARK4, PD1 | synuclein, alpha (non A4 component of amyloid precursor) |
C00004631
|
| 23642 | SNHG1, LINC00057, NCRNA00057, U22HG, UHG | small nucleolar RNA host gene 1 (non-protein coding) |
C00004631
|
| 283596 | SNHG10, C14orf62, LINC00063, NCRNA00063 | small nucleolar RNA host gene 10 (non-protein coding) |
C00004631
|
| 128439 | SNHG11, C20orf198, LINC00101, NCRNA00101 | small nucleolar RNA host gene 11 (non-protein coding) |
C00004631
|
| 100093630 | SNHG8, LINC00060, NCRNA00060 | small nucleolar RNA host gene 8 (non-protein coding) |
C00004631
|
| 684959 | SNORA25, ACA25 | small nucleolar RNA, H/ACA box 25 |
C00004631
|
| 677838 | SNORA61, ACA61 | small nucleolar RNA, H/ACA box 61 |
C00004631
|
| 26780 | SNORA68, RNU68, U68 | small nucleolar RNA, H/ACA box 68 |
C00004631
|
| 6641 | SNTB1, 59-DAP, A1B, BSYN2, DAPA1B, SNT2, SNT2B1, TIP-43 | syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
C00004631
|
| 57231 | SNX14, RGS-PX2 | sorting nexin 14 |
C00004631
|
| 28966 | SNX24, PRO1284 | sorting nexin 24 |
C00004631
|
| 51375 | SNX7 | sorting nexin 7 |
C00004631
|
| 29886 | SNX8, Mvp1 | sorting nexin 8 |
C00004631
|
| 6646 | SOAT1, ACACT, ACAT, ACAT-1, ACAT1, SOAT, STAT | sterol O-acyltransferase 1 (EC:2.3.1.26) |
C00004631
|
| 8435 | SOAT2, ACACT2, ACAT2, ARGP2 | sterol O-acyltransferase 2 (EC:2.3.1.26) |
C00004631
|
| 55084 | SOBP, JXC1, MRAMS | sine oculis binding protein homolog (Drosophila) |
C00004631
|
| 122809 | SOCS4, SOCS7 | suppressor of cytokine signaling 4 |
C00004631
|
| 9306 | SOCS6, CIS-4, CIS4, HSPC060, SOCS-4, SOCS-6, SOCS4, SSI4, STAI4, STATI4 | suppressor of cytokine signaling 6 |
C00004631
|
| 51166 | AADAT, KAT2, KATII | aminoadipate aminotransferase (EC:2.6.1.7 2.6.1.39) |
C00004631
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00004631
|
| 8470 | SORBS2, ARGBP2, PRO0618 | sorbin and SH3 domain containing 2 |
C00004631
|
| 6654 | SOS1, GF1, GGF1, GINGF, HGF, NS4 | son of sevenless homolog 1 (Drosophila) |
C00004631
|
| 11063 | SOX30 | SRY (sex determining region Y)-box 30 |
C00004631
|
| 6659 | SOX4, EVI16 | SRY (sex determining region Y)-box 4 |
C00004631
|
| 6667 | SP1 | Sp1 transcription factor |
C00004631
|
| 389058 | SP5 | Sp5 transcription factor |
C00004631
|
| 53340 | SPA17, CT22, SP17, SP17-1 | sperm autoantigenic protein 17 |
C00004631
|
| 6674 | SPAG1, CILD28, HSD-3.8, SP75, TPIS | sperm associated antigen 1 |
C00004631
|
| 79582 | SPAG16, PF20, WDR29 | sperm associated antigen 16 |
C00004631
|
| 6676 | SPAG4, CT127, SUN4 | sperm associated antigen 4 |
C00004631
|
| 10615 | SPAG5, DEEPEST, MAP126, hMAP126 | sperm associated antigen 5 |
C00004631
|
| 6683 | SPAST, ADPSP, FSP2, SPG4 | spastin (EC:3.6.4.3) |
C00004631
|
| 132671 | SPATA18, Mieap, SPETEX1 | spermatogenesis associated 18 |
C00004631
|
| 124044 | SPATA2L, C16orf76, tamo | spermatogenesis associated 2-like |
C00004631
|
| 124045 | SPATA33, C16orf55 | spermatogenesis associated 33 |
C00004631
|
| 57405 | SPC25, SPBC25, hSpc25 | SPC25, NDC80 kinetochore complex component |
C00004631
|
| 169981 | SPIN3, SPIN-3, bA445O16.1 | spindlin family, member 3 |
C00004631
|
| 139886 | SPIN4 | spindlin family, member 4 |
C00004631
|
| 6690 | SPINK1, PCTT, PSTI, Spink3, TATI, TCP | serine peptidase inhibitor, Kazal type 1 |
C00004631
|
| 90853 | SPOCD1 | SPOC domain containing 1 |
C00004631
|
| 6696 | SPP1, BNSP, BSPI, ETA-1, OPN | secreted phosphoprotein 1 |
C00004631
|
| 83932 | SPRTN, C1orf124, DDDL1880, DVC1, PRO4323, Spartan, dJ876B10.3 | SprT-like N-terminal domain |
C00004631
|
| 10252 | SPRY1, hSPRY1 | sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
C00004631
|
| 80176 | SPSB1, SSB-1, SSB1 | splA/ryanodine receptor domain and SOCS box containing 1 |
C00004631
|
| 6711 | SPTBN1, ELF, SPTB2, betaSpII | spectrin, beta, non-erythrocytic 1 |
C00004631
|
| 55304 | SPTLC3, C20orf38, LCB2B, LCB3, SPT3, SPTLC2L, dJ718P11, dJ718P11.1, hLCB2b | serine palmitoyltransferase, long chain base subunit 3 (EC:2.3.1.50) |
C00004631
|
| 6713 | SQLE | squalene epoxidase (EC:1.14.13.132) |
C00004631
|
| 58472 | SQRDL, PRO1975 | sulfide quinone reductase-like (yeast) |
C00004631
|
| 8878 | SQSTM1, A170, OSIL, PDB3, ZIP3, p60, p62, p62B | sequestosome 1 |
C00004631
|
| 6720 | SREBF1, SREBP-1c, SREBP1, bHLHd1 | sterol regulatory element binding transcription factor 1 |
C00004631
|
| 9901 | SRGAP3, ARHGAP14, MEGAP, SRGAP2, WRP | SLIT-ROBO Rho GTPase activating protein 3 |
C00004631
|
| 8406 | SRPX, DRS, ETX1, SRPX1 | sushi-repeat containing protein, X-linked |
C00004631
|
| 27286 | SRPX2, BPP, CBPS, PMGX, RESDX, SRPUL | sushi-repeat containing protein, X-linked 2 |
C00004631
|
| 10929 | SRSF8, DSM-1, SFRS2B, SRP46 | serine/arginine-rich splicing factor 8 |
C00004631
|
| 6742 | SSBP1, Mt-SSB, SOSS-B1, SSBP, mtSSB | single-stranded DNA binding protein 1, mitochondrial |
C00004631
|
| 6747 | SSR3, TRAPG | signal sequence receptor, gamma (translocon-associated protein gamma) |
C00004631
|
| 51066 | SSUH2, C3orf32, SSU-2, fls485 | ssu-2 homolog (C. elegans) |
C00004631
|
| 6480 | ST6GAL1, SIAT1, ST6GalI, ST6N | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (EC:2.4.99.1) |
C00004631
|
| 54879 | ST7L, FAM4B, ST7R, STLR | suppression of tumorigenicity 7 like |
C00004631
|
| 10274 | STAG1, SA1, SCC3A | stromal antigen 1 |
C00004631
|
| 90627 | STARD13, ARHGAP37, DLC2, GT650, LINC00464 | StAR-related lipid transfer (START) domain containing 13 |
C00004631
|
| 57519 | STARD9 | StAR-related lipid transfer (START) domain containing 9 |
C00004631
|
| 6772 | STAT1, CANDF7, ISGF-3, STAT91 | signal transducer and activator of transcription 1, 91kDa |
C00004631
|
| 27067 | STAU2, 39K2, 39K3 | staufen double-stranded RNA binding protein 2 |
C00004631
|
| 8987 | STBD1, GENEX3414 | starch binding domain 1 |
C00004631
|
| 6781 | STC1, STC | stanniocalcin 1 |
C00004631
|
| 8614 | STC2, STC-2, STCRP | stanniocalcin 2 |
C00004631
|
| 26872 | STEAP1, PRSS24, STEAP | six transmembrane epithelial antigen of the prostate 1 |
C00004631
|
| 261729 | STEAP2, IPCA1, PCANAP1, PUMPCn, STAMP1, STMP | STEAP family member 2, metalloreductase |
C00004631
|
| 6491 | STIL, MCPH7, SIL | SCL/TAL1 interrupting locus |
C00004631
|
| 57620 | STIM2 | stromal interaction molecule 2 |
C00004631
|
| 8576 | STK16, KRCT, MPSK, PKL12, TSF1 | serine/threonine kinase 16 (EC:2.7.11.1 2.7.10.2) |
C00004631
|
| 9263 | STK17A, DRAK1 | serine/threonine kinase 17a (EC:2.7.11.1) |
C00004631
|
| 9262 | STK17B, DRAK2 | serine/threonine kinase 17b (EC:2.7.11.1) |
C00004631
|
| 11329 | STK38, NDR, NDR1 | serine/threonine kinase 38 (EC:2.7.11.1) |
C00004631
|
| 3925 | STMN1, C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | stathmin 1 |
C00004631
|
| 201254 | STRA13, CENP-X, CENPX, D9, FAAP10, MHF2 | stimulated by retinoic acid 13 |
C00004631
|
| 55342 | STRBP, ILF3L, SPNR, p74 | spermatid perinuclear RNA binding protein |
C00004631
|
| 8675 | STX16, SYN16 | syntaxin 16 |
C00004631
|
| 6804 | STX1A, HPC-1, P35-1, STX1, SYN1A | syntaxin 1A (brain) |
C00004631
|
| 6809 | STX3, STX3A | syntaxin 3 |
C00004631
|
| 6811 | STX5, SED5, STX5A | syntaxin 5 |
C00004631
|
| 134957 | STXBP5, LGL3, LLGL3, Nbla04300 | syntaxin binding protein 5 (tomosyn) |
C00004631
|
| 6815 | STYX | serine/threonine/tyrosine interacting protein |
C00004631
|
| 10923 | SUB1, P15, PC4, p14 | SUB1 homolog (S. cerevisiae) |
C00004631
|
| 8801 | SUCLG2, GBETA | succinate-CoA ligase, GDP-forming, beta subunit (EC:6.2.1.4) |
C00004631
|
| 55959 | SULF2, HSULF-2 | sulfatase 2 |
C00004631
|
| 29974 | A1CF, ACF, ACF64, ACF65, APOBEC1CF, ASP | APOBEC1 complementation factor |
C00004631
|
| 6799 | SULT1A2, HAST4, P-PST, ST1A2, STP2, TSPST2 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (EC:2.8.2.1) |
C00004631
|
| 6818 | SULT1A3, HAST, HAST3, M-PST, ST1A3/ST1A4, ST1A5, STM, TL-PST | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (EC:2.8.2.1) |
C00004631
|
| 20888 |
C00004631
|
||
| 6819 | SULT1C2, ST1C1, ST1C2, SULT1C1, humSULTC2 | sulfotransferase family, cytosolic, 1C, member 2 (EC:2.8.2.-) |
C00004631
|
| 1 | A1BG, A1B, ABG, GAB, HYST2477 | alpha-1-B glycoprotein |
C00004631
|
| 6822 | SULT2A1, DHEA-ST, DHEAS, HST, ST2, ST2A1, ST2A3, STD, hSTa | sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 (EC:2.8.2.14) |
C00004631
|
| 6821 | SUOX | sulfite oxidase (EC:1.8.3.1) |
C00004631
|
| 203328 | SUSD3 | sushi domain containing 3 |
C00004631
|
| 258010 | SVIP | small VCP/p97-interacting protein |
C00004631
|
| 256126 | SYCE2, CESC1 | synaptonemal complex central element protein 2 |
C00004631
|
| 81493 | SYNC, SYNC1, SYNCOILIN | syncoilin, intermediate filament protein |
C00004631
|
| 163183 | SYNE4, C19orf46, Nesp4 | spectrin repeat containing, nuclear envelope family member 4 |
C00004631
|
| 9143 | SYNGR3 | synaptogyrin 3 |
C00004631
|
| 84958 | SYTL1, JFC1, SLP1 | synaptotagmin-like 1 |
C00004631
|
| 94121 | SYTL4, SLP4 | synaptotagmin-like 4 |
C00004631
|
| 255061 | TAC4, EK, HK-1, HK1, PPT-C | tachykinin 4 (hemokinin) |
C00004631
|
| 769871 |
C00004631
|
||
| 9015 | TAF1A, MGC:17061, RAFI48, SL1, TAFI48 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
C00004631
|
| 83860 | TAF3, TAF140, TAFII-140, TAFII140 | TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
C00004631
|
| 6877 | TAF5, TAF2D, TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
C00004631
|
| 27097 | TAF5L, PAF65B | TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
C00004631
|
| 6880 | TAF9, MGC:5067, STAF31/32, TAF2G, TAFII-31, TAFII-32, TAFII31, TAFII32, TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa (EC:2.7.4.3) |
C00004631
|
| 51616 | TAF9B, DN-7, DN7, TAF9L, TAFII31L, TFIID-31 | TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
C00004631
|
| 6876 | TAGLN, SM22, SMCC, TAGLN1, WS3-10 | transgelin |
C00004631
|
| 6888 | TALDO1, TAL, TAL-H, TALDOR, TALH | transaldolase 1 (EC:2.2.1.2) |
C00004631
|
| 128989 | TANGO2, C22orf25 | transport and golgi organization 2 homolog (Drosophila) |
C00004631
|
| 51347 | TAOK3, DPK, JIK, MAP3K18 | TAO kinase 3 (EC:2.7.11.1) |
C00004631
|
| 6890 | TAP1, ABC17, ABCB2, APT1, D6S114E, PSF-1, PSF1, RING4, TAP1*0102N, TAP1N | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
C00004631
|
| 6894 | TARBP1, TRM3, TRP-185, TRP185 | TAR (HIV-1) RNA binding protein 1 |
C00004631
|
| 51256 | TBC1D7, PIG51, TBC7, dJ257A7.3 | TBC1 domain family, member 7 |
C00004631
|
| 11138 | TBC1D8, AD3, HBLP1, TBC1D8A, VRP | TBC1 domain family, member 8 (with GRAM domain) |
C00004631
|
| 54885 | TBC1D8B | TBC1 domain family, member 8B (with GRAM domain) |
C00004631
|
| 23158 | TBC1D9, MDR1 | TBC1 domain family, member 9 (with GRAM domain) |
C00004631
|
| 93627 | TBCK, TBCKL | TBC1 domain containing kinase |
C00004631
|
| 7352 | UCP3, SLC25A9 | uncoupling protein 3 (mitochondrial, proton carrier) |
C00004565
|
| 6926 | TBX3, TBX3-ISO, UMS, XHL | T-box 3 |
C00004631
|
| 90843 | TCEAL8 | transcription elongation factor A (SII)-like 8 |
C00004631
|
| 6923 | TCEB2, ELOB, SIII | transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
C00004631
|
| 6938 | TCF12, CRS3, HEB, HTF4, HsT17266, bHLHb20 | transcription factor 12 |
C00004631
|
| 6941 | TCF19, SC1, TCF-19 | transcription factor 19 |
C00004631
|
| 6925 | TCF4, E2-2, ITF-2, ITF2, PTHS, SEF-2, SEF2, SEF2-1, SEF2-1A, SEF2-1B, TCF-4, bHLHb19 | transcription factor 4 |
C00004631
|
| 6934 | TCF7L2, TCF-4, TCF4 | transcription factor 7-like 2 (T-cell specific, HMG-box) |
C00004631
|
| 55346 | TCP11L1, dJ85M6.3 | t-complex 11, testis-specific-like 1 |
C00004631
|
| 6988 | TCTA | T-cell leukemia translocation altered |
C00004631
|
| 255758 | TCTEX1D2 | Tctex1 domain containing 2 |
C00004631
|
| 51567 | TDP2, AD022, EAP2, EAPII, TTRAP, dJ30M3.3, hTDP2 | tyrosyl-DNA phosphodiesterase 2 (EC:3.1.4.-) |
C00004631
|
| 81550 | TDRD3 | tudor domain containing 3 |
C00004631
|
| 23424 | TDRD7, CATC4, CTRCT36, PCTAIRE2BP, RP11-508D10.1, TRAP | tudor domain containing 7 |
C00004631
|
| 79736 | TEFM, C17orf42 | transcription elongation factor, mitochondrial |
C00004631
|
| 7011 | TEP1, TLP1, TP1, TROVE1, VAULT2, p240 | telomerase-associated protein 1 |
C00004631
|
| 7015 | TERT, CMM9, DKCA2, DKCB4, EST2, PFBMFT1, TCS1, TP2, TRT, hEST2, hTRT | telomerase reverse transcriptase (EC:2.7.7.49) |
C00004631
|
| 80312 | TET1, CXXC6, LCX, bA119F7.1 | tet methylcytosine dioxygenase 1 (EC:1.14.11.n2) |
C00004631
|
| 400629 | TEX19 | testis expressed 19 |
C00004631
|
| 374618 | TEX9 | testis expressed 9 |
C00004631
|
| 7018 | TF, PRO1557, PRO2086, TFQTL1 | transferrin |
C00004631
|
| 51106 | TFB1M, CGI75, mtTFB, mtTFB1 | transcription factor B1, mitochondrial (EC:2.1.1.-) |
C00004631
|
| 7029 | TFDP2, DP2 | transcription factor Dp-2 (E2F dimerization partner 2) |
C00004631
|
| 7032 | TFF2, SML1, SP | trefoil factor 2 |
C00004631
|
| 7035 | TFPI, EPI, LACI, TFI, TFPI1 | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
C00004631
|
| 7980 | TFPI2, PP5, REF1, TFPI-2 | tissue factor pathway inhibitor 2 |
C00004631
|
| 7036 | TFR2, HFE3, TFRC2 | transferrin receptor 2 |
C00004631
|
| 7037 | TFRC, CD71, T9, TFR, TFR1, TR, TRFR, p90 | transferrin receptor |
C00004631
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00004631
|
| 7041 | TGFB1I1, ARA55, HIC-5, HIC5, TSC-5 | transforming growth factor beta 1 induced transcript 1 |
C00004631
|
| 55331 | ACER3, APHC, PHCA | alkaline ceramidase 3 (EC:3.5.1.23) |
C00004631
|
| 60436 | TGIF2 | TGFB-induced factor homeobox 2 |
C00004631
|
| 56906 | THAP10 | THAP domain containing 10 |
C00004631
|
| 83591 | THAP2 | THAP domain containing, apoptosis associated protein 2 |
C00004631
|
| 7057 | THBS1, THBS, THBS-1, TSP, TSP-1, TSP1 | thrombospondin 1 |
C00004631
|
| 7058 | THBS2, TSP2 | thrombospondin 2 |
C00004631
|
| 100286461 |
C00004631
|
||
| 79896 | THNSL1, TSH1 | threonine synthase-like 1 (S. cerevisiae) |
C00004631
|
| 9984 | THOC1, HPR1, P84, P84N5 | THO complex 1 |
C00004631
|
| 29087 | THYN1, MDS012, MY105, THY28, THY28KD | thymocyte nuclear protein 1 |
C00004631
|
| 7072 | TIA1, TIA-1, WDM | TIA1 cytotoxic granule-associated RNA binding protein |
C00004631
|
| 148022 | TICAM1, IIAE6, MyD88-3, PRVTIRB, TICAM-1, TRIF | toll-like receptor adaptor molecule 1 |
C00004631
|
| 353376 | TICAM2, MyD88-4, TICAM-2, TIRAP3, TIRP, TRAM | toll-like receptor adaptor molecule 2 |
C00004631
|
| 92610 | TIFA, T2BP, T6BP, TIFAA | TRAF-interacting protein with forkhead-associated domain |
C00004631
|
| 200765 | TIGD1, EEYORE | tigger transposable element derived 1 |
C00004631
|
| 166815 | TIGD2 | tigger transposable element derived 2 |
C00004631
|
| 8914 | TIMELESS, TIM, TIM1, hTIM | timeless circadian clock |
C00004631
|
| 26515 | TIMM10B, FXC1, TIM10B, Tim9b | translocase of inner mitochondrial membrane 10 homolog B (yeast) |
C00004631
|
| 1678 | TIMM8A, DDP, DDP1, DFN1, MTS, TIM8 | translocase of inner mitochondrial membrane 8 homolog A (yeast) |
C00004631
|
| 7076 | TIMP1, CLGI, EPA, EPO, HCI, TIMP | TIMP metallopeptidase inhibitor 1 |
C00004631
|
| 7077 | TIMP2, CSC-21K, DDC8 | TIMP metallopeptidase inhibitor 2 |
C00004631
|
| 54962 | TIPIN | TIMELESS interacting protein |
C00004631
|
| 7082 | TJP1, ZO-1 | tight junction protein 1 |
C00004631
|
| 9414 | TJP2, C9DUPq21.11, DFNA51, DUP9q21.11, X104, ZO2 | tight junction protein 2 |
C00004631
|
| 7086 | TKT, TK, TKT1 | transketolase (EC:2.2.1.1) |
C00004631
|
| 9874 | TLK1, PKU-beta | tousled-like kinase 1 (EC:2.7.11.1) |
C00004631
|
| 7099 | TLR4, ARMD10, CD284, TLR-4, TOLL | toll-like receptor 4 |
C00004631
|
| 80213 | TM2D3, BLP2 | TM2 domain containing 3 |
C00004631
|
| 4071 | TM4SF1, H-L6, L6, M3S1, TAAL6 | transmembrane 4 L six family member 1 |
C00004631
|
| 51768 | TM7SF3 | transmembrane 7 superfamily member 3 |
C00004631
|
| 51643 | TMBIM4, GAAP, LFG4, S1R, ZPRO | transmembrane BAX inhibitor motif containing 4 |
C00004631
|
| 79905 | TMC7 | transmembrane channel-like 7 |
C00004631
|
| 23023 | TMCC1 | transmembrane and coiled-coil domain family 1 |
C00004631
|
| 146456 | TMED6, PRO34237, SPLL9146 | transmembrane emp24 protein transport domain containing 6 |
C00004631
|
| 8577 | TMEFF1, C9orf2, CT120.1, H7365, TR-1 | transmembrane protein with EGF-like and two follistatin-like domains 1 |
C00004631
|
| 113277 | TMEM106A | transmembrane protein 106A |
C00004631
|
| 84216 | TMEM117 | transmembrane protein 117 |
C00004631
|
| 84233 | TMEM126A, OPA7 | transmembrane protein 126A |
C00004631
|
| 55863 | TMEM126B | transmembrane protein 126B |
C00004631
|
| 85013 | TMEM128 | transmembrane protein 128 |
C00004631
|
| 65084 | TMEM135, PMP52 | transmembrane protein 135 |
C00004631
|
| 51524 | TMEM138 | transmembrane protein 138 |
C00004631
|
| 135932 | TMEM139 | transmembrane protein 139 |
C00004631
|
| 55281 | TMEM140 | transmembrane protein 140 |
C00004631
|
| 28978 | TMEM14A, C6orf73 | transmembrane protein 14A |
C00004631
|
| 80008 | TMEM156 | transmembrane protein 156 |
C00004631
|
| 57146 | TMEM159, PROMETHIN | transmembrane protein 159 |
C00004631
|
| 55858 | TMEM165, CDG2K, FT27, GDT1, TMPT27, TPARL | transmembrane protein 165 |
C00004631
|
| 64418 | TMEM168 | transmembrane protein 168 |
C00004631
|
| 100113407 | TMEM170B | transmembrane protein 170B |
C00004631
|
| 68027 |
C00004631
|
||
| 8269 | TMEM187, CXorf12, DXS9878E, ITBA1 | transmembrane protein 187 |
C00004631
|
| 55266 | TMEM19 | transmembrane protein 19 |
C00004631
|
| 201931 | TMEM192 | transmembrane protein 192 |
C00004631
|
| 23306 | TMEM194A, TMEM194 | transmembrane protein 194A |
C00004631
|
| 23670 | TMEM2 | transmembrane protein 2 |
C00004631
|
| 84065 | TMEM222, C1orf160 | transmembrane protein 222 |
C00004631
|
| 55026 | TMEM255A, FAM70A | transmembrane protein 255A |
C00004631
|
| 57393 | TMEM27, NX-17, NX17 | transmembrane protein 27 |
C00004631
|
| 440026 | TMEM41B | transmembrane protein 41B |
C00004631
|
| 55076 | TMEM45A, DERP7 | transmembrane protein 45A |
C00004631
|
| 757 | TMEM50B, C21orf4, HCVP7TP3 | transmembrane protein 50B |
C00004631
|
| 113452 | TMEM54, BCLP, CAC-1, CAC1 | transmembrane protein 54 |
C00004631
|
| 85025 | TMEM60, C7orf35 | transmembrane protein 60 |
C00004631
|
| 169200 | TMEM64 | transmembrane protein 64 |
C00004631
|
| 157378 | TMEM65 | transmembrane protein 65 |
C00004631
|
| 91147 | TMEM67, JBTS6, MECKELIN, MKS3, NPHP11, TNEM67 | transmembrane protein 67 |
C00004631
|
| 137695 | TMEM68 | transmembrane protein 68 |
C00004631
|
| 255043 | TMEM86B | transmembrane protein 86B (EC:3.3.2.2 3.3.2.5) |
C00004631
|
| 56674 | TMEM9B, C11orf15 | TMEM9 domain family, member B |
C00004631
|
| 164656 | TMPRSS6, IRIDA | transmembrane protease, serine 6 |
C00004631
|
| 9168 | TMSB10, MIG12, TB10 | thymosin beta 10 |
C00004631
|
| 54495 | TMX3, PDIA13, TXNDC10 | thioredoxin-related transmembrane protein 3 (EC:5.3.4.1) |
C00004631
|
| 25816 | TNFAIP8, GG2-1, MDC-3.13, NDED, SCC-S2, SCCS2 | tumor necrosis factor, alpha-induced protein 8 |
C00004631
|
| 8797 | TNFRSF10A, APO2, CD261, DR4, TRAILR-1, TRAILR1 | tumor necrosis factor receptor superfamily, member 10a |
C00004631
|
| 8795 | TNFRSF10B, CD262, DR5, KILLER, KILLER/DR5, TRAIL-R2, TRAILR2, TRICK2, TRICK2A, TRICK2B, TRICKB, ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b |
C00004631
|
| 8793 | TNFRSF10D, CD264, DCR2, TRAIL-R4, TRAILR4, TRUNDD | tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
C00004631
|
| 8792 | TNFRSF11A, CD265, FEO, LOH18CR1, ODFR, OFE, OPTB7, OSTS, PDB2, RANK, TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
C00004631
|
| 4982 | TNFRSF11B, OCIF, OPG, TR1 | tumor necrosis factor receptor superfamily, member 11b |
C00004631
|
| 51330 | TNFRSF12A, CD266, FN14, TWEAKR | tumor necrosis factor receptor superfamily, member 12A |
C00004631
|
| 8764 | TNFRSF14, ATAR, CD270, HVEA, HVEM, LIGHTR, TR2 | tumor necrosis factor receptor superfamily, member 14 |
C00004631
|
| 55504 | TNFRSF19, TAJ, TAJ-alpha, TRADE, TROY | tumor necrosis factor receptor superfamily, member 19 |
C00004631
|
| 7133 | TNFRSF1B, CD120b, TBPII, TNF-R-II, TNF-R75, TNFBR, TNFR1B, TNFR2, TNFR80, p75, p75TNFR | tumor necrosis factor receptor superfamily, member 1B |
C00004631
|
| 27242 | TNFRSF21, BM-018, CD358, DR6 | tumor necrosis factor receptor superfamily, member 21 |
C00004631
|
| 7019 | TFAM, MTTF1, MTTFA, TCF6, TCF6L1, TCF6L2, TCF6L3 | transcription factor A, mitochondrial |
C00004565
|
| 7292 | TNFSF4, CD134L, CD252, GP34, OX-40L, OX4OL, TXGP1 | tumor necrosis factor (ligand) superfamily, member 4 |
C00004631
|
| 8658 | TNKS, ARTD5, PARP-5a, PARP5A, PARPL, TIN1, TINF1, TNKS1, pART5 | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase (EC:2.4.2.30) |
C00004631
|
| 7134 | TNNC1, CMD1Z, CMH13, TN-C, TNC, TNNC | troponin C type 1 (slow) |
C00004631
|
| 7136 | TNNI2, AMCD2B, DA2B, FSSV, fsTnI | troponin I type 2 (skeletal, fast) |
C00004631
|
| 7145 | TNS1, MST091, MST122, MST127, MSTP122, MSTP127, MXRA6, TNS | tensin 1 |
C00004631
|
| 56993 | TOMM22, 1C9-2, MST065, MSTP065, TOM22 | translocase of outer mitochondrial membrane 22 homolog (yeast) |
C00004631
|
| 4796 | TONSL, IKBR, NFKBIL2 | tonsoku-like, DNA repair protein |
C00004631
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00004631
|
| 7155 | TOP2B, TOPIIB, top2beta | topoisomerase (DNA) II beta 180kDa (EC:5.99.1.3) |
C00004631
|
| 26092 | TOR1AIP1, LAP1, LAP1B | torsin A interacting protein 1 |
C00004631
|
| 27433 | TOR2A, TORP1 | torsin family 2, member A |
C00004631
|
| 7159 | TP53BP2, 53BP2, ASPP2, BBP, P53BP2, PPP1R13A | tumor protein p53 binding protein, 2 |
C00004631
|
| 94241 | TP53INP1, SIP, TP53DINP1, TP53INP1A, TP53INP1B, Teap, p53DINP1 | tumor protein p53 inducible nuclear protein 1 |
C00004631
|
| 112858 | TP53RK, BUD32, C20orf64, Nori-2, Nori-2p, PRPK, dJ101A2 | TP53 regulating kinase (EC:2.7.11.1) |
C00004631
|
| 11257 | TP53TG1, LINC00096, NCRNA00096, P53TG1, P53TG1-D, TP53AP1 | TP53 target 1 (non-protein coding) |
C00004631
|
| 7162 | TPBG, 5T4, 5T4AG, M6P1 | trophoblast glycoprotein |
C00004631
|
| 43582 |
C00004631
|
||
| 7167 | TPI1, TIM, TPI, TPID | triosephosphate isomerase 1 (EC:5.3.1.1) |
C00004631
|
| 7168 | TPM1, C15orf13, CMD1Y, CMH3, HTM-alpha, LVNC9, TMSA | tropomyosin 1 (alpha) |
C00004631
|
| 7171 | TPM4 | tropomyosin 4 |
C00004631
|
| 1200 | TPP1, CLN2, LPIC, TPP-1 | tripeptidyl peptidase I (EC:3.4.14.9) |
C00004631
|
| 8459 | TPST2, TANGO13B | tyrosylprotein sulfotransferase 2 (EC:2.8.2.20) |
C00004631
|
| 29896 | TRA2A, AWMS1, HSU53209 | transformer 2 alpha homolog (Drosophila) |
C00004631
|
| 8717 | TRADD, Hs.89862 | TNFRSF1A-associated via death domain |
C00004631
|
| 9618 | TRAF4, CART1, MLN62, RNF83 | TNF receptor-associated factor 4 |
C00004631
|
| 133022 | TRAM1L1 | translocation associated membrane protein 1-like 1 |
C00004631
|
| 7109 | TRAPPC10, EHOC-1, EHOC1, GT334, TMEM1, TRS130, TRS30 | trafficking protein particle complex 10 |
C00004631
|
| 60684 | TRAPPC11, C4orf41, FOIGR, GRY, LGMD2S | trafficking protein particle complex 11 |
C00004631
|
| 6399 | TRAPPC2, MIP2A, SEDL, SEDT, TRAPPC2P1, TRS20, ZNF547L, hYP38334 | trafficking protein particle complex 2 |
C00004631
|
| 79090 | TRAPPC6A, TRS33 | trafficking protein particle complex 6A |
C00004631
|
| 54209 | TREM2, TREM-2, Trem2a, Trem2b, Trem2c | triggering receptor expressed on myeloid cells 2 |
C00004631
|
| 51499 | TRIAP1, MDM35, P53CSV, WF-1 | TP53 regulated inhibitor of apoptosis 1 |
C00004631
|
| 57761 | TRIB3, C20orf97, NIPK, SINK, SKIP3, TRB3 | tribbles pseudokinase 3 |
C00004631
|
| 81559 | TRIM11, BIA1, RNF92 | tripartite motif containing 11 |
C00004631
|
| 6737 | TRIM21, RNF81, RO52, SSA, SSA1 | tripartite motif containing 21 |
C00004631
|
| 10346 | TRIM22, GPSTAF50, RNF94, STAF50 | tripartite motif containing 22 |
C00004631
|
| 8805 | TRIM24, PTC6, RNF82, TF1A, TIF1, TIF1A, TIF1ALPHA, hTIF1 | tripartite motif containing 24 |
C00004631
|
| 10475 | TRIM38, RNF15, RORET | tripartite motif containing 38 |
C00004631
|
| 89122 | TRIM4, RNF87 | tripartite motif containing 4 |
C00004631
|
| 91107 | TRIM47, GOA, RNF100 | tripartite motif containing 47 |
C00004631
|
| 84851 | TRIM52, RNF102 | tripartite motif containing 52 |
C00004631
|
| 286827 | TRIM59, MRF1, RNF104, TRIM57, TSBF1 | tripartite motif containing 59 |
C00004631
|
| 11078 | TRIOBP, DFNB28, TARA, dJ37E16.4 | TRIO and F-actin binding protein |
C00004631
|
| 9320 | TRIP12, TRIP-12, ULF | thyroid hormone receptor interactor 12 |
C00004631
|
| 9319 | TRIP13, 16E1BP | thyroid hormone receptor interactor 13 |
C00004631
|
| 60487 | TRMT11, C6orf75, MDS024, TRM11, TRMT11-1 | tRNA methyltransferase 11 homolog (S. cerevisiae) (EC:2.1.1.-) |
C00004631
|
| 55006 | TRMT61B | tRNA methyltransferase 61 homolog B (S. cerevisiae) (EC:2.1.1.220) |
C00004631
|
| 55687 | TRMU, MTO2, MTU1, TRMT, TRMT1, TRNT1 | tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (EC:2.1.1.61) |
C00004631
|
| 7442 | TRPV1, VR1 | transient receptor potential cation channel, subfamily V, member 1 |
C00004631
|
| 51393 | TRPV2, VRL, VRL-1, VRL1 | transient receptor potential cation channel, subfamily V, member 2 |
C00004631
|
| 8295 | TRRAP, PAF350/400, PAF400, STAF40, TR-AP, Tra1 | transformation/transcription domain-associated protein |
C00004631
|
| 7248 | TSC1, LAM, TSC | tuberous sclerosis 1 |
C00004631
|
| 7249 | TSC2, LAM, TSC4 | tuberous sclerosis 2 |
C00004631
|
| 116461 | TSEN15, C1orf19, sen15 | TSEN15 tRNA splicing endonuclease subunit |
C00004631
|
| 10102 | TSFM, EFTS, EFTSMT | Ts translation elongation factor, mitochondrial |
C00004631
|
| 7251 | TSG101, TSG10, VPS23 | tumor susceptibility 101 |
C00004631
|
| 10194 | TSHZ1, CAA, NY-CO-33, SDCCAG33, TSH1 | teashirt zinc finger homeobox 1 |
C00004631
|
| 10099 | TSPAN3, TM4-A, TM4SF8, TSPAN-3 | tetraspanin 3 |
C00004631
|
| 7106 | TSPAN4, NAG-2, NAG2, TETRASPAN, TM4SF7, TSPAN-4 | tetraspanin 4 |
C00004631
|
| 10098 | TSPAN5, NET-4, NET4, TM4SF9, TSPAN-5 | tetraspanin 5 |
C00004631
|
| 7103 | TSPAN8, CO-029, TM4SF3 | tetraspanin 8 |
C00004631
|
| 64061 | TSPYL2, CDA1, CINAP, CTCL, DENTT, HRIHFB2216, NP79, SE204, TSPX | TSPY-like 2 |
C00004631
|
| 23270 | TSPYL4, dJ486I3.2 | TSPY-like 4 |
C00004631
|
| 283629 | TSSK4, C14orf20, STK22E, TSK-4, TSK4, TSSK5 | testis-specific serine kinase 4 (EC:2.7.11.1) |
C00004631
|
| 7263 | TST, RDS | thiosulfate sulfurtransferase (rhodanese) (EC:2.8.1.1) |
C00004631
|
| 151613 | TTC14, DRDL5813, PRO19630 | tetratricopeptide repeat domain 14 |
C00004631
|
| 55761 | TTC17 | tetratricopeptide repeat domain 17 |
C00004631
|
| 55622 | TTC27 | tetratricopeptide repeat domain 27 |
C00004631
|
| 150737 | TTC30B, IFT70, fleer | tetratricopeptide repeat domain 30B |
C00004631
|
| 23548 | TTC33, OSRF | tetratricopeptide repeat domain 33 |
C00004631
|
| 125488 | TTC39C, C18orf17, HsT2697 | tetratricopeptide repeat domain 39C |
C00004631
|
| 145567 | TTC7B, TTC7L1, c14_5685 | tetratricopeptide repeat domain 7B |
C00004631
|
| 7272 | TTK, CT96, ESK, MPH1, MPS1, MPS1L1, PYT | TTK protein kinase (EC:2.7.12.1) |
C00004631
|
| 26140 | TTLL3, HOTTL | tubulin tyrosine ligase-like family, member 3 |
C00004631
|
| 7276 | TTR, CTS, CTS1, HsT2651, PALB, TBPA | transthyretin |
C00004631
|
| 80727 | TTYH3 | tweety family member 3 |
C00004631
|
| 7846 | TUBA1A, B-ALPHA-1, LIS3, TUBA3 | tubulin, alpha 1a |
C00004631
|
| 203068 | TUBB, M40, OK/SW-cl.56, TUBB1, TUBB5 | tubulin, beta class I |
C00004631
|
| 81027 | TUBB1 | tubulin, beta 1 class VI |
C00004631
|
| 347733 | TUBB2B, PMGYSA, bA506K6.1 | tubulin, beta 2B class IIb |
C00004631
|
| 84617 | TUBB6, HsT1601, TUBB-5 | tubulin, beta 6 class V |
C00004631
|
| 643224 | TUBBP5 | tubulin, beta pseudogene 5 |
C00004631
|
| 51174 | TUBD1, TUBD | tubulin, delta 1 |
C00004631
|
| 10426 | TUBGCP3, 104p, GCP3, Grip104, SPBC98, Spc98, Spc98p | tubulin, gamma complex associated protein 3 |
C00004631
|
| 85378 | TUBGCP6, GCP-6, GCP6, MCCRP, MCPHCR | tubulin, gamma complex associated protein 6 |
C00004631
|
| 7284 | TUFM, COXPD4, EF-TuMT, EFTU, P43 | Tu translation elongation factor, mitochondrial |
C00004631
|
| 7286 | TUFT1 | tuftelin 1 |
C00004631
|
| 7991 | TUSC3, D8S1992, M33, MRT22, MRT7, N33, OST3A | tumor suppressor candidate 3 |
C00004631
|
| 200081 | TXLNA, IL14, RP4-622L5.4, TXLN | taxilin alpha |
C00004631
|
| 79770 | TXNDC15, C5orf14, UNQ335 | thioredoxin domain containing 15 |
C00004631
|
| 57544 | TXNDC16, ERp90, KIAA1344 | thioredoxin domain containing 16 |
C00004631
|
| 10628 | TXNIP, EST01027, HHCPA78, THIF, VDUP1 | thioredoxin interacting protein |
C00004631
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00004631
|
| 7298 | TYMS, HST422, TMS, TS | thymidylate synthetase (EC:2.1.1.45) |
C00004631
|
| 7299 | TYR, ATN, CMM8, OCA1, OCA1A, OCAIA, SHEP3 | tyrosinase (EC:1.14.18.1) |
C00004631
|
| 127253 | TYW3, C1orf171 | tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) (EC:2.1.1.282) |
C00004631
|
| 91373 | UAP1L1, RP11-229P13.18 | UDP-N-acetylglucosamine pyrophosphorylase 1-like 1 |
C00004631
|
| 9039 | UBA3, NAE2, UBE1C, hUBA3 | ubiquitin-like modifier activating enzyme 3 |
C00004631
|
| 79876 | UBA5, THIFP1, UBE1DC1 | ubiquitin-like modifier activating enzyme 5 |
C00004631
|
| 390595 | UBAP1L, UBAP-1L | ubiquitin associated protein 1-like |
C00004631
|
| 55833 | UBAP2, UBAP-2 | ubiquitin associated protein 2 |
C00004631
|
| 84959 | UBASH3B, STS-1, STS1, TULA-2, p70 | ubiquitin associated and SH3 domain containing B (EC:3.1.3.48) |
C00004631
|
| 10537 | UBD, FAT10, GABBR1, UBD-3 | ubiquitin D |
C00004631
|
| 70348 |
C00004631
|
||
| 7325 | UBE2E2, UBCH8 | ubiquitin-conjugating enzyme E2E 2 (EC:6.3.2.19) |
C00004631
|
| 7332 | UBE2L3, E2-F1, L-UBC, UBCH7, UbcM4 | ubiquitin-conjugating enzyme E2L 3 (EC:6.3.2.19) |
C00004631
|
| 92912 | UBE2Q2 | ubiquitin-conjugating enzyme E2Q family member 2 (EC:6.3.2.19) |
C00004631
|
| 89910 | UBE3B, BPIDS | ubiquitin protein ligase E3B (EC:6.3.2.-) |
C00004631
|
| 5412 | UBL3, HCG-1, PNSC1 | ubiquitin-like 3 |
C00004631
|
| 23352 | UBR4, RBAF600, ZUBR1, p600 | ubiquitin protein ligase E3 component n-recognin 4 |
C00004631
|
| 7347 | UCHL3, UCH-L3 | ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) (EC:3.4.19.12) |
C00004631
|
| 7349 | UCN, UI, UROC | urocortin |
C00004631
|
| 7351 | UCP2, BMIQ4, SLC25A8, UCPH | uncoupling protein 2 (mitochondrial, proton carrier) |
C00004631
|
| 7353 | UFD1L, UFD1 | ubiquitin fusion degradation 1 like (yeast) |
C00004631
|
| 51569 | UFM1, BM-002, C13orf20 | ubiquitin-fold modifier 1 |
C00004631
|
| 55325 | UFSP2, C4orf20 | UFM1-specific peptidase 2 (EC:3.4.22.-) |
C00004631
|
| 7357 | UGCG, GCS, GLCT1 | UDP-glucose ceramide glucosyltransferase (EC:2.4.1.80) |
C00004631
|
| 7360 | UGP2, UDPG, UDPGP, UDPGP2, UGP1, UGPP1, UGPP2, pHC379 | UDP-glucose pyrophosphorylase 2 (EC:2.7.7.9) |
C00004631
|
| 54578 | UGT1A6, GNT1, HLUGP, HLUGP1, UDPGT, UDPGT_1-6, UGT1, UGT1A6S, UGT1F | UDP glucuronosyltransferase 1 family, polypeptide A6 (EC:2.4.1.17) |
C00004631
|
| 79799 | UGT2A3 | UDP glucuronosyltransferase 2 family, polypeptide A3 (EC:2.4.1.17) |
C00004631
|
| 8202 | NCOA3, ACTR, AIB-1, AIB1, CAGH16, CTG26, KAT13B, RAC3, SRC-3, SRC3, TNRC14, TNRC16, TRAM-1, bHLHe42, pCIP | nuclear receptor coactivator 3 (EC:2.3.1.48) |
C00004565
|
| 7363 | UGT2B4, HLUG25, UDPGT2B4, UDPGTH1, UGT2B11 | UDP glucuronosyltransferase 2 family, polypeptide B4 (EC:2.4.1.17) |
C00004631
|
| 127933 | UHMK1, KIS, KIST, P-CIP2 | U2AF homology motif (UHM) kinase 1 (EC:2.7.11.1) |
C00004631
|
| 25972 | UNC50, GMH1, UNCL, URP | unc-50 homolog (C. elegans) |
C00004631
|
| 222643 | UNC5CL, MUXA, ZUD | unc-5 homolog C (C. elegans)-like |
C00004631
|
| 65109 | UPF3B, HUPF3B, MRXS14, RENT3B, UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) |
C00004631
|
| 7380 | UPK3A, UP3A, UPIII, UPIIIA, UPK3 | uroplakin 3A |
C00004631
|
| 55245 | UQCC1, BFZB, C20orf44, CBP3, UQCC | ubiquinol-cytochrome c reductase complex assembly factor 1 |
C00004631
|
| 9816 | URB2, KIAA0133, NET10, NPA2 | URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
C00004631
|
| 7389 | UROD, PCT, UPD | uroporphyrinogen decarboxylase (EC:4.1.1.37) |
C00004631
|
| 8975 | USP13, ISOT3, IsoT-3 | ubiquitin specific peptidase 13 (isopeptidase T-3) (EC:3.4.19.12) |
C00004631
|
| 57558 | USP35 | ubiquitin specific peptidase 35 (EC:3.4.19.12) |
C00004631
|
| 7375 | USP4, UNP, Unph | ubiquitin specific peptidase 4 (proto-oncogene) (EC:3.4.19.12) |
C00004631
|
| 124739 | USP43 | ubiquitin specific peptidase 43 (EC:3.4.19.12) |
C00004631
|
| 64854 | USP46 | ubiquitin specific peptidase 46 (EC:3.4.19.12) |
C00004631
|
| 9712 | USP6NL, RNTRE, TRE2NL, USP6NL-IT1 | USP6 N-terminal like |
C00004631
|
| 84135 | UTP15, NET21 | UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
C00004631
|
| 51096 | UTP18, CGI-48, WDR50 | UTP18 small subunit (SSU) processome component homolog (yeast) |
C00004631
|
| 27340 | UTP20, DRIM | UTP20, small subunit (SSU) processome component, homolog (yeast) |
C00004631
|
| 81839 | VANGL1, KITENIN, LPP2, STB2, STBM2 | VANGL planar cell polarity protein 1 |
C00004631
|
| 114990 | VASN, SLITL2 | vasorin |
C00004631
|
| 10493 | VAT1, VATI | vesicle amine transport 1 |
C00004631
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00004565
|
| 7416 | VDAC1, PORIN, VDAC-1 | voltage-dependent anion channel 1 |
C00004631
|
| 9687 | GREB1 | growth regulation by estrogen in breast cancer 1 |
C00004565
|
| 7428 | VHL, HRCA1, RCA1, VHL1, pVHL | von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
C00004631
|
| 7429 | VIL1, D2S1471, VIL | villin 1 |
C00004631
|
| 7433 | VIPR1, HVR1, II, PACAP-R-2, PACAP-R2, RDC1, V1RG, VAPC1, VIP-R-1, VIPR, VIRG, VPAC1, VPAC1R, VPCAP1R | vasoactive intestinal peptide receptor 1 |
C00004631
|
| 7436 | VLDLR, CARMQ1, CHRMQ1, VLDLRCH | very low density lipoprotein receptor |
C00004631
|
| 8876 | VNN1, HDLCQ8, Tiff66 | vanin 1 (EC:3.5.1.92) |
C00004631
|
| 64601 | VPS16, hVPS16 | vacuolar protein sorting 16 homolog (S. cerevisiae) |
C00004631
|
| 51028 | VPS36, C13orf9, EAP45 | vacuolar protein sorting 36 homolog (S. cerevisiae) |
C00004631
|
| 51542 | VPS54, HCC8, SLP-8p, VPS54L, WR, hVps54L | vacuolar protein sorting 54 homolog (S. cerevisiae) |
C00004631
|
| 7443 | VRK1, PCH1, PCH1A | vaccinia related kinase 1 (EC:2.7.11.1) |
C00004631
|
| 7444 | VRK2 | vaccinia related kinase 2 (EC:2.7.11.1) |
C00004631
|
| 147645 | VSIG10L | V-set and immunoglobulin domain containing 10 like |
C00004631
|
| 23584 | VSIG2, 2210413P10Rik, CTH, CTXL | V-set and immunoglobulin domain containing 2 |
C00004631
|
| 7447 | VSNL1, HLP3, HPCAL3, HUVISL1, VILIP, VILIP-1 | visinin-like 1 |
C00004631
|
| 7448 | VTN, V75, VN, VNT | vitronectin |
C00004631
|
| 375260 | WASH2P, FAM39B | WAS protein family homolog 2 pseudogene |
C00004631
|
| 54838 | WBP1L, C10orf26, OPA1L, OPAL1 | WW domain binding protein 1-like |
C00004631
|
| 57590 | WDFY1, FENS-1, WDF1, ZFYVE17 | WD repeat and FYVE domain containing 1 |
C00004631
|
| 11169 | WDHD1, AND-1, CHTF4, CTF4 | WD repeat and HMG-box DNA binding protein 1 |
C00004631
|
| 55759 | WDR12, YTM1 | WD repeat domain 12 |
C00004631
|
| 89891 | WDR34, DIC5, FAP133, bA216B9.3 | WD repeat domain 34 |
C00004631
|
| 55255 | WDR41 | WD repeat domain 41 |
C00004631
|
| 54521 | WDR44, RAB11BP, RPH11 | WD repeat domain 44 |
C00004631
|
| 22911 | WDR47 | WD repeat domain 47 |
C00004631
|
| 348793 | WDR53 | WD repeat domain 53 |
C00004631
|
| 84058 | WDR54 | WD repeat domain 54 |
C00004631
|
| 54554 | WDR5B | WD repeat domain 5B |
C00004631
|
| 80349 | WDR61, REC14, SKI8 | WD repeat domain 61 |
C00004631
|
| 144406 | WDR66 | WD repeat domain 66 |
C00004631
|
| 23335 | WDR7, TRAG | WD repeat domain 7 |
C00004631
|
| 151525 | WDSUB1, UBOX6, WDSAM1 | WD repeat, sterile alpha motif and U-box domain containing 1 |
C00004631
|
| 7465 | WEE1, WEE1A, WEE1hu | WEE1 G2 checkpoint kinase (EC:2.7.10.2) |
C00004631
|
| 84305 | WIBG, PYM | within bgcn homolog (Drosophila) |
C00004631
|
| 7481 | WNT11, HWNT11 | wingless-type MMTV integration site family, member 11 |
C00004631
|
| 7485 | WRB, CHD5 | tryptophan rich basic protein |
C00004631
|
| 26118 | WSB1, SWIP1, WSB-1 | WD repeat and SOCS box containing 1 |
C00004631
|
| 23286 | WWC1, HBEBP3, HBEBP36, KIBRA | WW and C2 domain containing 1 |
C00004631
|
| 80014 | WWC2, BOMB | WW and C2 domain containing 2 |
C00004631
|
| 25937 | WWTR1, TAZ | WW domain containing transcription regulator 1 |
C00004631
|
| 7494 | XBP1, TREB5, XBP-1, XBP2 | X-box binding protein 1 |
C00004631
|
| 27113 | BBC3, JFY-1, JFY1, PUMA | BCL2 binding component 3 |
C00004565
|
| 7508 | XPC, RAD4, XP3, XPCC | xeroderma pigmentosum, complementation group C |
C00004631
|
| 7512 | XPNPEP2, APP2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (EC:3.4.11.9) |
C00004631
|
| 63929 | XPNPEP3, APP3, NPHPL1 | X-prolyl aminopeptidase (aminopeptidase P) 3, putative (EC:3.4.11.9) |
C00004631
|
| 7514 | XPO1, CRM1, emb, exp1 | exportin 1 (CRM1 homolog, yeast) |
C00004631
|
| 11260 | XPOT, XPO3 | exportin, tRNA |
C00004631
|
| 7517 | XRCC3, CMM6 | X-ray repair complementing defective repair in Chinese hamster cells 3 |
C00004631
|
| 91419 | XRCC6BP1, KUB3 | XRCC6 binding protein 1 (EC:3.4.24.-) |
C00004631
|
| 57002 | YAE1D1, C7orf36 | Yae1 domain containing 1 |
C00004631
|
| 10413 | YAP1, YAP, YAP2, YAP65, YKI | Yes-associated protein 1 |
C00004631
|
| 51067 | YARS2, MLASA2, MT-TYRRS, TYRRS | tyrosyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.1) |
C00004631
|
| 388403 | YPEL2 | yippee-like 2 (Drosophila) |
C00004631
|
| 79693 | YRDC, DRIP3, IRIP, RP11-109P14.4, SUA5 | yrdC N(6)-threonylcarbamoyltransferase domain containing |
C00004631
|
| 84327 | ZBED3 | zinc finger, BED-type containing 3 |
C00004631
|
| 58486 | ZBED5, Buster1 | zinc finger, BED-type containing 5 |
C00004631
|
| 22890 | ZBTB1, ZNF909 | zinc finger and BTB domain containing 1 |
C00004631
|
| 65986 | ZBTB10, RINZF | zinc finger and BTB domain containing 10 |
C00004631
|
| 7541 | ZBTB14, ZF5, ZFP-161, ZFP-5, ZFP161, ZNF478 | zinc finger and BTB domain containing 14 |
C00004631
|
| 57621 | ZBTB2, ZNF437 | zinc finger and BTB domain containing 2 |
C00004631
|
| 49854 | ZBTB21, ZNF295 | zinc finger and BTB domain containing 21 |
C00004631
|
| 253461 | ZBTB38, CIBZ, ZNF921 | zinc finger and BTB domain containing 38 |
C00004631
|
| 79696 | ZC2HC1C, C14orf140, FAM164C | zinc finger, C2HC-type containing 1C |
C00004631
|
| 376940 | ZC3H6, ZC3HDC6 | zinc finger CCCH-type containing 6 |
C00004631
|
| 29066 | ZC3H7A, ZC3H7, ZC3HDC7 | zinc finger CCCH-type containing 7A |
C00004631
|
| 54877 | ZCCHC2, C18orf49 | zinc finger, CCHC domain containing 2 |
C00004631
|
| 84186 | ZCCHC7, AIR1, RP11-397D12.1 | zinc finger, CCHC domain containing 7 |
C00004631
|
| 51201 | ZDHHC2, DHHC2, ZNF372 | zinc finger, DHHC-type containing 2 (EC:2.3.1.225) |
C00004631
|
| 340481 | ZDHHC21, 9130404H11Rik, DHHC-21, DHHC21, DNZ1, HSPC097 | zinc finger, DHHC-type containing 21 (EC:2.3.1.225) |
C00004631
|
| 254887 | ZDHHC23, NIDD | zinc finger, DHHC-type containing 23 (EC:2.3.1.225) |
C00004631
|
| 7771 | ZNF112, ZFP112, ZNF228 | zinc finger protein 112 |
C00004631
|
| 7538 | ZFP36, G0S24, GOS24, NUP475, RNF162A, TIS11, TTP, zfp-36 | ZFP36 ring finger protein |
C00004631
|
| 677 | ZFP36L1, BRF1, Berg36, ERF-1, ERF1, RNF162B, TIS11B, cMG1 | ZFP36 ring finger protein-like 1 |
C00004631
|
| 7539 | ZFP37, ZNF906, zfp-37 | ZFP37 zinc finger protein |
C00004631
|
| 286128 | ZFP41, ZNF753, zfp-41 | ZFP41 zinc finger protein |
C00004631
|
| 339559 | ZFP69, RP11-656D10.2, ZFP69A, ZKSCAN23A, ZNF642, ZSCAN54A | ZFP69 zinc finger protein |
C00004631
|
| 65243 | ZFP69B, RP11-656D10.1, ZKSCAN23B, ZNF643, ZSCAN54B | ZFP69 zinc finger protein B |
C00004631
|
| 7542 | ZFPL1, D11S750, MCG4 | zinc finger protein-like 1 |
C00004631
|
| 7543 | ZFX, ZNF926 | zinc finger protein, X-linked |
C00004631
|
| 53349 | ZFYVE1, DFCP1, SR3, TAFF1, ZNFN2A1 | zinc finger, FYVE domain containing 1 |
C00004631
|
| 9765 | ZFYVE16, PPP1R69 | zinc finger, FYVE domain containing 16 |
C00004631
|
| 84936 | ZFYVE19, MPFYVE | zinc finger, FYVE domain containing 19 |
C00004631
|
| 124220 | ZG16B, EECP, HRPE773, JCLN2, PAUF, PRO1567 | zymogen granule protein 16B |
C00004631
|
| 22882 | ZHX2, AFR1, RAF | zinc fingers and homeoboxes 2 |
C00004631
|
| 7546 | ZIC2, HPE5 | Zic family member 2 |
C00004631
|
| 64393 | ZMAT3, PAG608, WIG-1, WIG1 | zinc finger, matrin-type 3 |
C00004631
|
| 10269 | ZMPSTE24, FACE-1, FACE1, HGPS, PRO1, STE24, Ste24p | zinc metallopeptidase STE24 (EC:3.4.24.84) |
C00004631
|
| 9204 | ZMYM6, Buster2, MYM, ZNF198L4, ZNF258 | zinc finger, MYM-type 6 |
C00004631
|
| 163071 | ZNF114 | zinc finger protein 114 |
C00004631
|
| 51351 | ZNF117, H-plk, HPF9 | zinc finger protein 117 |
C00004631
|
| 7696 | ZNF137P, ZNF137, pHZ-30 | zinc finger protein 137, pseudogene |
C00004631
|
| 7697 | ZNF138, pHZ-32 | zinc finger protein 138 |
C00004631
|
| 7718 | ZNF165, CT53, LD65, ZSCAN7 | zinc finger protein 165 |
C00004631
|
| 7738 | ZNF184 | zinc finger protein 184 |
C00004631
|
| 7743 | ZNF189 | zinc finger protein 189 |
C00004631
|
| 10520 | ZNF211, C2H2-25, CH2H2-25, ZNF-25, ZNFC25 | zinc finger protein 211 |
C00004631
|
| 7764 | ZNF217, ZABC1 | zinc finger protein 217 |
C00004631
|
| 7768 | ZNF225 | zinc finger protein 225 |
C00004631
|
| 7770 | ZNF227 | zinc finger protein 227 |
C00004631
|
| 7571 | ZNF23, KOX16, ZNF359, ZNF612, Zfp612 | zinc finger protein 23 |
C00004631
|
| 57209 | ZNF248, bA162G10.3 | zinc finger protein 248 |
C00004631
|
| 339324 | ZNF260, OZRF1, PEX1, ZFP260 | zinc finger protein 260 |
C00004631
|
| 129025 | ZNF280A, 3'OY11.1, SUHW1, ZNF280, ZNF636 | zinc finger protein 280A |
C00004631
|
| 26974 | ZNF285, ZNF285A | zinc finger protein 285 |
C00004631
|
| 90075 | ZNF30, KOX28 | zinc finger protein 30 |
C00004631
|
| 26152 | ZNF337 | zinc finger protein 337 |
C00004631
|
| 7582 | ZNF33B, KOX2, KOX31, ZNF11B | zinc finger protein 33B |
C00004631
|
| 149076 | ZNF362, RN, lin-29 | zinc finger protein 362 |
C00004631
|
| 100129482 | ZNF37BP, KOX21, ZNF37B | zinc finger protein 37B, pseudogene |
C00004631
|
| 151126 | ZNF385B, ZNF533 | zinc finger protein 385B |
C00004631
|
| 55893 | ZNF395, HDBP-2, HDBP2, HDRF-2, PBF, PRF-1, PRF1, Si-1-8-14 | zinc finger protein 395 |
C00004631
|
| 90594 | ZNF439 | zinc finger protein 439 |
C00004631
|
| 51710 | ZNF44, GIOT-2, KOX7, ZNF, ZNF504, ZNF55, ZNF58 | zinc finger protein 44 |
C00004631
|
| 126070 | ZNF440 | zinc finger protein 440 |
C00004631
|
| 126068 | ZNF441 | zinc finger protein 441 |
C00004631
|
| 90333 | ZNF468 | zinc finger protein 468 |
C00004631
|
| 130557 | ZNF513, HMFT0656, RP58 | zinc finger protein 513 |
C00004631
|
| 9849 | ZNF518A, ZNF518 | zinc finger protein 518A |
C00004631
|
| 79818 | ZNF552 | zinc finger protein 552 |
C00004631
|
| 148254 | ZNF555 | zinc finger protein 555 |
C00004631
|
| 79230 | ZNF557 | zinc finger protein 557 |
C00004631
|
| 84527 | ZNF559, NBLA00121 | zinc finger protein 559 |
C00004631
|
| 93134 | ZNF561 | zinc finger protein 561 |
C00004631
|
| 147929 | ZNF565 | zinc finger protein 565 |
C00004631
|
| 126295 | ZNF57, ZNF424 | zinc finger protein 57 |
C00004631
|
| 126231 | ZNF573 | zinc finger protein 573 |
C00004631
|
| 51157 | ZNF580 | zinc finger protein 580 |
C00004631
|
| 51545 | ZNF581 | zinc finger protein 581 |
C00004631
|
| 84622 | ZNF594 | zinc finger protein 594 |
C00004631
|
| 162966 | ZNF600, KR-ZNF1 | zinc finger protein 600 |
C00004631
|
| 57507 | ZNF608, NY-REN-36 | zinc finger protein 608 |
C00004631
|
| 57547 | ZNF624 | zinc finger protein 624 |
C00004631
|
| 51193 | ZNF639, 6230400O18Rik, ANC-2H01, ANC_2H01, ZASC1 | zinc finger protein 639 |
C00004631
|
| 22834 | ZNF652 | zinc finger protein 652 |
C00004631
|
| 127396 | ZNF684 | zinc finger protein 684 |
C00004631
|
| 90592 | ZNF700 | zinc finger protein 700 |
C00004631
|
| 7562 | ZNF708, KOX8, ZNF15, ZNF15L1 | zinc finger protein 708 |
C00004631
|
| 7552 | ZNF711, CMPX1, MRX97, ZNF4, ZNF5, ZNF6, Zfp711, dJ75N13.1 | zinc finger protein 711 |
C00004631
|
| 7627 | ZNF75A | zinc finger protein 75a |
C00004631
|
| 388507 | ZNF788 | zinc finger family member 788 |
C00004631
|
| 285989 | ZNF789 | zinc finger protein 789 |
C00004631
|
| 7633 | ZNF79, pT7 | zinc finger protein 79 |
C00004631
|
| 7637 | ZNF84, HPF2 | zinc finger protein 84 |
C00004631
|
| 342892 | ZNF850, ZNF850P | zinc finger protein 850 |
C00004631
|
| 30834 | ZNRD1, HTEX-6, Rpa12, TEX6, ZR14, hZR14, tctex-6 | zinc ribbon domain containing 1 (EC:2.7.7.6) |
C00004631
|
| 84133 | ZNRF3, BK747E2.3, RNF203 | zinc and ring finger 3 |
C00004631
|
| 7589 | ZSCAN21, NY-REN-21, ZNF38, Zipro1 | zinc finger and SCAN domain containing 21 |
C00004631
|
| 7741 | ZSCAN26, SRE-ZBP, ZNF187 | zinc finger and SCAN domain containing 26 |
C00004631
|
| 64288 | ZSCAN31, ZNF20-Lp, ZNF310P, ZNF323, dJ874C20.2 | zinc finger and SCAN domain containing 31 |
C00004631
|
| 201516 | ZSCAN4, ZNF494 | zinc finger and SCAN domain containing 4 |
C00004631
|
| 57643 | ZSWIM5 | zinc finger, SWIM-type containing 5 |
C00004631
|
| 57688 | ZSWIM6 | zinc finger, SWIM-type containing 6 |
C00004631
|
| 9183 | ZW10, HZW10, KNTC1AP | zw10 kinetochore protein |
C00004631
|
| 55055 | ZWILCH, KNTC1AP, hZwilch | zwilch kinetochore protein |
C00004631
|
| 158586 | ZXDB, ZNF905, dJ83L6.1 | zinc finger, X-linked, duplicated B |
C00004631
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00003817
|
| 1586 | CYP17A1, CPT7, CYP17, P450C17, S17AH | cytochrome P450, family 17, subfamily A, polypeptide 1 (EC:1.14.99.9 4.1.2.30) |
C00003817
|
| 1589 | CYP21A2, CA21H, CAH1, CPS1, CYP21, CYP21B, P450c21B | cytochrome P450, family 21, subfamily A, polypeptide 2 (EC:1.14.99.10) |
C00003817
|
| 3284 | HSD3B2, HSD3B, HSDB, SDR11E2 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 (EC:1.1.1.145 5.3.3.1) |
C00003817
|
| 472 | ATM, AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | ataxia telangiectasia mutated (EC:2.7.11.1) |
C00004565
|
| 5562 | PRKAA1, AMPK, AMPKa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit (EC:2.7.11.1 2.7.11.26 2.7.11.27 2.7.11.31) |
C00003817
|
| 6198 | RPS6KB1, PS6K, S6K, S6K-beta-1, S6K1, STK14A, p70_S6KA, p70(S6K)-alpha, p70-S6K, p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 (EC:2.7.11.1) |
C00003817
|
| 329 | BIRC2, API1, HIAP2, Hiap-2, MIHB, RNF48, c-IAP1, cIAP1 | baculoviral IAP repeat containing 2 |
C00000674
|
| 2354 | FOSB, AP-1, G0S3, GOS3, GOSB | FBJ murine osteosarcoma viral oncogene homolog B |
C00000674
|
| 5602 | MAPK10, JNK3, JNK3A, PRKM10, SAPK1b, p493F12, p54bSAPK | mitogen-activated protein kinase 10 (EC:2.7.11.24) |
C00000674
|
| 9572 | NR1D1, EAR1, THRA1, THRAL, ear-1, hRev | nuclear receptor subfamily 1, group D, member 1 |
C00000674
|
| 7128 | TNFAIP3, A20, OTUD7C, TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 (EC:3.4.19.12) |
C00000674
|
| 10451 | VAV3 | vav 3 guanine nucleotide exchange factor |
C00000674
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (160)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #615224 | Advanced sleep phase syndrome, familial, 2; fasps2 |
P48730
|
| #615214 | Agammaglobulinemia 7, autosomal recessive; agm7 |
P27986
|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #103780 | Alcohol dependence |
P47869
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #608584 | Asthma-related traits, susceptibility to, 2 |
Q6W5P4
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #613985 | Beta-thalassemia |
P68871
|
| #603902 | Beta-thalassemia, dominant inclusion body type |
P68871
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #614490 | Blood group, junior system; jr |
Q9UNQ0
|
| #210900 | Bloom syndrome; blm |
P54132
|
| %606641 | Body mass index; bmi |
P37231
|
| #114480 | Breast cancer |
P31749
P38398 P42336 |
| #604370 | Breast-ovarian cancer, familial, susceptibility to, 1; brovca1 |
P38398
|
| #300615 | Brunner syndrome |
P21397
|
| #615279 | Cardiofaciocutaneous syndrome 3; cfc3 |
Q02750
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #606595 | Charcot-marie-tooth disease, axonal, type 2f; cmt2f |
P04792
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #613641 | Charcot-marie-tooth disease, recessive intermediate b; cmtrib |
Q15046
|
| #114500 | Colorectal cancer; crc |
P18054
P31749 P42336 P84022 Q14191 |
| #612918 | Congenital lipomatous overgrowth, vascular malformations, and epidermal nevi |
P42336
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #604931 | Cortisone reductase deficiency 1; cortrd1 |
P28845
|
| #615108 | Cowden syndrome 5; cws5 |
P42336
|
| #615109 | Cowden syndrome 6; cws6 |
P31749
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #613916 | Deafness, autosomal recessive 89; dfnb89 |
Q15046
|
| #304800 | Diabetes insipidus, nephrogenic, x-linked |
P30518
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #607208 | Dravet syndrome |
P18507
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #237500 | Dubin-johnson syndrome; djs |
Q92887
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #607681 | Epilepsy, childhood absence, susceptibility to, 2; eca2 |
P18507
|
| #612269 | Epilepsy, childhood absence, susceptibility to, 5; eca5 |
P28472
|
| #613060 | Epilepsy, idiopathic generalized, susceptibility to, 10; eig10 |
O14764
|
| #611136 | Epilepsy, juvenile myoclonic, susceptibility to, 5; ejm5 |
P14867
|
| #133239 | Esophageal cancer |
P04637
P18054 |
| #615363 | Estrogen resistance; estrr |
P03372
|
| #301500 | Fabry disease |
P06280
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #230200 | Galactokinase deficiency |
P51570
|
| #608013 | Gaucher disease, perinatal lethal |
P04062
|
| #230800 | Gaucher disease, type i |
P04062
|
| #230900 | Gaucher disease, type ii |
P04062
|
| #231000 | Gaucher disease, type iii |
P04062
|
| #231005 | Gaucher disease, type iiic |
P04062
|
| #609446 | Generalized epilepsy and paroxysmal dyskinesia; gepd |
Q12791
|
| #604233 | Generalized epilepsy with febrile seizures plus, type 1; gefsp1 |
P18507
|
| #611277 | Generalized epilepsy with febrile seizures plus, type 3; gefsp3 |
P18507
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #137750 | Glaucoma 1, open angle, a; glc1a |
Q16678
|
| #231300 | Glaucoma 3, primary congenital, a; glc3a |
Q16678
|
| #137760 | Glaucoma, primary open angle; poag |
Q16678
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
P37231 |
| #232300 | Glycogen storage disease ii |
P10253
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #140700 | Heinz body anemias |
P68871
|
| #114550 | Hepatocellular carcinoma |
P42336
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237450 | Hyperbilirubinemia, rotor type; hblrr |
Q9NPD5
Q9Y6L6 |
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #182000 | Keratosis, seborrheic |
P42336
|
| #601626 | Leukemia, acute myeloid; aml |
P36888
|
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #211980 | Lung cancer |
P00533
P04637 |
| #300635 | Lymphoproliferative syndrome, x-linked, 2; xlp2 |
P98170
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #602501 | Megalencephaly-capillary malformation-polymicrogyria syndrome; mcap |
P42336
|
| #603387 | Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome; mpph |
P42336
|
| #609048 | Melanoma, cutaneous malignant, susceptibility to, 3; cmm3 |
P11802
|
| #614104 | Mental retardation, autosomal dominant 7; mrd7 |
Q13627
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #253220 | Mucopolysaccharidosis, type vii; mps7 |
P08236
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #254500 | Myeloma, multiple |
P24385
|
| #254600 | Myeloperoxidase deficiency; mpod |
P05164
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #300539 | Nephrogenic syndrome of inappropriate antidiuresis; nsiad |
P30518
|
| #608634 | Neuronopathy, distal hereditary motor, type iib; hmn2b |
P04792
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #257220 | Niemann-pick disease, type c1; npc1 |
O15118
|
| #601665 | Obesity |
P37231
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #167000 | Ovarian cancer |
P38398
P42336 |
| #614320 | Pancreatic cancer, susceptibility to, 4; pnca4 |
P38398
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #168600 | Parkinson disease, late-onset; pd |
P04062
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #614674 | Periodic fever, menstrual cycle-dependent |
P08908
|
| #604229 | Peters anomaly |
Q16678
|
| #172700 | Pick disease of brain |
P10636
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #176920 | Proteus syndrome |
P31749
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
| #269880 | Short syndrome |
P27986
|
| #603903 | Sickle cell anemia |
P68871
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #602111 | Spondyloepimetaphyseal dysplasia, missouri type |
P45452
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
O14763
P04637 |
| %612223 | Stature quantitative trait locus 11; stqtl11 |
Q00534
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #187950 | Thrombocythemia 1; thcyt1 |
P40225
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #138900 | Uric acid concentration, serum, quantitative trait locus 1; uaqtl1 |
Q9UNQ0
|
| #614916 | Ventricular tachycardia, catecholaminergic polymorphic, 4; cpvt4 |
P62158
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #277700 | Werner syndrome; wrn |
Q14191
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (130)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00783 | Febrile seizures |
O14764
(related)
P18507 (related) |
| H00136 | Niemann-Pick disease type C (NPC) |
O15118
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
P04637 (marker) Q16790 (marker) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) P24385 (related) P24385 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P04637 (related) P04637 (marker) P24385 (related) P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
P04637 (related) |
| H00022 | Bladder cancer |
P00533
(related)
P04637 (related) P68871 (marker) |
| H00028 | Choriocarcinoma |
P00533
(related)
P03956 (related) P04637 (related) P08253 (related) |
| H00030 | Cervical cancer |
P00533
(related)
P11802 (related) |
| H00042 | Glioma |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) P11802 (related) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) P24385 (related) P24385 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
Q01453 (related) Q15046 (related) |
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
P37231 (related) |
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04637 (related) Q92731 (marker) |
| H00066 | Lewy body dementia (LBD) |
P04062
(related)
|
| H00126 | Gaucher disease |
P04062
(related)
|
| H00426 | Defects in the degradation of ganglioside |
P04062
(related)
|
| H00810 | Progressive myoclonic epilepsy (PME) |
P04062
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P04150
(related)
P11511 (related) |
| H00004 | Chronic myeloid leukemia (CML) |
P04637
(related)
Q01196 (related) |
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
P28907 (marker) |
| H00006 | Hairy-cell leukemia |
P04637
(related)
P24385 (related) |
| H00008 | Burkitt lymphoma |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00010 | Multiple myeloma |
P04637
(related)
P24385 (related) |
| H00013 | Small cell lung cancer |
P04637
(related)
|
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00019 | Pancreatic cancer |
P04637
(related)
P04637 (marker) |
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) P68871 (marker) |
| H00025 | Penile cancer |
P04637
(related)
P04637 (marker) P08253 (related) P14780 (related) P35354 (related) |
| H00027 | Ovarian cancer |
P04637
(related)
P38398 (related) P42336 (related) |
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00031 | Breast cancer |
P04637
(related)
P24385 (related) P38398 (related) |
| H00032 | Thyroid cancer |
P04637
(related)
P27487 (marker) P37231 (related) |
| H00036 | Osteosarcoma |
P04637
(related)
P08684 (marker) |
| H00038 | Malignant melanoma |
P04637
(related)
P11802 (related) P14679 (marker) |
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00041 | Kaposi's sarcoma |
P04637
(related)
|
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00046 | Cholangiocarcinoma |
P04637
(related)
P35354 (related) |
| H00047 | Gallbladder cancer |
P04637
(related)
P24385 (marker) |
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00856 | Distal hereditary motor neuropathies (dHMN) |
P04792
(related)
|
| H00101 | Other phagocyte defects |
P05164
(related)
|
| H00003 | Acute myeloid leukemia (AML) |
P05164
(marker)
P36888 (related) Q01196 (related) Q01196 (marker) Q13951 (marker) |
| H00093 | Combined immunodeficiencies (CIDs) |
P06239
(related)
|
| H00125 | Fabry disease |
P06280
(related)
|
| H00132 | Mucopolysaccharidosis type VII (MPS7) |
P08236
(related)
|
| H00421 | Mucopolysaccharidosis (MPS) |
P08236
(related)
|
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00024 | Prostate cancer |
P10275
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
P45452 (related) |
| H00808 | Idiopathic generalized epilepsies (IGEs) |
P14867
(related)
P18507 (related) |
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
Q92887 (related) |
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H00559 | von Hippel-Lindau syndrome |
P24385
(related)
|
| H01111 | Cortisone reductase deficiency (CRD) |
P28845
(related)
|
| H00252 | Congenital nephrogenic diabetes insipidus (NDI) |
P30518
(related)
|
| H01294 | Nephrogenic syndrome of inappropriate antidiuresis (NSIAD) |
P30518
(related)
|
| H00539 | PTEN hamartoma tumor syndrome (PHTS) |
P31749
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00227 | Congenital amegakaryocytic thrombocytopenia (CAMT) |
P40225
(marker)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00070 | Galactosemia |
P51570
(related)
|
| H00606 | Early infantile epileptic encephalopathy |
P53779
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
Q14191 (related) |
| H00228 | Thalassemia |
P68871
(related)
|
| H00229 | Sickle cell anemia (SCA) |
P68871
(related)
|
| H00107 | Other well-defined immunodeficiency syndromes |
P98170
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00523 | Noonan syndrome and related disorders |
Q02750
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H01258 | Generalized epilepsy and paroxysmal dyskinesia (GEPD) |
Q12791
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00612 | Primary open angle glaucoma |
Q16678
(related)
|
| H01075 | Peters anomaly |
Q16678
(related)
|
| H01159 | Anterior segment dysgenesis (ASD) |
Q16678
(related)
|
| H01203 | Primary congenital glaucoma (PCG) |
Q16678
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
Q9NUW8 (related) |
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00932 | Tropical calcific pancreatitis |
Q99895
(related)
|
| H00933 | Hereditary pancreatitis |
Q99895
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
70 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D001943 | Breast Neoplasms |
C00004565
C00004631 C00003817 |
| D009202 | Cardiomyopathies |
C00004565
C00004631 C00000674 |
| D010051 | Ovarian Neoplasms |
C00004631
C00000674 |
| D011041 | Poisoning |
C00004631
C00003817 |
| D006943 | Hyperglycemia |
C00004565
C00004631 |
| D007938 | Leukemia |
C00004565
C00004631 |
| D011471 | Prostatic Neoplasms |
C00004631
C00003817 |
| D011297 | Prenatal Exposure Delayed Effects |
C00004565
C00004631 |
| D009362 | Neoplasm Metastasis |
C00004631
C00003817 |
| D006529 | Hepatomegaly |
C00004631
C00000674 |
| D005483 | Flushing |
C00004631
C00000674 |
| D056486 | Drug-Induced Liver Injury |
C00004631
C00003817 |
| D003110 | Colonic Neoplasms |
C00004631
C00003817 |
| D009369 | Neoplasms |
C00004631
|
| D006332 | Cardiomegaly |
C00004631
|
| D002312 | Cardiomyopathy, Hypertrophic |
C00004631
|
| D002375 | Catalepsy |
C00004631
|
| D002471 | Cell Transformation, Neoplastic |
C00004631
|
| D003072 | Cognition Disorders |
C00004631
|
| D006528 | Carcinoma, Hepatocellular |
C00004631
|
| D015179 | Colorectal Neoplasms |
C00004631
|
| D003556 | Cystitis |
C00004631
|
| D003921 | Diabetes Mellitus, Experimental |
C00004631
|
| D002105 | Cadmium Poisoning |
C00004631
|
| D004409 | Dyskinesia, Drug-Induced |
C00004631
|
| D004942 | Esophagitis, Peptic |
C00004631
|
| D005234 | Fatty Liver |
C00004631
|
| D005355 | Fibrosis |
C00004631
|
| D001327 | Autoimmune Diseases |
C00004631
|
| D005909 | Glioblastoma |
C00004631
|
| D005910 | Glioma |
C00004631
|
| D006461 | Hemolysis |
C00004631
|
| D001260 | Ataxia Telangiectasia |
C00004631
|
| D006930 | Hyperalgesia |
C00004631
|
| D006973 | Hypertension |
C00004631
|
| D007249 | Inflammation |
C00004631
|
| D007251 | Influenza, Human |
C00004631
|
| D007333 | Insulin Resistance |
C00004631
|
| D007627 | Keloid |
C00004631
|
| D007674 | Kidney Diseases |
C00004631
|
| D008106 | Liver Cirrhosis, Experimental |
C00004631
|
| D008107 | Liver Diseases |
C00004631
|
| D008113 | Liver Neoplasms |
C00004631
|
| D008114 | Liver Neoplasms, Experimental |
C00004631
|
| D008175 | Lung Neoplasms |
C00004631
|
| D015674 | Mammary Neoplasms, Animal |
C00004631
|
| D008569 | Memory Disorders |
C00004631
|
| D028361 | Mitochondrial Diseases |
C00004631
|
| D009203 | Myocardial Infarction |
C00004631
|
| D015428 | Myocardial Reperfusion Injury |
C00004631
|
| D009336 | Necrosis |
C00004631
|
| D000743 | Anemia, Hemolytic |
C00004631
|
| D021441 | Carcinoma, Pancreatic Ductal |
C00004631
|
| D009374 | Neoplasms, Experimental |
C00004631
|
| D009410 | Nerve Degeneration |
C00004631
|
| D020078 | Neurogenic Inflammation |
C00004631
|
| D015663 | OSTEOPOROSIS, POSTMENOPAUSAL |
C00004631
|
| D010024 | Osteoporosis |
C00004565
|
| D010190 | Pancreatic Neoplasms |
C00004631
|
| D002318 | Cardiovascular Diseases |
C00004565
|
| D011230 | Precancerous Conditions |
C00004631
|
| D050197 | Atherosclerosis |
C00004565
|
| D011507 | Proteinuria |
C00004631
|
| D012554 | Schistosomiasis japonica |
C00004631
|
| D013274 | Stomach Neoplasms |
C00004631
|
| D001749 | Urinary Bladder Neoplasms |
C00004631
|
| D000257 | Adenoviridae Infections |
C00003817
|
| D009361 | Neoplasm Invasiveness |
C00003817
|
| D002386 | Cataract |
C00000674
|
| D008103 | Liver Cirrhosis |
C00000674
|