Euodia xanthoxyloides
Species
KNApSAcK Entry
| Organism name | Euodia xanthoxyloides |
|---|---|
| Genus | Euodia |
| Family | Rutaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Euodia |
|---|---|
| Linked NCBI taxonomy ID | 354493 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Rutaceae |
|---|---|
| ID | 23513 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (3)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
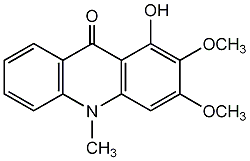
C00002137
|
Arborinine
|
CHEMBL349609
|
C022784
|
1 / 0 / 0 | No. 257 | No. 7 |
|
|
|
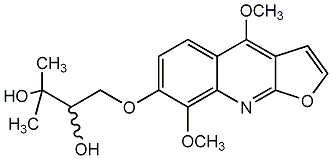
C00002158
|
Evoxine
/ 7-(2,3-Dihydroxy-3-methyl-butyl)oxy-8-methyldictamnine |
CHEMBL1317300
CHEMBL1395597 CHEMBL1416006 |
14 / 9 / 9 | No. 600 | No. 7 |
|
||
|
C00002157
|
Evoxanthidine
|
No. 2197 | No. 7 |

|
Human Protein / Gene in interactions
15 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00002158 | 1 / 0 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00002137 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00002158 | 1 / 1 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00002158 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00002158 | 0 / 1 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002158 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00002158 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00002158 | 0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00002158 | 3 / 3 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00002158 | 2 / 2 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00002158 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00002158 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002158 | 0 / 1 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00002158 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00002158 | 1 / 0 |
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (9)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
KEGG DISEASE (9)
| KEGG | name | UniProt |
|---|---|---|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|