Impatiens balsamina
Species
KNApSAcK Entry
| Organism name | Impatiens balsamina |
|---|---|
| Genus | Impatiens |
| Family | Balsaminaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Impatiens balsamina |
|---|---|
| Linked NCBI taxonomy ID | 63779 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Balsaminaceae |
|---|---|
| ID | 25692 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (10)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
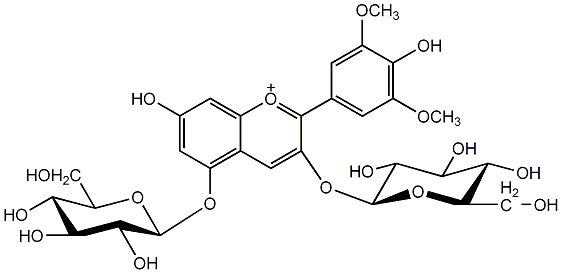
C00002384
|
Malvin
/ Malvidin diglucoside / Malvidin 3,5-diglucoside |
CHEMBL2009656
|
No. 1 | No. 15 |
|
|||
|
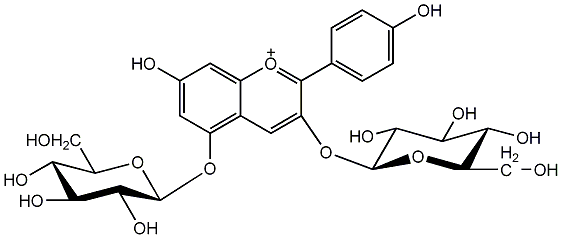
C00002387
|
Pelargonin
/ Pelargonidin 3,5-di-beta-D-glucoside |
No. 1 | No. 15 |
|
||||
|
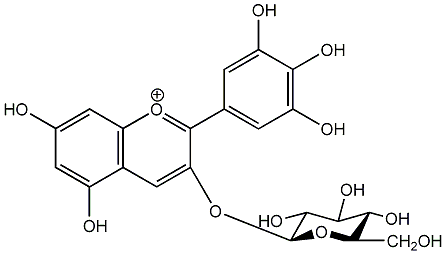
C00006698
|
Myrtillin
/ Delphinidin 3-O-beta-D-glucopyranoside |
CHEMBL518846
|
No. 2 | No. 15 |
|
|||
|
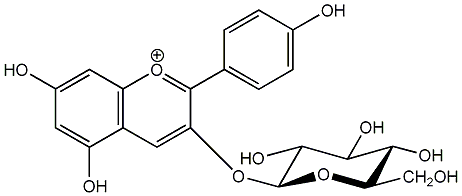
C00006630
|
Callistephin
/ Pelargonidin 3-O-beta-D-glucopyranoside |
C078485
|
No. 2 | No. 15 |
|
|||
|
C00005213
|
Kaempferol 3-(3G-glucosylneohesperidoside)
/ Kaempferol 3-rhamnosyl-(1->2)-[glucosyl-(1->3)-glucoside] / 3-[(O-6-Deoxy-alpha-L-mannopyranosyl-(1->2)-O-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl)oxy]-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
No. 5 | No. 15 |
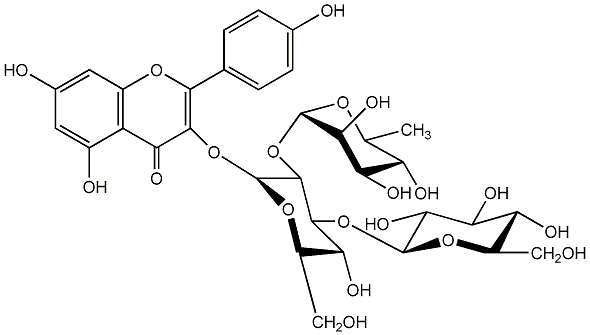
|
||||
|
C00002837
|
Lawsone
|
CHEMBL240963
|
C005090
|
16 / 12 / 13 | 1 / 3 | No. 1047 | No. 80 |
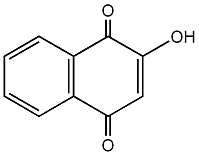
|
|
C00049940
|
2-Methoxy-1,4-naphthoquinone
|
CHEMBL106562
|
C096118
|
13 / 5 / 5 | No. 1047 | No. 80 |

|
|
|
C00002674
|
Syringic acid
/ 4-Hydroxy-3,5-dimethoxybenzoic acid |
CHEMBL1414
|
C001945
|
12 / 3 / 5 | No. 1073 |
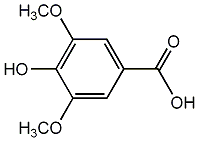
|
||
|
C00050009
|
Balsaminone A
|
No. 5829 |

|
|||||
|
C00050010
|
Balsaminone B
|
CHEMBL463739
|
No. 6510 |

|
Human Protein / Gene in interactions
40 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P14902 | Indoleamine 2,3-dioxygenase 1 | Enzyme | C00002837 C00049940 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00049940 | 1 / 1 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00002674 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00002674 | 0 / 0 |
| P22894 | Neutrophil collagenase | M10A | C00002837 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00002837 | 2 / 2 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00002674 | 1 / 2 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00002837 | 2 / 3 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00002674 | 0 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00002674 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002837 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00002674 | 1 / 2 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00002837 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00049940 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00049940 | 2 / 0 |
| O75496 | Geminin | Unclassified protein | C00049940 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00002674 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00002674 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00049940 | 0 / 0 |
| P03956 | Interstitial collagenase | M10A | C00002837 | 0 / 1 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00002674 | 0 / 0 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | C00049940 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00002837 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00002837 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00002674 | 0 / 1 |
| P08253 | 72 kDa type IV collagenase | M10A | C00002837 | 1 / 3 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00002837 | 2 / 2 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00049940 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002837 | 0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00049940 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00002837 | 4 / 3 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00049940 | 0 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00002674 | 0 / 0 |
| P08254 | Stromelysin-1 | M10A | C00002837 | 1 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00002674 | 1 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002837 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00002837 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00049940 | 1 / 0 |
| P01215 | Glycoprotein hormones alpha chain | Unclassified protein | C00049940 | 0 / 3 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00049940 | 1 / 1 |
1 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00002837
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (20)
| OMIM | preferred title | UniProt |
|---|---|---|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #114500 | Colorectal cancer; crc |
P84022
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (22)
| KEGG | name | UniProt |
|---|---|---|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00081 | Hashimoto's thyroiditis |
P01215
(marker)
|
| H00082 | Graves' disease |
P01215
(marker)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P01215
(marker)
|
| H00028 | Choriocarcinoma |
P03956
(related)
P08253 (related) |
| H00025 | Penile cancer |
P08253
(related)
P14780 (related) |
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
Q13148 (related) |
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|