Verbena littoralis
Species
KNApSAcK Entry
| Organism name | Verbena littoralis |
|---|---|
| Genus | Verbena |
| Family | Verbenaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Verbena |
|---|---|
| Linked NCBI taxonomy ID | 22002 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Verbenaceae |
|---|---|
| ID | 21910 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (9)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
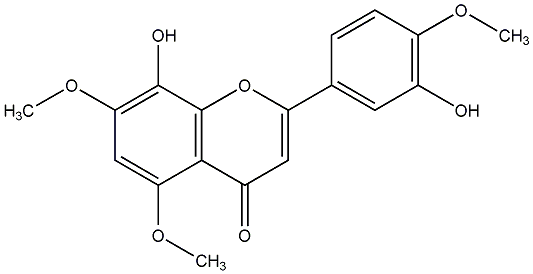
C00013321
|
8,3'-Dihydroxy-5,7,4'-trimethoxyflavone
/ 8-Hydroxy-2-(3-hydroxy-4-methoxyphenyl)-5,7-dimethoxy-4H-1-benzopyran-4-one |
No. 3 | No. 15 |
|
||||
|
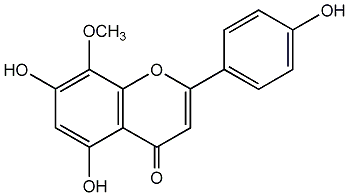
C00003850
|
8-Methoxyapigenin
/ 4'-Hydroxywogonin / Isoscutellarein 8-methyl ether / 5,7,4'-Trihydroxy-8-methoxyflavone |
CHEMBL245712
|
C093385
|
No. 3 | No. 15 |
|
||
|
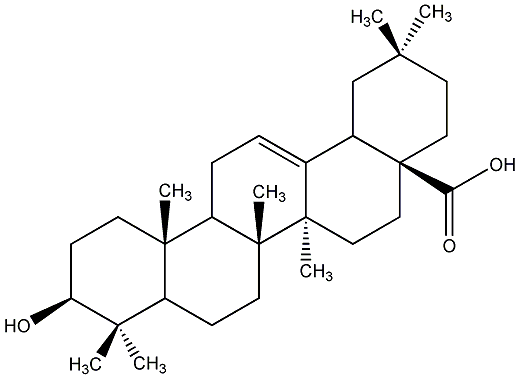
C00019064
|
Oleanolic acid
/ Astrantiagenin C / Virgaureagenin B / 3beta-Hydroxyolean-12-en-28-oic acid |
CHEMBL56615
CHEMBL168 CHEMBL180553 CHEMBL365375 CHEMBL486382 CHEMBL1413646 CHEMBL1436454 |
D009828
|
30 / 8 / 12 | 21 / 15 | No. 13 | No. 51 |
|
|
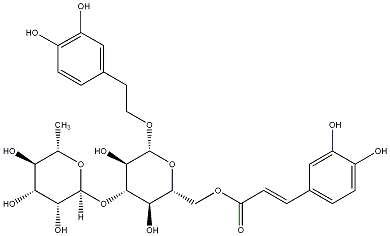
C00030516
|
Isoacteoside
/ Isoverbascoside |
CHEMBL504873
|
C064683
|
6 / 5 / 4 | No. 33 |
|
||
|
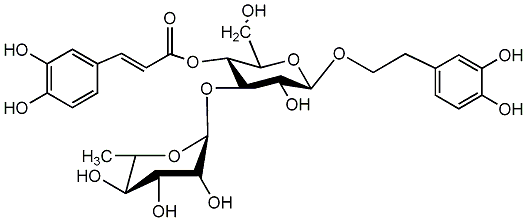
C00002783
|
Kusaginin
/ Acteoside / Verbascoside |
CHEMBL231853
CHEMBL444478 CHEMBL1364101 |
C058956
|
12 / 1 / 1 | 34 / 3 | No. 33 |
|
|
|
C00045086
|
Stigmast-5-ene 3beta,4beta,7alpha,22alpha-tetraol
|
CHEMBL468236
|
No. 53 | No. 11 |

|
|||
|
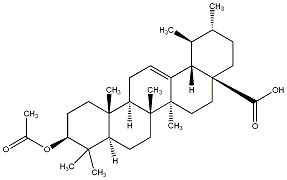
C00029633
|
Ursolic acid
/ Acetylursolic acid |
CHEMBL55086
CHEMBL410525 |
4 / 2 / 2 | No. 177 |
|
|||
|
C00014636
|
Littorachalcone
|
No. 375 |

|
|||||
|
C00014637
|
Verbenachalcone
|
CHEMBL519611
|
No. 375 |

|
Human Protein / Gene in interactions
46 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002783 C00019064 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00019064 C00029633 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002783 C00019064 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00019064 C00029633 | 2 / 2 |
| O75496 | Geminin | Unclassified protein | C00002783 C00019064 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00019064 C00029633 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00019064 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00002783 | 0 / 0 |
| P39900 | Macrophage metalloelastase | M10A | C00030516 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00030516 | 2 / 2 |
| P24666 | Low molecular weight phosphotyrosine protein phosphatase | Tyr | C00019064 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00019064 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00019064 | 0 / 0 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00019064 | 0 / 0 |
| P10586 | Receptor-type tyrosine-protein phosphatase F | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00019064 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002783 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00019064 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00019064 | 0 / 3 |
| P15121 | Aldose reductase | Enzyme | C00019064 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00019064 | 0 / 0 |
| P03956 | Interstitial collagenase | M10A | C00030516 | 0 / 1 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00002783 | 0 / 0 |
| Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | Tyr | C00019064 | 4 / 2 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00019064 | 0 / 0 |
| P45452 | Collagenase 3 | M10A | C00030516 | 1 / 1 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00019064 | 0 / 0 |
| P18433 | Receptor-type tyrosine-protein phosphatase alpha | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00019064 | 0 / 0 |
| P08253 | 72 kDa type IV collagenase | M10A | C00030516 | 1 / 3 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00019064 | 0 / 0 |
| P35228 | Nitric oxide synthase, inducible | Enzyme | C00019064 | 1 / 1 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002783 | 1 / 1 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00002783 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00002783 | 0 / 0 |
| P17706 | Tyrosine-protein phosphatase non-receptor type 2 | Tyr | C00019064 | 0 / 1 |
| P08254 | Stromelysin-1 | M10A | C00030516 | 1 / 0 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | Tyr | C00019064 | 0 / 0 |
| P04054 | Phospholipase A2 | Enzyme | C00019064 | 0 / 0 |
| P23469 | Receptor-type tyrosine-protein phosphatase epsilon | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002783 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00029633 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002783 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00019064 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00019064 | 1 / 4 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002783 | 0 / 0 |
49 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002783
C00019064
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002783
C00019064
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002783
C00019064
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002783
C00019064
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002783
C00019064
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00002783
C00019064
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002783
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00002783
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00002783
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00002783
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002783
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00002783
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00002783
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002783
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002783
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00002783
|
| 896 | CCND3 | cyclin D3 |
C00002783
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00002783
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002783
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002783
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00002783
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00002783
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00002783
|
| 27035 | NOX1, GP91-2, MOX1, NOH-1, NOH1 | NADPH oxidase 1 |
C00002783
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00002783
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00002783
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002783
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002783
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00002783
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00002783
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00002783
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00002783
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002783
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00002783
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00019064
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00019064
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00019064
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00019064
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00019064
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00019064
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00019064
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00019064
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00019064
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00019064
|
| 5052 | PRDX1, MSP23, NKEF-A, NKEFA, PAG, PAGA, PAGB, PRX1, PRXI, TDPX2 | peroxiredoxin 1 (EC:1.11.1.15) |
C00019064
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00019064
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00019064
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00019064
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00019064
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (14)
| OMIM | preferred title | UniProt |
|---|---|---|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #607785 | Juvenile myelomonocytic leukemia; jmml |
Q06124
|
| #151100 | Leopard syndrome 1 |
Q06124
|
| #611162 | Malaria, susceptibility to |
P35228
|
| #156250 | Metachondromatosis; metcds |
Q06124
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #163950 | Noonan syndrome 1; ns1 |
Q06124
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #602111 | Spondyloepimetaphyseal dysplasia, missouri type |
P45452
|
KEGG DISEASE (16)
| KEGG | name | UniProt |
|---|---|---|
| H00028 | Choriocarcinoma |
P03956
(related)
P08253 (related) |
| H00025 | Penile cancer |
P08253
(related)
P14780 (related) P35354 (related) |
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
P45452 (related) |
| H00408 | Type I diabetes mellitus |
P17706
(related)
|
| H00017 | Esophageal cancer |
P35228
(related)
P35354 (related) |
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00523 | Noonan syndrome and related disorders |
Q06124
(related)
|
| H01018 | Metachondromatosis |
Q06124
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
Diseases related to CTD interactions
17 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D007249 | Inflammation |
C00002783
C00019064 |
| D007674 | Kidney Diseases |
C00019064
|
| D014947 | Wounds and Injuries |
C00002783
|
| D002252 | Carbon Tetrachloride Poisoning |
C00019064
|
| D056486 | Drug-Induced Liver Injury |
C00019064
|
| D050171 | Dyslipidemias |
C00019064
|
| D018149 | Glucose Intolerance |
C00019064
|
| D006949 | Hyperlipidemias |
C00019064
|
| D007938 | Leukemia |
C00002783
|
| D008103 | Liver Cirrhosis |
C00019064
|
| D008106 | Liver Cirrhosis, Experimental |
C00019064
|
| D008107 | Liver Diseases |
C00019064
|
| D017202 | Myocardial Ischemia |
C00019064
|
| D009369 | Neoplasms |
C00019064
|
| D009765 | Obesity |
C00019064
|
| D011041 | Poisoning |
C00019064
|
| D011230 | Precancerous Conditions |
C00019064
|