KCF-S cluster No. 2076 (5 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| Liliopsida | 15 |
| rosids | 14 |
| asterids | 8 |
| eudicotyledons | 6 |
| Magnoliophyta | 3 |
| Spermatophyta | 2 |
| Euphyllophyta | 2 |
| Embryophyta | 1 |
| Viridiplantae | 1 |
Cumulative family count (Top 20)
| class name | count |
|---|---|
|
Rutaceae
|
4 |
|
Orchidaceae
|
4 |
|
Zingiberaceae
|
3 |
|
Hymenochaetaceae
|
3 |
|
Lauraceae
|
3 |
|
Poaceae
|
3 |
|
Asteraceae
|
3 |
|
Posidoniaceae
|
2 |
|
Euphorbiaceae
|
2 |
|
Pteridaceae
|
2 |
|
Viscaceae
|
2 |
|
Fabaceae
|
2 |
|
Cupressaceae
|
2 |
|
Caryophyllaceae
|
2 |
|
Begoniaceae
|
1 |
|
Rubiaceae
|
1 |
|
Asparagaceae
|
1 |
|
Cardiopteridaceae
|
1 |
|
Lycopodiaceae
|
1 |
|
Styelidae
|
1 |
KEGG BRITE br08003
Categories (0)
| br08003 Category | # of metabolite |
|---|
metabolites link (0)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|
Metabolite list (5)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
figure |
|---|---|---|---|---|---|---|
|
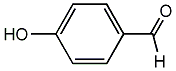
C00002657
|
p-Formylphenol
/ 4-Hydroxybenzaldehyde / p-Hydroxybenzaldehyde |
CHEMBL14193
|
C011483
|
3 / 2 / 2 |
|
|
|
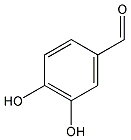
C00018099
|
Catechaldehyde
/ Protocatechualdehyde / 3,4-Dihydroxybenzaldehyde |
CHEMBL222021
|
C005581
|
7 / 0 / 0 | 5 / 2 |
|
|
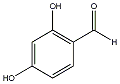
C00029435
|
2,4-Dihydroxybenzaldehyde
|
CHEMBL243587
|
2 / 0 / 0 |
|
||
|
C00029760
|
Atranol
|
CHEMBL223288
|
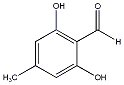
|
|||
|
C00031273
|
Salicylaldehyde
|
CHEMBL108925
|
C013243
|
2 / 4 / 2 |
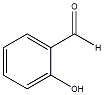
|
Human Protein / Gene in interactions
12 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00018099 C00029435 | 0 / 0 |
| O00519 | Fatty-acid amide hydrolase 1 | Enzyme | C00029435 C00031273 | 0 / 0 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00002657 | 1 / 1 |
| P51151 | Ras-related protein Rab-9A | Unclassified protein | C00018099 | 0 / 0 |
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00002657 | 1 / 1 |
| P14679 | Tyrosinase | Oxidoreductase | C00031273 | 4 / 2 |
| P14902 | Indoleamine 2,3-dioxygenase 1 | Enzyme | C00002657 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00018099 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00018099 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00018099 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00018099 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00018099 | 0 / 0 |
5 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00018099
|
| 6285 | S100B, NEF, S100, S100-B, S100beta | S100 calcium binding protein B |
C00018099
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00018099
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00018099
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00018099
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (6)
| OMIM | preferred title | UniProt |
|---|---|---|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|