Bruguiera sexangula var.rhynchopetala
Species
KNApSAcK Entry
| Organism name | Bruguiera sexangula var.rhynchopetala |
|---|---|
| Genus | Bruguiera |
| Family | Rhizophoraceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Bruguiera |
|---|---|
| Linked NCBI taxonomy ID | 39983 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Rhizophoraceae |
|---|---|
| ID | 40029 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (14)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00005737
|
Myricetin 3-rutinoside
/ Myricetin-3-O-rutinoside |
No. 1 | No. 15 |
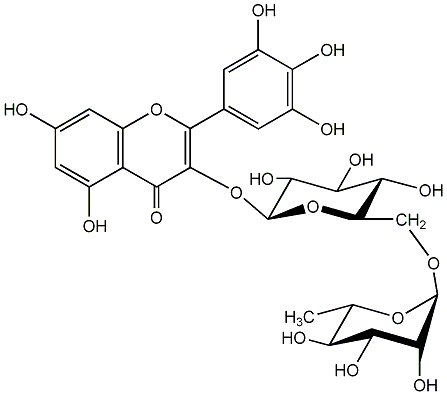
|
||||
|
C00005169
|
Nicotiflorin
/ Nicotifloroside / Kaempferol 3-O-rutinoside / Kaempferol 3-O-(alpha-L-rhamnopyranosyl(1->6)-beta-D-glucopyranoside / (-)-Kaempferol 3-O-(alpha-L-rhamnopyranosyl(1->6)-beta-D-glucopyranoside |
CHEMBL431610
CHEMBL79790 CHEMBL255020 CHEMBL501550 CHEMBL498879 CHEMBL1419228 CHEMBL1875691 |
22 / 10 / 12 | No. 1 | No. 15 |
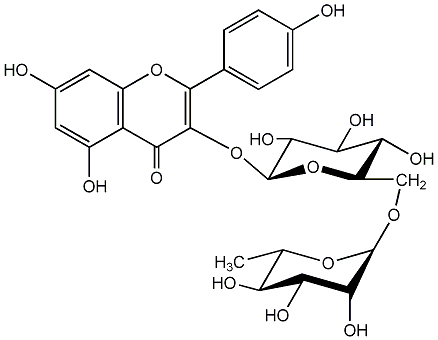
|
||
|
C00005413
|
Rutin
/ Birutan / 3-Rutinosylquercetin / Quercetin 3-O-rutinoside / (+)-Quercetin 3-O-rutinoside / Quercetin 3-O-beta-rutinoside / (+)-Quercetin 3-O-beta-rutinoside / 3,3',4',5,7-Pentahydroxyflavone 3-rutinoside / Quercetin 3-O-alpha-L-rhamnopyranosyl-(1->6)-beta-D-glucopyranoside |
CHEMBL32579
CHEMBL310754 CHEMBL182108 CHEMBL226335 CHEMBL502782 CHEMBL1436093 CHEMBL1532989 |
D012431
|
25 / 18 / 16 | 29 / 8 | No. 1 | No. 15 |
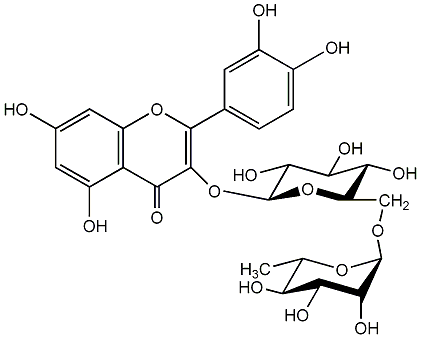
|
|
C00004444
|
Tricin 7-glucoside
/ Tricin 7-O-glucoside / Tricin 7-O-beta-D-glucopyranoside |
No. 2 | No. 15 |
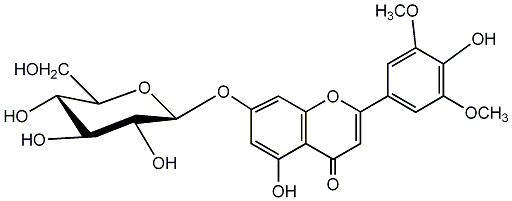
|
||||
|
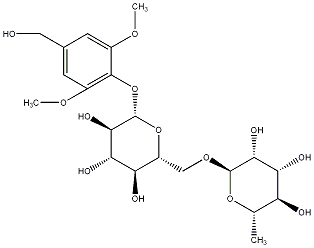
C00031169
|
Rhyncoside D
/ (-)-Rhyncoside D |
No. 128 | No. 72 |
|
||||
|
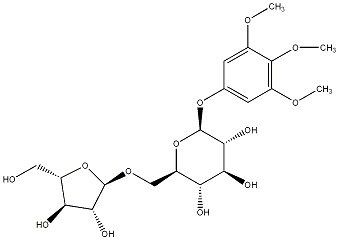
C00031168
|
Rhyncoside C
/ (-)-Rhyncoside C |
No. 128 | No. 72 |
|
||||
|
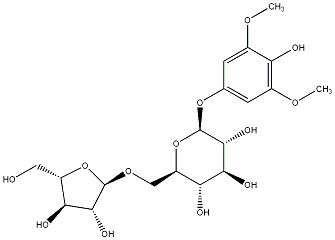
C00031167
|
Rhyncoside B
/ (-)-Rhyncoside B |
No. 128 | No. 72 |
|
||||
|
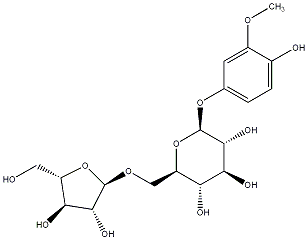
C00031166
|
Rhyncoside A
|
No. 128 | No. 72 |
|
||||
|
C00030699
|
Lyoniside
/ (+)-Lyoniside |
CHEMBL466738
CHEMBL583884 |
No. 174 | No. 22 |

|
|||
|
C00030465
|
Hedyotisol C
|
CHEMBL388779
CHEMBL1761832 |
No. 907 |

|
||||
|
C00030464
|
Hedyotisol B
|
CHEMBL388779
CHEMBL1761832 |
No. 907 |

|
||||
|
C00030463
|
Hedyotisol A
|
CHEMBL388779
CHEMBL1761832 |
No. 907 |

|
||||
|
C00031170
|
Rhyncoside E
|
No. 907 |

|
|||||
|
C00031171
|
Rhyncoside F
|
No. 907 |

|
Human Protein / Gene in interactions
41 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P06746 | DNA polymerase beta | Enzyme | C00005169 C00005413 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00005169 C00005413 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00005169 C00005413 | 0 / 0 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00005169 C00005413 | 1 / 0 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00005169 C00005413 | 1 / 1 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00005169 C00005413 | 1 / 1 |
| P14679 | Tyrosinase | Oxidoreductase | C00005413 | 4 / 2 |
| P33527 | Multidrug resistance-associated protein 1 | drugs | C00005413 | 0 / 0 |
| P18825 | Alpha-2C adrenergic receptor | Adrenergic receptor | C00005413 | 0 / 0 |
| P07237 | Protein disulfide-isomerase | Enzyme | C00005169 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00005413 | 0 / 3 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00005413 | 0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00005169 | 2 / 3 |
| P39748 | Flap endonuclease 1 | Enzyme | C00005169 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00005169 | 0 / 0 |
| P08913 | Alpha-2A adrenergic receptor | Adrenergic receptor | C00005413 | 0 / 0 |
| P27487 | Dipeptidyl peptidase 4 | S9B | C00005413 | 0 / 1 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00005169 | 1 / 1 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00005169 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00005413 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00005169 | 0 / 0 |
| P06276 | Cholinesterase | Hydrolase | C00005413 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00005413 | 0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00005413 | 3 / 3 |
| Q9UNQ0 | ATP-binding cassette sub-family G member 2 | ATP binding cassette | C00005413 | 2 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00005413 | 1 / 1 |
| P22303 | Acetylcholinesterase | Hydrolase | C00005413 | 1 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00005413 | 0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00005169 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00005169 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00005169 | 1 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00005413 | 4 / 3 |
| P48147 | Prolyl endopeptidase | S9A | C00005413 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00005169 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00005413 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00005169 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00005413 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00005169 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00005169 | 1 / 4 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00005169 | 0 / 0 |
| Q14191 | Werner syndrome ATP-dependent helicase | Enzyme | C00005169 | 2 / 1 |
29 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00005413
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00005413
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00005413
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00005413
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00005413
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00005413
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00005413
|
| 873 | CBR1, CBR, SDR21C1, hCBR1 | carbonyl reductase 1 (EC:1.1.1.189 1.1.1.197 1.1.1.184) |
C00005413
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00005413
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00005413
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00005413
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00005413
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00005413
|
| 2936 | GSR | glutathione reductase (EC:1.8.1.7) |
C00005413
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00005413
|
| 3480 | IGF1R, CD221, IGFIR, IGFR, JTK13 | insulin-like growth factor 1 receptor (EC:2.7.10.1) |
C00005413
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00005413
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00005413
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00005413
|
| 3667 | IRS1, HIRS-1 | insulin receptor substrate 1 |
C00005413
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00005413
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00005413
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00005413
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00005413
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00005413
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00005413
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00005413
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00005413
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00005413
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (25)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #614490 | Blood group, junior system; jr |
Q9UNQ0
|
| #114500 | Colorectal cancer; crc |
Q14191
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #138900 | Uric acid concentration, serum, quantitative trait locus 1; uaqtl1 |
Q9UNQ0
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #277700 | Werner syndrome; wrn |
Q14191
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (26)
| KEGG | name | UniProt |
|---|---|---|
| H00026 | Endometrial Cancer |
P03372
(marker)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00038 | Malignant melanoma |
P14679
(marker)
|
| H00032 | Thyroid cancer |
P27487
(marker)
|
| H00017 | Esophageal cancer |
P35354
(related)
|
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00296 | Defects in RecQ helicases |
Q14191
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
8 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D001424 | Bacterial Infections |
C00005413
|
| D003092 | Colitis |
C00005413
|
| D004409 | Dyskinesia, Drug-Induced |
C00005413
|
| D015212 | Inflammatory Bowel Diseases |
C00005413
|
| D007674 | Kidney Diseases |
C00005413
|
| D028361 | Mitochondrial Diseases |
C00005413
|
| D010243 | Paralysis |
C00005413
|
| D013276 | Stomach Ulcer |
C00005413
|