Enantia chlorantha
Species
KNApSAcK Entry
| Organism name | Enantia chlorantha |
|---|---|
| Genus | Enantia |
| Family | Annonaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Enantia |
|---|---|
| Linked NCBI taxonomy ID | 320276 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Pieridae |
|---|---|
| ID | 7114 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Metazoa |
|---|---|
| ID | 33208 |
Plant class
| Plant class | |
|---|---|
| ID |
Metabolite list (11)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00029094
|
Xylopinine
/ Tetrahydropseudopalmatine |
CHEMBL465824
CHEMBL2357282 |
1 / 0 / 0 | No. 37 | No. 4 |

|
||
|
C00026149
|
Corypalmine
/ Tetrahydrojatrorrhizine |
CHEMBL2334885
CHEMBL2334886 |
4 / 3 / 0 | No. 37 | No. 4 |
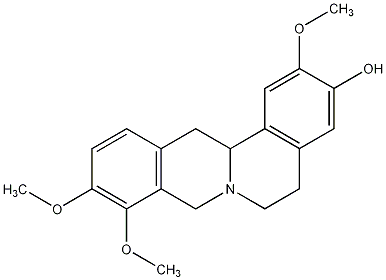
|
||
|
C00026150
|
Tetrahydropalmatine
/ dl-Tetrahydropalmatine / (+/-)-Tetrahydropalmatine |
CHEMBL187892
CHEMBL487182 CHEMBL2334889 |
8 / 5 / 2 | No. 37 | No. 4 |
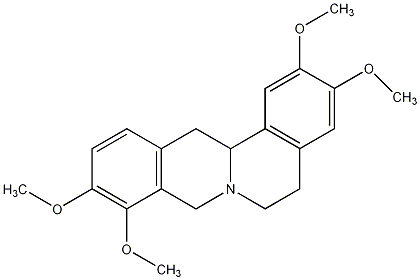
|
||
|
C00025632
|
Isocorypalmine
/ (-)-Isocorypalmine / Tetrahydrocolumbamine / O10-Methylstepholidine |
CHEMBL2334891
CHEMBL2334892 |
4 / 3 / 0 | No. 37 | No. 4 |

|
||
|
C00025890
|
Liridine
/ Homomoschatoline / O-Methylmoschatoline / O-Methylisomoschatoline |
CHEMBL227533
|
No. 74 |

|
||||
|
C00001878
|
Liriodenine
/ Oxoushinsunine / Spermatheridine |
CHEMBL37736
|
C026980
|
5 / 3 / 1 | No. 74 |

|
||
|
C00025935
|
Lysicamine
/ Oxonuciferine |
CHEMBL510090
|
C069090
|
No. 74 |
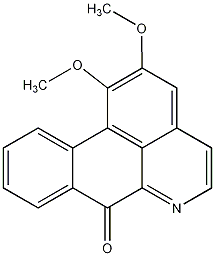
|
|||
|
C00026143
|
Govanine
/ Pseudocolumbamine |
CHEMBL1197451
|
No. 155 | No. 4 |

|
|||
|
C00025342
|
Pseudopalmatine
/ 5,6-Dihydro-8-demethylcoralyne |
CHEMBL376300
|
No. 155 | No. 4 |

|
|||
|
C00027159
|
Depiline
/ Palmatine / Berbericinine / O,O-Dimethyldemethyleneberberine |
CHEMBL206106
|
C005413
|
8 / 13 / 13 | 8 / 0 | No. 155 | No. 4 |

|
|
C00001874
|
Neprotin
/ Neprotine / Yatrorizine / Jatrorrhizine |
CHEMBL251055
|
C055785
|
14 / 27 / 20 | 1 / 0 | No. 155 | No. 4 |

|
Human Protein / Gene in interactions
32 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00001874 C00026150 C00027159 | 0 / 1 |
| P08908 | 5-hydroxytryptamine receptor 1A | Serotonin receptor | C00025632 C00026149 C00026150 | 1 / 0 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00025632 C00026149 C00026150 | 0 / 0 |
| P14416 | D(2) dopamine receptor | Dopamine receptor | C00025632 C00026149 C00026150 | 2 / 0 |
| P28223 | 5-hydroxytryptamine receptor 2A | Serotonin receptor | C00025632 C00026149 C00026150 | 0 / 0 |
| P43351 | DNA repair protein RAD52 homolog | Unclassified protein | C00001874 C00029094 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00001874 C00027159 | 11 / 10 |
| P39748 | Flap endonuclease 1 | Enzyme | C00001874 | 0 / 0 |
| P30926 | Neuronal acetylcholine receptor subunit beta-4 | CHRN beta | C00001878 | 0 / 0 |
| P32297 | Neuronal acetylcholine receptor subunit alpha-3 | CHRN alpha | C00001878 | 1 / 0 |
| P17787 | Neuronal acetylcholine receptor subunit beta-2 | CHRN beta | C00001878 | 1 / 1 |
| P43681 | Neuronal acetylcholine receptor subunit alpha-4 | CHRN alpha | C00001878 | 1 / 1 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00001874 | 1 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00001874 | 1 / 1 |
| Q14761 | Protein tyrosine phosphatase receptor type C-associated protein | Enzyme | C00001878 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00027159 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00027159 | 0 / 1 |
| Q99720 | Sigma non-opioid intracellular receptor 1 | Membrane receptor | C00026150 | 1 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00027159 | 0 / 0 |
| P13726 | Tissue factor | Membrane receptor | C00026150 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00027159 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00026150 | 1 / 1 |
| P04062 | Glucosylceramidase | Enzyme | C00001874 | 6 / 4 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00027159 | 1 / 1 |
| P06746 | DNA polymerase beta | Enzyme | C00001874 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00001874 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00001874 | 4 / 3 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00001874 | 4 / 1 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00001874 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00001874 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00001874 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00027159 | 1 / 0 |
8 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00001874
C00027159
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00027159
|
| 6348 | CCL3, G0S19-1, LD78ALPHA, MIP-1-alpha, MIP1A, SCYA3 | chemokine (C-C motif) ligand 3 |
C00027159
|
| 6351 | CCL4, ACT2, AT744.1, G-26, HC21, LAG-1, LAG1, MIP-1-beta, MIP1B, MIP1B1, SCYA2, SCYA4 | chemokine (C-C motif) ligand 4 |
C00027159
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00027159
|
| 3570 | IL6R, CD126, IL-6R-1, IL-6RA, IL6Q, IL6RA, IL6RQ, gp80 | interleukin 6 receptor |
C00027159
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00027159
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00027159
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (37)
| OMIM | preferred title | UniProt |
|---|---|---|
| #103780 | Alcohol dependence |
P14416
|
| #614373 | Amyotrophic lateral sclerosis 16, juvenile; als16 |
Q99720
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #600513 | Epilepsy, nocturnal frontal lobe, 1; enfl1 |
P43681
|
| #605375 | Epilepsy, nocturnal frontal lobe, 3; enfl3 |
P17787
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #608013 | Gaucher disease, perinatal lethal |
P04062
|
| #230800 | Gaucher disease, type i |
P04062
|
| #230900 | Gaucher disease, type ii |
P04062
|
| #231000 | Gaucher disease, type iii |
P04062
|
| #231005 | Gaucher disease, type iiic |
P04062
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #159900 | Myoclonic dystonia |
P14416
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #168600 | Parkinson disease, late-onset; pd |
P04062
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #614674 | Periodic fever, menstrual cycle-dependent |
P08908
|
| #172700 | Pick disease of brain |
P10636
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #612052 | Smoking as a quantitative trait locus 3; sqtl3 |
P32297
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
KEGG DISEASE (24)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
|
| H00066 | Lewy body dementia (LBD) |
P04062
(related)
|
| H00126 | Gaucher disease |
P04062
(related)
|
| H00426 | Defects in the degradation of ganglioside |
P04062
(related)
|
| H00810 | Progressive myoclonic epilepsy (PME) |
P04062
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00807 | Autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) |
P17787
(related)
P43681 (related) |
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|