Curcuma comosa
Species
KNApSAcK Entry
| Organism name | Curcuma comosa |
|---|---|
| Genus | Curcuma |
| Family | Zingiberaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Curcuma comosa |
|---|---|
| Linked NCBI taxonomy ID | 199633 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Zingiberaceae |
|---|---|
| ID | 4642 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | Liliopsida |
|---|---|
| ID | 4447 |
Metabolite list (3)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
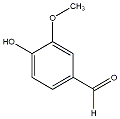
C00029531
|
Vanillic aldehyde
/ 4-Hydroxy-3-methoxybenzaldehyde |
CHEMBL13883
|
C100058
|
18 / 8 / 9 | 33 / 1 | No. 1003 |
|
|
|
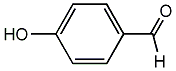
C00002657
|
p-Formylphenol
/ 4-Hydroxybenzaldehyde / p-Hydroxybenzaldehyde |
CHEMBL14193
|
C011483
|
3 / 2 / 2 | No. 2076 |
|
||
|
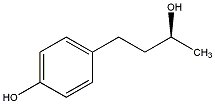
C00029306
|
(+)-Rhododendrol
|
CHEMBL108014
CHEMBL1086681 CHEMBL1778763 |
1 / 0 / 0 | No. 4115 |
|
Human Protein / Gene in interactions
20 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00002657 C00029531 | 1 / 1 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00002657 C00029531 | 1 / 1 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00029306 | 0 / 0 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00029531 | 0 / 0 |
| P34969 | 5-hydroxytryptamine receptor 7 | Serotonin receptor | C00029531 | 0 / 0 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00029531 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00029531 | 0 / 0 |
| P14902 | Indoleamine 2,3-dioxygenase 1 | Enzyme | C00002657 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00029531 | 0 / 0 |
| O00519 | Fatty-acid amide hydrolase 1 | Enzyme | C00029531 | 0 / 0 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00029531 | 0 / 0 |
| P78527 | DNA-dependent protein kinase catalytic subunit | Atypical serine/threonine protein kinase PIKK subfamily | C00029531 | 0 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00029531 | 0 / 0 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00029531 | 2 / 2 |
| Q9Y4X1 | UDP-glucuronosyltransferase 2A1 | Enzyme | C00029531 | 0 / 0 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00029531 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00029531 | 0 / 0 |
| P10275 | Androgen receptor | NR3C4 | C00029531 | 3 / 4 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00029531 | 1 / 1 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00029531 | 0 / 0 |
33 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1646 | AKR1C2, AKR1C-pseudo, BABP, DD, DD2, DDH2, HAKRD, HBAB, MCDR2, SRXY8 | aldo-keto reductase family 1, member C2 (EC:1.3.1.20 1.1.1.357) |
C00029531
|
| 759 | CA1, CA-I, CAB, Car1 | carbonic anhydrase I (EC:4.2.1.1) |
C00029531
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00029531
|
| 1196 | CLK2, hCLK2 | CDC-like kinase 2 (EC:2.7.12.1) |
C00029531
|
| 1490 | CTGF, CCN2, HCS24, IGFBP8, NOV2 | connective tissue growth factor |
C00029531
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00029531
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00029531
|
| 1789 | DNMT3B, ICF, ICF1, M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta (EC:2.1.1.37) |
C00029531
|
| 1848 | DUSP6, HH19, MKP3, PYST1 | dual specificity phosphatase 6 (EC:3.1.3.16 3.1.3.48) |
C00029531
|
| 2263 | FGFR2, BBDS, BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 | fibroblast growth factor receptor 2 (EC:2.7.10.1) |
C00029531
|
| 2535 | FZD2, Fz2, fz-2, fzE2, hFz2 | frizzled family receptor 2 |
C00029531
|
| 2729 | GCLC, GCL, GCS, GLCL, GLCLC | glutamate-cysteine ligase, catalytic subunit (EC:6.3.2.2) |
C00029531
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00029531
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00029531
|
| 3251 | HPRT1, HGPRT, HPRT | hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8) |
C00029531
|
| 3304 | HSPA1B, HSP70-1B, HSP70-2 | heat shock 70kDa protein 1B |
C00029531
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00029531
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00029531
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00029531
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00029531
|
| 57787 | MARK4, MARK4L, MARK4S, MARKL1, MARKL1L, PAR-1D | MAP/microtubule affinity-regulating kinase 4 (EC:2.7.11.1) |
C00029531
|
| 4502 | MT2A, MT2 | metallothionein 2A |
C00029531
|
| 5514 | PPP1R10, CAT53, FB19, PNUTS, PP1R10, R111, p99 | protein phosphatase 1, regulatory subunit 10 |
C00029531
|
| 11046 | SLC35D2, HFRC1, SQV7L, UGTrel8, hfrc | solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
C00029531
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00029531
|
| 7041 | TGFB1I1, ARA55, HIC-5, HIC5, TSC-5 | transforming growth factor beta 1 induced transcript 1 |
C00029531
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00029531
|
| 10626 | TRIM16, EBBP | tripartite motif containing 16 |
C00029531
|
| 7360 | UGP2, UDPG, UDPGP, UDPGP2, UGP1, UGPP1, UGPP2, pHC379 | UDP-glucose pyrophosphorylase 2 (EC:2.7.7.9) |
C00029531
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00029531
|
| 54659 | UGT1A3, UDPGT, UDPGT_1-3, UGT-1C, UGT1-03, UGT1.3, UGT1C | UDP glucuronosyltransferase 1 family, polypeptide A3 (EC:2.4.1.17) |
C00029531
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00029531
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00029531
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (8)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
KEGG DISEASE (9)
| KEGG | name | UniProt |
|---|---|---|
| H00024 | Prostate cancer |
P10275
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q9NUW8
(related)
|