KEGG Mapping
KEGG mapping as a set operation
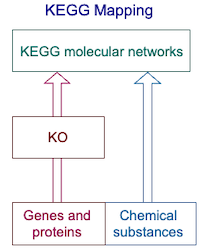 KEGG mapping is the process to map molecular objects (genes, proteins, small molecules, etc.) to molecular network objects (KEGG pathway maps, BRITE hierarchies and KEGG modules). It is not simply an enrichment process; rather it is a set operation to generate a new set. From the beginning of the KEGG project, the basic idea was to automatically generate organism-specific pathways by the set operation between manually created pathway maps (called reference pathway maps) and an annotated set of genes in the genome, originally (until 1999) annotated with EC numbers and then (since 2000) annotated with KO (KEGG Orthology) identifiers or K numbers. Thus, the KEGG mapping set operation has played a role to expand the KEGG knowledge base by converting KOs to individual gene IDs in many organisms (KO expansion). In addition, the KEGG mapping procedure has been used to assist integration and interpretation of users' datasets, especially large-scale datasets generated by high-throughput technologies (see: KEGG Mapper tools).
KEGG mapping is the process to map molecular objects (genes, proteins, small molecules, etc.) to molecular network objects (KEGG pathway maps, BRITE hierarchies and KEGG modules). It is not simply an enrichment process; rather it is a set operation to generate a new set. From the beginning of the KEGG project, the basic idea was to automatically generate organism-specific pathways by the set operation between manually created pathway maps (called reference pathway maps) and an annotated set of genes in the genome, originally (until 1999) annotated with EC numbers and then (since 2000) annotated with KO (KEGG Orthology) identifiers or K numbers. Thus, the KEGG mapping set operation has played a role to expand the KEGG knowledge base by converting KOs to individual gene IDs in many organisms (KO expansion). In addition, the KEGG mapping procedure has been used to assist integration and interpretation of users' datasets, especially large-scale datasets generated by high-throughput technologies (see: KEGG Mapper tools).
Here the network objects of pathway maps and Brite hierarchies are explained.
KEGG Pathway Maps
Graphical map objects
The KEGG pathway map is a moleculalr interaction/reaction network diagram represented in terms of KO functional orthologs, so that experimental evidence in specific organisms can be generalized to other organisms. Each map is manually drawn with in-house software called KegSketch, which generates the KGML+ file, an XML file with SVG graphics data. Basic graphics objects in the regular reference pathway maps are:
Convention of map number prefix
- boxes - KOs identified by K numbers and enzymatic reactions identified by R numbers
- circles - other molecules, mostly chemical substances identified by C/G/D numbers, but including DNAs, peptides, etc. that are not subject to KO expansion
- lines - non-enzymatic reactions identified by R numbers
- lines - KOs identified by K numbers and reactions identified by R numbers
- circles - chemical substances identified by C/G numbers
Each pathway map is identified by the combination of 2-4 letter code and 5 digit number (see KEGG Identifiers). The prefix has the following meaning:
As shown here, "map" pathways are not colored, "ko/ec/rn" pathways are colored blue, and organism-specific pathways are colored green, where coloring indicates that map objects exist and are linked to corresponding entries.
For global metabolism maps, "map" pathways are fully colored, so that "ko/ec/rn" pathways and organism-specific pathways are generated by reducing the coloring indicating the absence of corresponding entries.
About KGML files
- map - Reference pathway
- ko - Reference pathway (KO)
- ec - Reference pathway (EC)
- rn - Reference pathway (Reaction)
- org - Organism-specific pathway map
map00010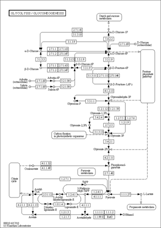 |
ko00010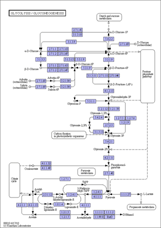 |
hsa00010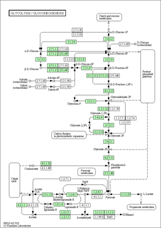 |
As shown here, "map" pathways are not colored, "ko/ec/rn" pathways are colored blue, and organism-specific pathways are colored green, where coloring indicates that map objects exist and are linked to corresponding entries.
For global metabolism maps, "map" pathways are fully colored, so that "ko/ec/rn" pathways and organism-specific pathways are generated by reducing the coloring indicating the absence of corresponding entries.
KGML is an exchange format of KEGG pathway maps. It is meant for outside users and is not used in any service or database update procedure within KEGG. KGML files, which are computationally generated from the manually defined KGML+ file, contain information about entries (KEGG objects) and two types of relationships.
- relations - relationships between boxes
- reactions - relationships between circles
BRITE Functional Hierarchies
BRITE hierarchy files
The KEGG BRITE database is a collection of BRITE hierarchy files, called htext (hierarchical text) files, with additional files for binary relations. The htext file is manually created with in-house software called KegHierEditor. The htext file contains "A", "B", "C", etc. at the first column to indicate the hierarchy level.
Each BRITE hierarchy file represents a classification system of KEGG objects identified by the KEGG Identifiers; for example, pathway-based gene classification or protein family classification by the K numbers, compound classification by C numbers, drug classification by D numbers, and disease classification by H numbers.
The binary relation files contain the relationship between KEGG objects and attributes, which can be dynamically added to the hierarchy file as additional columns using the join feature of the Brite hierarchy viewer. Many binary relation files are computationally generated from the KEGG database contents and shown in the left panel of the Brite hierarchy viewer.
Convention of brite number prefix
A Metabolism B Carbohydrate Metabolism C 00010 Glycolysis / Gluconeogenesis [PATH:ko00010] D K00844 HK; hexokinase [EC:2.7.1.1] D K12407 GCK; glucokinase [EC:2.7.1.2] D K00845 glk; glucokinase [EC:2.7.1.2] D ......
The binary relation files contain the relationship between KEGG objects and attributes, which can be dynamically added to the hierarchy file as additional columns using the join feature of the Brite hierarchy viewer. Many binary relation files are computationally generated from the KEGG database contents and shown in the left panel of the Brite hierarchy viewer.
Each BRITE hierarchy file is identified by the combination of 2-4 letter code and 5 digit number (see KEGG Identifiers). The prefix has the following meaning:
- ko - Reference hierarchy (KO)
- org - Organism-specific hierarchy
- br - Non-KO hierarchy
- jp - Non-KO hierarchy in Japanese
Last updated: November 7, 2025
