Chemical Reactions
KEGG REACTION is a database of chemical reactions, mostly enzymatic reactions, containing all reactions that appear in the KEGG metabolic pathway maps and additional reactions that appear only in the Enzyme Nomenclature.
Each reaction is identified by the R number, such as R00259 for the acetylation of L-glutamate.
Reactions are linked to enzyme KOs as defined by the KO database, enabling integrated analysis of genomic (enzyme genes) and chemical (compound pairs) information.
Reaction Class
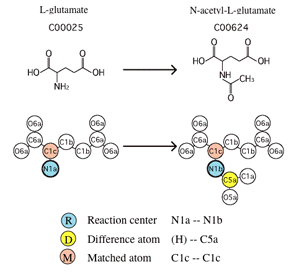
KEGG RCLASS contains classification of reactions based on the chemical structure transformation patterns of substrate-product pairs (reactant pairs), which are represented by the so-called RDM patterns. For example, R00259 is a reaction defined by:
- Acetyl-CoA + L-Glutamate <=> CoA + N-Acetyl-L-glutamate
In order to distinguish functional groups and microenvironments of atoms, atomic species of C, N, O, S, and P are classified into 68 types, called KEGG atom types. They were first introduced for detecting biochemical similarities by graph-based chemical structure comparison [1].
The RDM pattern is defined as KEGG atom type changes at the reaction center (R), the difference region (D), and the matched region (M) for each reactant pair. It characterizes chemical structure transformation patterns associated with enzymatic reactions [2].
Currently, the reaction class is defined for each unique RDM pattern or a unique combination of RDM patters when more than one reaction center is identified for a reactant pair.
Currently, the reaction class is defined for each unique RDM pattern or a unique combination of RDM patters when more than one reaction center is identified for a reactant pair.
Reaction Module
The reaction class represents an identical reaction (in the sense of the identical RDM pattern) with varying overall structures of substrates and products. By the systematic analysis of all the KEGG metabolic pathways, conserved sequences of identical reaction classes were identified [3]. They are called the reaction modules.
Reaction Pathway Search and Prediction
For any sequence of reactions or reaction classes, the following tools may be used to search similar reaction sequences.
- PathSearch: search for similar reaction pathways
- KEGG Mapper: Search
- E-zyme: automatic assignment of EC numbers
- PathPred: prediction of biodegradation/biosynthetic pathways
References
- Hattori, M., Okuno, Y., Goto, S., and Kanehisa, M.; Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J. Am. Chem. Soc. 125, 11853-11865 (2003). [pubmed]
- Kotera, M., Okuno, Y., Hattori, M., Goto, S., and Kanehisa, M.; Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J. Am. Chem. Soc. 126, 16487-16498 (2004). [pubmed]
- Muto, A., Kotera, M., Tokimatsu, T., Nakagawa, Z., Goto, S., and Kanehisa, M.; Modular architecture of metabolic pathways revealed by conserved sequences of reactions. J. Chem. Inf. Model. 53, 613-622 (2013). [pubmed] [pdf]
- Yamanishi, Y., Hattori, M., Kotera, M., Goto, S., and Kanehisa, M.; E-zyme: predicting potential EC numbers from the chemical transformation pattern of substrate-product pairs. Bioinformatics 25, i79-i86 (2009). [pubmed] [pdf]
- Oh, M., Yamada, T., Hattori, M., Goto, S., and Kanehisa, M.; Systematic analysis of enzyme-catalyzed reaction patterns and prediction of microbial biodegradation pathways. J. Chem. Inf. Model. 47, 1702-1712 (2007). [pubmed]
- Moriya, Y., Shigemizu, D., Hattori, M., Tokimatsu, T., Kotera, M., Goto, S., and Kanehisa, M.; PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 38, W138-W143 (2010). [pubmed] [pdf]
Last updated: September 8, 2023