phytochemical cluster No. 27 (47 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| Liliopsida | 79 |
| rosids | 18 |
| Embryophyta | 4 |
| eudicotyledons | 4 |
Cumulative family count
| class name | count |
|---|---|
| Orchidaceae | 52 |
| Dioscoreaceae | 27 |
| Combretaceae | 17 |
| Papaveraceae | 4 |
| Marchantiaceae | 4 |
| Betulaceae | 1 |
KEGG BRITE br08003
Categories (1)
| br08003 Category | # of metabolite |
|---|---|
| Phenanthrenes | 3 |
metabolites link (3)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|---|---|
| Phenanthrenes | C10246 | C00000325 |
| Phenanthrenes | C10244 | C00002869 |
| Phenanthrenes | C10265 | C00002886 |
KCF-S cluster (1)
| KCF-S ID | # of metabolite |
|---|---|
| No. 104 | 47 |
Metabolite list (47)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
figure |
|---|---|---|---|---|---|---|---|
|
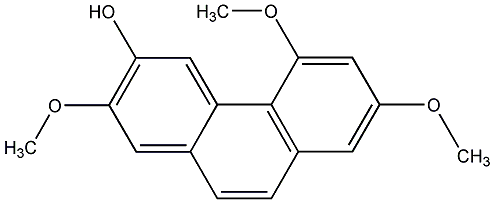
C00000325
|
2,3-Dimethoxy-7-hydroxy-phenanthrere
|
No. 104 |
|
||||
|
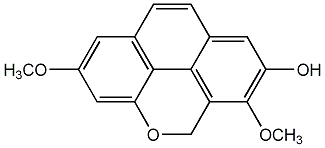
C00002869
|
2-Hydroxy-3,6-dimethoxyphenanthrene
|
No. 104 |
|
||||
|
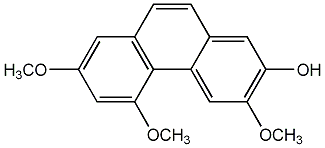
C00002886
|
3-Hydroxy-2,5-dimethoxyphenanthrene
|
No. 104 |
|
||||
|
C00015200
|
alpha-Thebaol
/ 3,6-Dimethoxy-4-hydroxyphenanthrene |
No. 104 |

|
||||
|
C00015210
|
3,7-Dimethoxy-5-hydroxyphenanthrene
|
No. 104 |
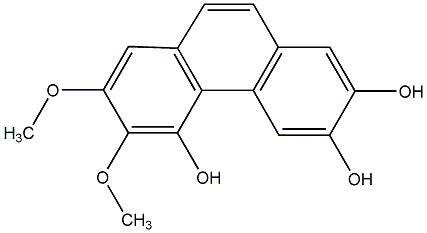
|
||||
|
C00015211
|
2,7-Dihydroxy-4,6-dimethoxyphenanthrene
|
CHEMBL509668
|
No. 104 |
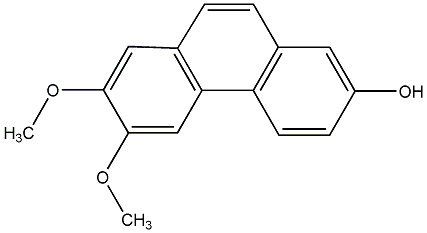
|
|||
|
C00015212
|
6,7-Dihydroxy-2,4-dimethoxyphenanthrene
|
No. 104 |
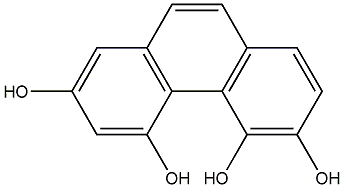
|
||||
|
C00015242
|
Batatasin I
/ 6-Hydroxy-2,4,7-trimethoxyphenanthrene |
No. 104 |
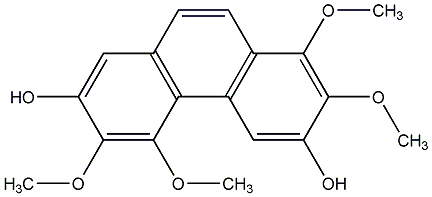
|
||||
|
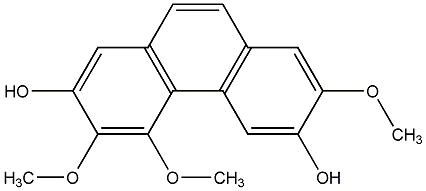
C00015244
|
Isobatatasin I
|
No. 104 |
|
||||
|
C00015257
|
O-Methylthebaol
/ O-Methyl-alpha-thebaol / 3,4,6-Trimethoxyphenanthrene |
No. 104 |
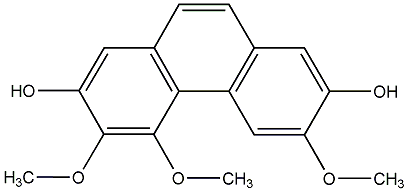
|
||||
|
C00015258
|
Flavanthrinin
|
CHEMBL447210
|
No. 104 |
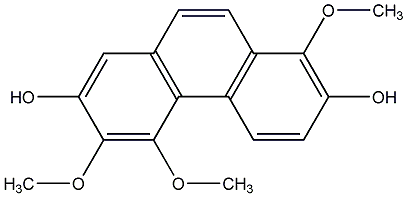
|
|||
|
C00015259
|
Lusianthrin
|
No. 104 |

|
||||
|
C00015260
|
2,7-Dihydroxy-3-methoxyphenanthrene
|
No. 104 |
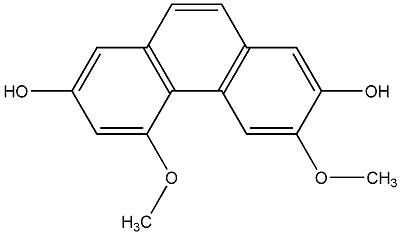
|
||||
|
C00015264
|
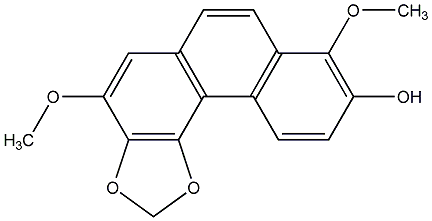
Moscatin
/ Plicatol B |
CHEMBL448260
|
C091085
|
No. 104 |
|
||
|
C00015273
|
4,6,7-Trihydroxy-2,3-dimethoxyphenanthrene
/ 2,3-Dimethoxy-4,6,7-trihydroxyphenanthrene |
No. 104 |

|
||||
|
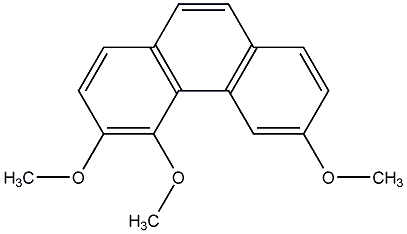
C00015302
|
4,6-Dimethoxy,2,3,7-trihydroxyphenanthrene
|
No. 104 |
|
||||
|
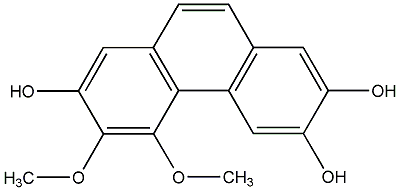
C00015318
|
3,4-Dimethoxy-2,6,7-trihydroxyphenanthrene
|
No. 104 |
|
||||
|
C00015402
|
Coeloginanthrin
|
No. 104 |

|
||||
|
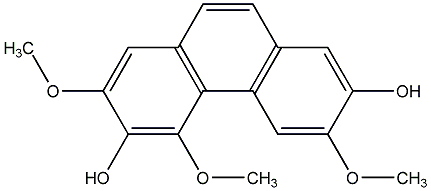
C00015403
|
4-Methoxyphenanthrene-2,3,6,7-tetrol
|
No. 104 |
|
||||
|
C00015404
|
3,7-Dihydroxy-2,4-dimethoxyphenanthrene
|
CHEMBL1083093
|
No. 104 |
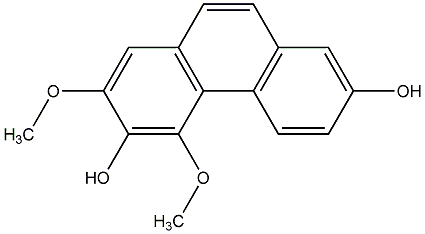
|
|||
|
C00015423
|
4,7-Dihydroxy-2,3-dimethoxyphenanthrene
|
No. 104 |

|
||||
|
C00015427
|
Nudol
|
CHEMBL253983
|
No. 104 |

|
|||
|
C00015539
|
Bulbophyllanthrin
/ 3,5-Dihydroxy-2,4-dimethoxyphenanthrene |
CHEMBL253981
|
No. 104 |
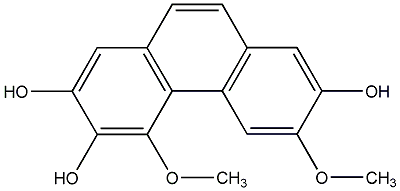
|
|||
|
C00015542
|
4-Methoxyphenanthrene-2,3,7-triol
|
No. 104 |
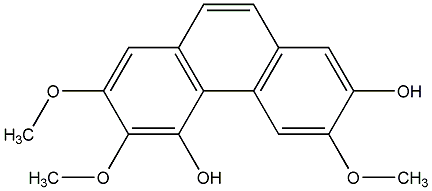
|
||||
|
C00015543
|
Fimbriol B
|
CHEMBL254188
|
No. 104 |
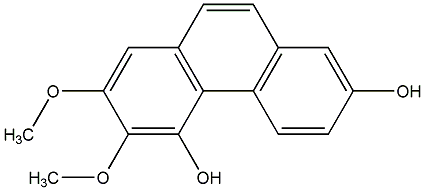
|
|||
|
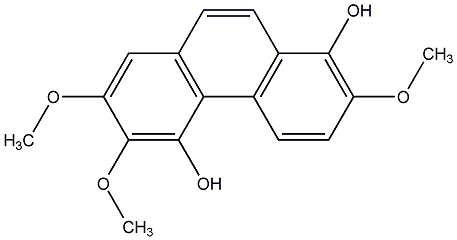
C00015547
|
2,4,5,6-Tetrahydroxyphenanthrene
|
No. 104 |
|
||||
|
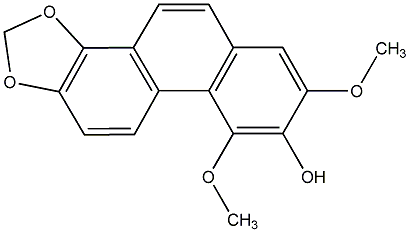
C00015590
|
2,7-Dihydroxy-3,4,6-trimethoxyphenanthrene
|
No. 104 |
|
||||
|
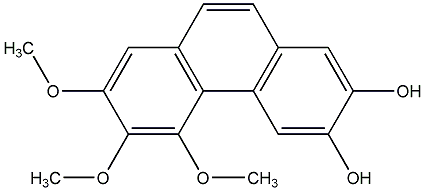
C00015608
|
6,7-Dihydroxy-2,3,4-trimethoxyphenanthrene
|
No. 104 |
|
||||
|
C00015609
|
2,6-Dihydroxy-3,4,7-trimethoxyphenanthrene
|
No. 104 |

|
||||
|
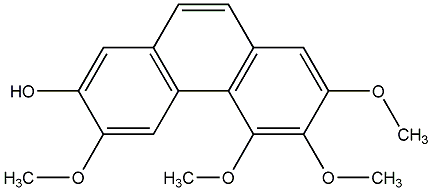
C00015685
|
3,7-Dihydroxy-2,4,6-trimethoxyphenanthrene
|
No. 104 |
|
||||
|
C00015690
|
4,7-dihydroxy-2,3,6-trimethoxyphenanthrene
|
CHEMBL1346456
|
34 / 20 / 13 | No. 104 |
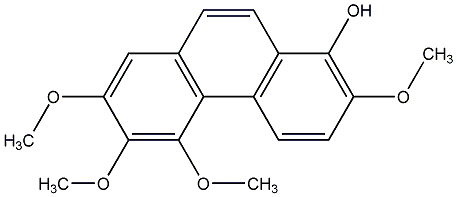
|
||
|
C00015897
|
Gymnopusin
/ 7,9-Dihydroxy-2,3,4-trimethoxyphenanthrene |
CHEMBL2208383
|
No. 104 |
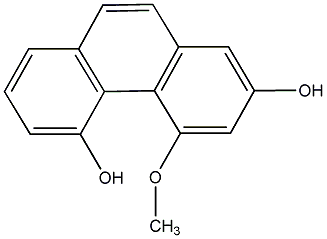
|
|||
|
C00015898
|
Ta IX
/ 4,8-Dihydroxy-2,3,7trimethoxyphenanthrene |
No. 104 |
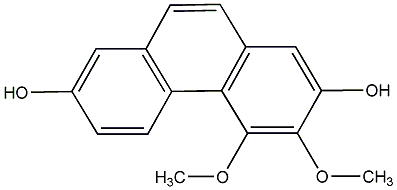
|
||||
|
C00015900
|
Denthyrsinin
|
CHEMBL469491
|
No. 104 |

|
|||
|
C00015902
|
Confusarin
/ 2,7-Dihydroxy-3,4,8-trimethoxy-9,10-dihydrophenanthrene |
No. 104 |
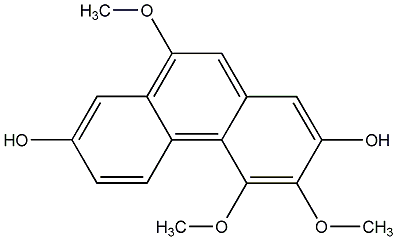
|
||||
|
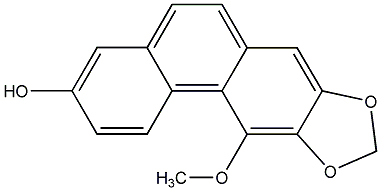
C00015903
|
Fimbriol A
|
CHEMBL2208382
|
No. 104 |
|
|||
|
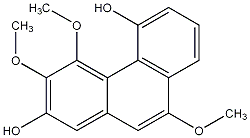
C00024152
|
Plicatol A
|
CHEMBL438339
|
No. 104 |
|
|||
|
C00024153
|
7-Hydroxy-2,3,4,6-tetramethoxyphenanthrene
|
CHEMBL421968
|
No. 104 |

|
|||
|
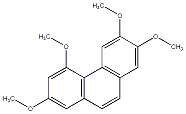
C00033691
|
8-Hydroxy-2,3,4,7-tetramethoxyphenanthrene
|
No. 104 |
|
||||
|
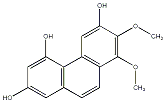
C00033729
|
Confusaridin
/ 2,6-Dihydroxy-3,4,7,8-tetramethoxy-9,10-dihydrophenanthrene |
No. 104 |
|
||||
|
C00033765
|
Agrostophyllin
|
No. 104 |
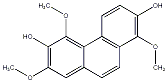
|
||||
|
C00033847
|
2,8-Dimethoxy-7-hydroxy-3,4,-methylenedioxyphenanthrene
|
No. 104 |

|
||||
|
C00034034
|
Ta IV
/ 5,7-Dimethoxy-6-hydroxy-1,2-methylenedioxyphenanthrene |
No. 104 |

|
||||
|
C00034086
|
Cirrhopetalin
/ 7-Hydroxy-4-methoxy-2,3-methylenedioxyphenanthrene |
No. 104 |

|
||||
|
C00034157
|
Ochrolic acid
|
No. 104 |
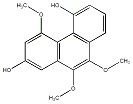
|
||||
|
C00040831
|
Callosuminin
|
No. 104 |
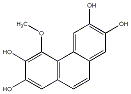
|
||||
|
C00040832
|
2,3,4,6,7-Pentamethoxyphenanthrene
|
No. 104 |

|
Human Protein / Gene in interactions
34 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q07820 | Induced myeloid leukemia cell differentiation protein Mcl-1 | Other cytosolic protein | C00015542 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00015542 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00015542 | 1 / 1 |
| P14618 | Pyruvate kinase PKM | Enzyme | C00015542 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00015542 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00015542 | 1 / 1 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00015542 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00015542 | 0 / 0 |
| P37840 | Alpha-synuclein | Unclassified protein | C00015542 | 4 / 2 |
| Q00535 | Cyclin-dependent kinase 5 | Cdk5 | C00015542 | 0 / 0 |
| Q15078 | Cyclin-dependent kinase 5 activator 1 | REG serine/threonine protein kinase family | C00015542 | 0 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00015542 | 1 / 2 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00015542 | 1 / 0 |
| P17405 | Sphingomyelin phosphodiesterase | Enzyme | C00015542 | 2 / 2 |
| P39748 | Flap endonuclease 1 | Enzyme | C00015542 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00015542 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00015542 | 1 / 1 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00015542 | 0 / 0 |
| P98073 | Enteropeptidase | Enzyme | C00015542 | 1 / 1 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00015542 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00015542 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00015542 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00015542 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00015542 | 4 / 3 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00015542 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00015542 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00015542 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00015542 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00015542 | 1 / 0 |
| Q9UBT2 | SUMO-activating enzyme subunit 2 | Enzyme | C00015542 | 0 / 0 |
| Q9UBE0 | SUMO-activating enzyme subunit 1 | Unclassified protein | C00015542 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00015542 | 0 / 0 |
| Q14191 | Werner syndrome ATP-dependent helicase | Enzyme | C00015542 | 2 / 1 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00015542 | 1 / 1 |
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (20)
| OMIM | preferred title | UniProt |
|---|---|---|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #114500 | Colorectal cancer; crc |
Q14191
|
| #127750 | Dementia, lewy body; dlb |
P37840
|
| #226200 | Enterokinase deficiency |
P98073
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #257200 | Niemann-pick disease, type a |
P17405
|
| #607616 | Niemann-pick disease, type b |
P17405
|
| #168601 | Parkinson disease 1, autosomal dominant; park1 |
P37840
|
| #605543 | Parkinson disease 4, autosomal dominant; park4 |
P37840
|
| #168600 | Parkinson disease, late-onset; pd |
P37840
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #277700 | Werner syndrome; wrn |
Q14191
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (13)
| KEGG | name | UniProt |
|---|---|---|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
Q13148 (related) |
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00137 | Niemann-Pick disease (NPD) typeA and B |
P17405
(related)
|
| H00424 | Defects in the degradation of sphingomyelin |
P17405
(related)
|
| H00057 | Parkinson's disease (PD) |
P37840
(related)
|
| H00066 | Lewy body dementia (LBD) |
P37840
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
Q14191 (related) |
| H00988 | Enterokinase deficiency |
P98073
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|