Isopyrum thalictroides
Species
KNApSAcK Entry
| Organism name | Isopyrum thalictroides |
|---|---|
| Genus | Isopyrum |
| Family | Ranunculaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Isopyrum thalictroides |
|---|---|
| Linked NCBI taxonomy ID | 432643 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Ranunculaceae |
|---|---|
| ID | 3440 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | eudicotyledons |
|---|---|
| ID | 71240 |
Metabolite list (14)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00005514
|
Rhamnetin 3-rutinoside
|
No. 1 | No. 15 |

|
||||
|
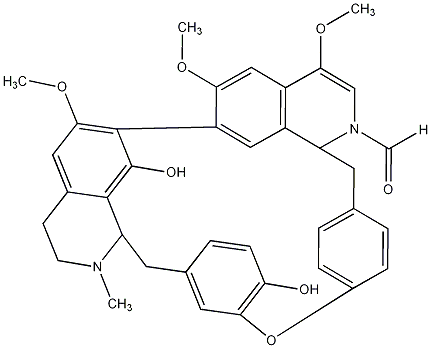
C00027396
|
Isothalmidine
/ (+)-Isothalmidine |
No. 10 | No. 4 |
|
||||
|
C00001872
|
Isocorydine
/ (+)-Isocorydine / L-(+)-Isocorydine / (S)-(+)-Isocorydine |
CHEMBL489525
CHEMBL1376826 |
14 / 7 / 10 | No. 20 | No. 4 |

|
||
|
C00039457
|
Isothalicrine
/ (+)-Isothalicrine |
No. 89 | No. 4 |

|
||||
|
C00039459
|
Isothalirine
|
No. 89 | No. 4 |

|
||||
|
C00027159
|
Depiline
/ Palmatine / Berbericinine / O,O-Dimethyldemethyleneberberine |
CHEMBL206106
|
C005413
|
8 / 13 / 13 | 8 / 0 | No. 155 | No. 4 |

|
|
C00024667
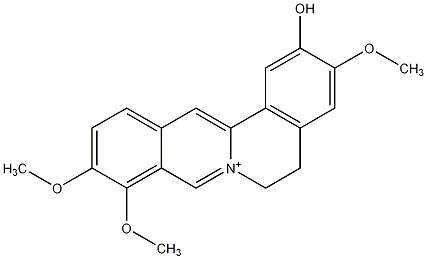
|
Columbamine
/ Dehydroisocorypalmine |
CHEMBL400345
|
C055786
|
1 / 0 / 1 | 8 / 1 | No. 155 | No. 4 |
|
|
C00026132
|
Isocoptisine
/ Pseudocoptisine |
No. 155 | No. 4 |

|
||||
|
C00001910
|
Reticuline
/ (+)-Reticuline |
CHEMBL235212
CHEMBL401501 CHEMBL464734 |
C003298
|
1 / 0 / 0 | No. 345 | No. 4 |

|
|
|
C00027393
|
Isopythaldine
|
No. 2328 |

|
|||||
|
C00027392
|
Isopyruthaldine
|
No. 2328 |

|
|||||
|
C00027146
|
Isothaliphine
/ (-)-Isothaliphine |
No. 2328 |

|
|||||
|
C00039458
|
Isothalictrine
|
No. 2328 |

|
|||||
|
C00027168
|
Thaliphine
|
No. 2328 |

|
Human Protein / Gene in interactions
18 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00001872 C00024667 C00027159 | 0 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00001872 C00027159 | 1 / 1 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00001872 C00027159 | 1 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00001872 C00027159 | 0 / 1 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00001872 C00027159 | 0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00001872 | 3 / 3 |
| O75496 | Geminin | Unclassified protein | C00001872 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00027159 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00001872 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00027159 | 0 / 0 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00001910 | 0 / 0 |
| P24941 | Cyclin-dependent kinase 2 | Cdc2 | C00001872 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00027159 | 11 / 10 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00001872 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00001872 | 1 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00001872 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00001872 | 1 / 4 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00001872 | 0 / 0 |
16 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00027159
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00027159
|
| 6348 | CCL3, G0S19-1, LD78ALPHA, MIP-1-alpha, MIP1A, SCYA3 | chemokine (C-C motif) ligand 3 |
C00027159
|
| 6351 | CCL4, ACT2, AT744.1, G-26, HC21, LAG-1, LAG1, MIP-1-beta, MIP1B, MIP1B1, SCYA2, SCYA4 | chemokine (C-C motif) ligand 4 |
C00027159
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00027159
|
| 3570 | IL6R, CD126, IL-6R-1, IL-6RA, IL6Q, IL6RA, IL6RQ, gp80 | interleukin 6 receptor |
C00027159
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00027159
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00027159
|
| 1003 | CDH5, 7B4, CD144 | cadherin 5, type 2 (vascular endothelium) |
C00024667
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00024667
|
| 1437 | CSF2, GMCSF | colony stimulating factor 2 (granulocyte-macrophage) |
C00024667
|
| 2048 | EPHB2, CAPB, DRT, EK5, EPHT3, ERK, Hek5, PCBC, Tyro5 | EPH receptor B2 (EC:2.7.10.1) |
C00024667
|
| 9500 | MAGED1, DLXIN-1, NRAGE | melanoma antigen family D, 1 |
C00024667
|
| 4313 | MMP2, CLG4, CLG4A, MMP-II, MONA, TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) |
C00024667
|
| 64321 | SOX17, VUR3 | SRY (sex determining region Y)-box 17 |
C00024667
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00024667
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (18)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
KEGG DISEASE (20)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|