Lippia dulcis TREV.
Species
KNApSAcK Entry
| Organism name | Lippia dulcis TREV. |
|---|---|
| Genus | Lippia |
| Family | Verbenaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Lippia |
|---|---|
| Linked NCBI taxonomy ID | 320344 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Verbenaceae |
|---|---|
| ID | 21910 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (12)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00003894
|
Eupatorin
/ 3',5-Dihydroxy-4',6,7-trimethoxyflavone / 5-Hydroxy-2-(3-hydroxy-4-methoxyphenyl)-6,7-dimethoxy-4H-1-benzopyran-4-one |
CHEMBL487402
|
C103110
|
5 / 5 / 4 | 5 / 0 | No. 3 | No. 15 |
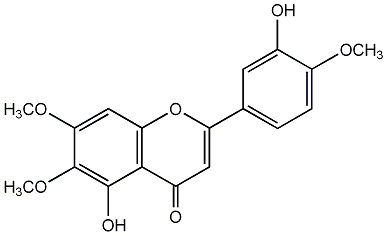
|
|
C00003837
|
Cirsimaritin
|
CHEMBL348436
|
C007072
|
13 / 18 / 14 | No. 3 | No. 15 |

|
|
|
C00003947
|
5,3'-Dihydroxy-6,7,4',5'-tetramethoxyflavone
/ 5-Hydroxy-2-(3-hydroxy-4,5-dimethoxyphenyl)-6,7-dimethoxy-4H-1-benzopyran-4-one |
No. 8 | No. 15 |
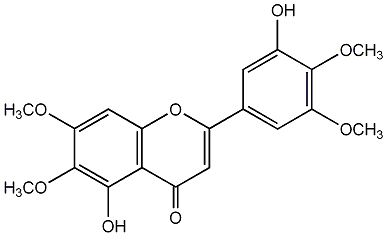
|
||||
|
C00003898
|
5-Hydroxy-6,7,3',4'-tetramethoxyflavone
/ 6-Hydroxyluteolin 6,7,3',4'-tetramethyl ether / 2-(3,4-Dimethoxyphenyl)-5-hydroxy-6,7-dimethoxy-4H-1-benzopyran-4-one |
CHEMBL226508
|
C045324
|
No. 8 | No. 15 |
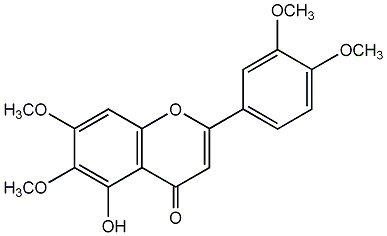
|
||
|
C00002783
|
Kusaginin
/ Acteoside / Verbascoside |
CHEMBL231853
CHEMBL444478 CHEMBL1364101 |
C058956
|
12 / 1 / 1 | 34 / 3 | No. 33 |
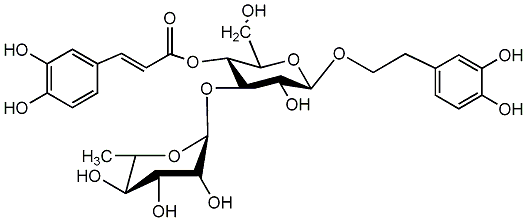
|
|
|
C00030516
|
Isoacteoside
/ Isoverbascoside |
CHEMBL504873
|
C064683
|
6 / 5 / 4 | No. 33 |
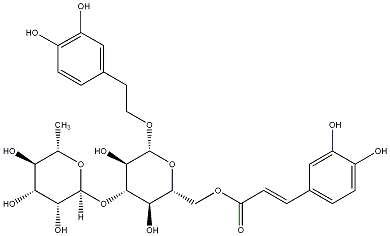
|
||
|
C00003840
|
Salvigenin
|
CHEMBL376644
|
C014049
|
No. 35 | No. 15 |
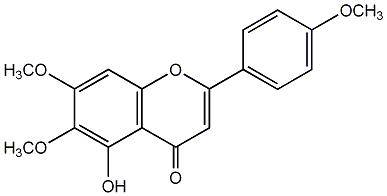
|
||
|
C00010590
|
Lamiide
|
No. 56 | No. 36 |

|
||||
|
C00035981
|
8-Epiloganin
|
CHEMBL1081584
CHEMBL1081586 CHEMBL1589904 CHEMBL2135791 |
2 / 1 / 1 | No. 56 | No. 36 |

|
||
|
C00030098
|
Verbasoside
/ (-)-Verbasoside / Decaffeoylverbascoside / (-)-Decaffeoylverbascoside |
CHEMBL1779146
|
No. 128 | No. 72 |
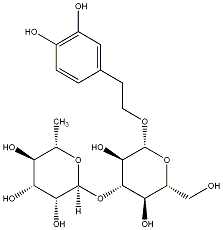
|
|||
|
C00031768
|
Epilippidulcine A
/ (-)-Epilippidulcine A |
No. 3312 |

|
|||||
|
C00031981
|
Lippidulcine A
|
No. 3312 |

|
Human Protein / Gene in interactions
35 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00002783 C00003837 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002783 C00003837 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00002783 C00035981 | 0 / 0 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00003837 | 7 / 3 |
| P06746 | DNA polymerase beta | Enzyme | C00003837 | 0 / 0 |
| P04062 | Glucosylceramidase | Enzyme | C00003837 | 6 / 4 |
| P17252 | Protein kinase C alpha type | Alpha | C00002783 | 0 / 0 |
| P39900 | Macrophage metalloelastase | M10A | C00030516 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00030516 | 2 / 2 |
| O14965 | Aurora kinase A | Aur | C00003894 | 0 / 0 |
| P04798 | Cytochrome P450 1A1 | Cytochrome P450 1A1 | C00003894 | 0 / 0 |
| P60033 | CD81 antigen | Unclassified protein | C00035981 | 1 / 1 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00003837 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002783 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00003837 | 0 / 3 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00003837 | 0 / 0 |
| P03956 | Interstitial collagenase | M10A | C00030516 | 0 / 1 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00003837 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00003837 | 1 / 1 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00002783 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002783 | 0 / 0 |
| Q16678 | Cytochrome P450 1B1 | Cytochrome P450 1B1 | C00003894 | 4 / 4 |
| Q96GD4 | Aurora kinase B | Aur | C00003894 | 0 / 0 |
| P35968 | Vascular endothelial growth factor receptor 2 | Vegfr | C00003894 | 1 / 0 |
| P45452 | Collagenase 3 | M10A | C00030516 | 1 / 1 |
| P08253 | 72 kDa type IV collagenase | M10A | C00030516 | 1 / 3 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00003837 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002783 | 1 / 1 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00002783 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00003837 | 4 / 3 |
| P08254 | Stromelysin-1 | M10A | C00030516 | 1 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002783 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00003837 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002783 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002783 | 0 / 0 |
38 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002783
C00003894
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002783
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002783
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002783
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00002783
|
| 896 | CCND3 | cyclin D3 |
C00002783
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00002783
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00002783
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00002783
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00002783
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002783
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00002783
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00002783
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00002783
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002783
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00002783
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00002783
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00002783
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002783
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00002783
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00002783
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002783
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002783
|
| 27035 | NOX1, GP91-2, MOX1, NOH-1, NOH1 | NADPH oxidase 1 |
C00002783
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002783
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00002783
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002783
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002783
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00002783
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00002783
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00002783
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00002783
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002783
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002783
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00003894
|
| 1557 | CYP2C19, CPCJ, CYP2C, P450C2C, P450IIC19 | cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00003894
|
| 1565 | CYP2D6, CPD6, CYP2D, CYP2D7AP, CYP2D7BP, CYP2D7P2, CYP2D8P2, CYP2DL1, CYPIID6, P450-DB1, P450C2D, P450DB1 | cytochrome P450, family 2, subfamily D, polypeptide 6 (EC:1.14.14.1) |
C00003894
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00003894
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (30)
| OMIM | preferred title | UniProt |
|---|---|---|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #608013 | Gaucher disease, perinatal lethal |
P04062
|
| #230800 | Gaucher disease, type i |
P04062
|
| #230900 | Gaucher disease, type ii |
P04062
|
| #231000 | Gaucher disease, type iii |
P04062
|
| #231005 | Gaucher disease, type iiic |
P04062
|
| #137750 | Glaucoma 1, open angle, a; glc1a |
Q16678
|
| #231300 | Glaucoma 3, primary congenital, a; glc3a |
Q16678
|
| #137760 | Glaucoma, primary open angle; poag |
Q16678
|
| #602089 | Hemangioma, capillary infantile |
P35968
|
| #613496 | Immunodeficiency, common variable, 6; cvid6 |
P60033
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #168600 | Parkinson disease, late-onset; pd |
P04062
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #604229 | Peters anomaly |
Q16678
|
| #172700 | Pick disease of brain |
P10636
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #602111 | Spondyloepimetaphyseal dysplasia, missouri type |
P45452
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (23)
| KEGG | name | UniProt |
|---|---|---|
| H00028 | Choriocarcinoma |
P03956
(related)
P08253 (related) |
| H00066 | Lewy body dementia (LBD) |
P04062
(related)
|
| H00126 | Gaucher disease |
P04062
(related)
|
| H00426 | Defects in the degradation of ganglioside |
P04062
(related)
|
| H00810 | Progressive myoclonic epilepsy (PME) |
P04062
(related)
|
| H00025 | Penile cancer |
P08253
(related)
P14780 (related) P35354 (related) |
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
P45452 (related) |
| H00017 | Esophageal cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00088 | Common variable immunodeficiency (CVID) |
P60033
(related)
|
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H00612 | Primary open angle glaucoma |
Q16678
(related)
|
| H01075 | Peters anomaly |
Q16678
(related)
|
| H01159 | Anterior segment dysgenesis (ASD) |
Q16678
(related)
|
| H01203 | Primary congenital glaucoma (PCG) |
Q16678
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|