Lippia alba
Species
KNApSAcK Entry
| Organism name | Lippia alba |
|---|---|
| Genus | Lippia |
| Family | Verbenaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Lippia alba |
|---|---|
| Linked NCBI taxonomy ID | 320345 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Verbenaceae |
|---|---|
| ID | 21910 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Natural Activity
List (33)
| Species | Activity |
|---|---|
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Analgesic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antemetic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antimutagenic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antioxidant |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antiradicular |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antiseptic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antispasmodic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antiulcer |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Antiviral |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Astringent |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Bactericide |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Candidicide |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Carminative |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Cicatrizant |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Cytotoxic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Decongestant |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Digestive |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Emmenagogue |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Expectorant |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Febrifuge |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Fungicide |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Hypotensive |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Insecticide |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Myorelaxant |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Nervine |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Oxytocic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Pectoral |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Sedative |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Sialagogue |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Stomachic |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Sudorific |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Tranquilizer |
| Lippia alba (Mill.) N.E. Br. Ex Britton & P. Wilson | Vulnerary |
Metabolite list (18)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
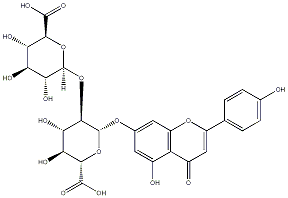
C00035270
|
Clerodendrin
|
C060994
|
No. 1 | No. 15 |
|
|||
|
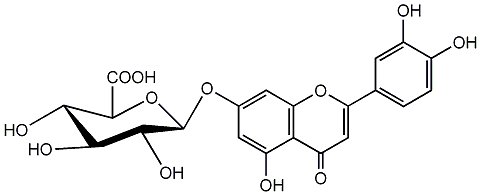
C00004268
|
Luteolin 7-glucuronide
/ Luteolin 7-O-glucuronide / Luteolin 7-O-beta-D-glucuronide |
CHEMBL464224
|
C075406
|
No. 2 | No. 15 |
|
||
|
C00004144
|
Apigenin 7-glucuronide
/ Apigenin 7-O-beta-D-glucuronide / (-)-Apigenin 7-O-beta-D-glucuronide |
CHEMBL254213
CHEMBL1980748 |
No. 2 | No. 15 |
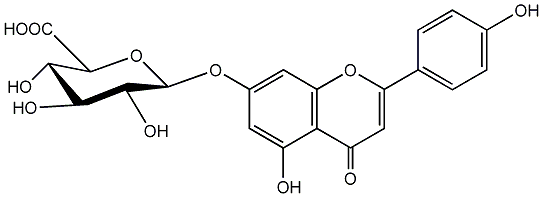
|
|||
|
C00033687
|
Calceolarioside E
|
No. 33 |

|
|||||
|
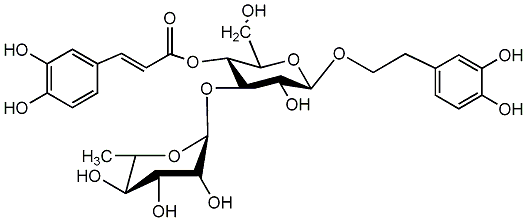
C00002783
|
Kusaginin
/ Acteoside / Verbascoside |
CHEMBL231853
CHEMBL444478 CHEMBL1364101 |
C058956
|
12 / 1 / 1 | 34 / 3 | No. 33 |
|
|
|
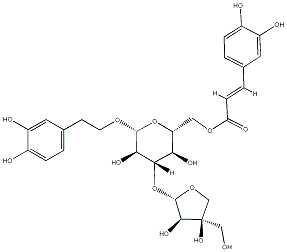
C00033958
|
Isonuomioside A
|
No. 33 |
|
|||||
|
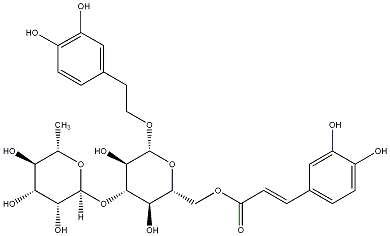
C00030516
|
Isoacteoside
/ Isoverbascoside |
CHEMBL504873
|
C064683
|
6 / 5 / 4 | No. 33 |
|
||
|
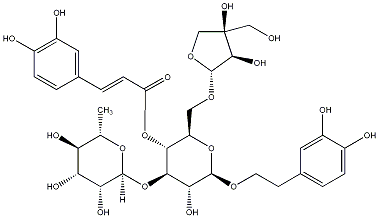
C00030317
|
Forsythoside B
|
CHEMBL393292
|
1 / 0 / 3 | No. 33 |
|
|||
|
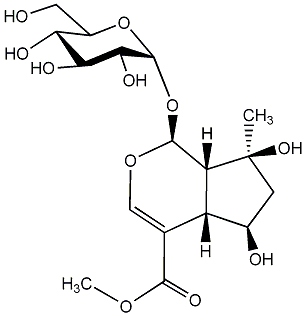
C00010687
|
Shanzhiside methyl ester
|
CHEMBL1081414
|
1 / 1 / 1 | No. 56 | No. 36 |
|
||
|
C00010595
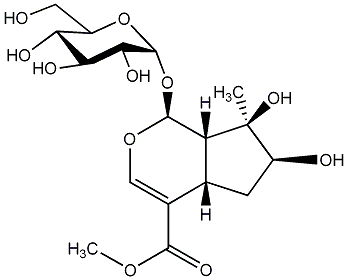
|
Caryoptoside
|
No. 56 | No. 36 |
|
||||
|
C00010630
|
Mussaenoside
|
CHEMBL2355776
|
1 / 1 / 1 | No. 56 | No. 36 |

|
||
|
C00010719
|
Theveside
|
No. 100 | No. 36 |

|
||||
|
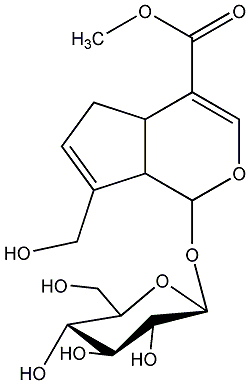
C00000320
|
Geniposide
|
CHEMBL462894
CHEMBL1417490 CHEMBL1718933 |
C007835
|
3 / 2 / 3 | 3 / 0 | No. 100 | No. 36 |
|
|
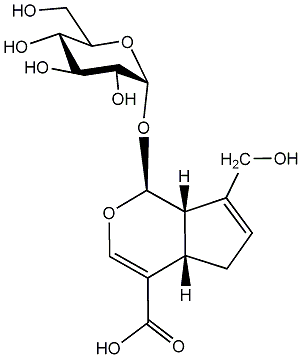
C00010564
|
Geniposidic acid
|
CHEMBL460031
|
C058966
|
No. 100 | No. 36 |
|
||
|
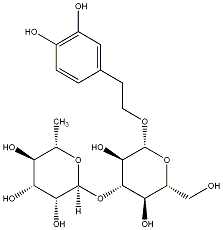
C00030098
|
Verbasoside
/ (-)-Verbasoside / Decaffeoylverbascoside / (-)-Decaffeoylverbascoside |
CHEMBL1779146
|
No. 128 | No. 72 |
|
|||
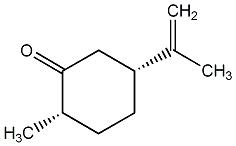
|
C00010947
|
(1S,4R)-(+)-Isodihydrocarvone
|
No. 2206 | No. 35 |
|
||||
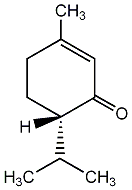
|
C00003054
|
(+)-Piperitone
|
No. 4185 | No. 35 |
|
||||
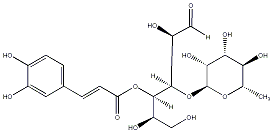
|
C00035259
|
Cistanoside F
|
No. 6662 |
|
Human Protein / Gene in interactions
23 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| O75496 | Geminin | Unclassified protein | C00000320 C00002783 | 0 / 0 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00002783 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00030317 | 0 / 3 |
| P17252 | Protein kinase C alpha type | Alpha | C00002783 | 0 / 0 |
| P39900 | Macrophage metalloelastase | M10A | C00030516 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00030516 | 2 / 2 |
| P60033 | CD81 antigen | Unclassified protein | C00010687 | 1 / 1 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00000320 | 2 / 3 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002783 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00002783 | 0 / 0 |
| P03956 | Interstitial collagenase | M10A | C00030516 | 0 / 1 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002783 | 1 / 1 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002783 | 0 / 0 |
| P45452 | Collagenase 3 | M10A | C00030516 | 1 / 1 |
| P08253 | 72 kDa type IV collagenase | M10A | C00030516 | 1 / 3 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00000320 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002783 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00002783 | 0 / 0 |
| P08254 | Stromelysin-1 | M10A | C00030516 | 1 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002783 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002783 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002783 | 0 / 0 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00010630 | 1 / 1 |
37 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00000320
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00000320
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00000320
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00002783
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00002783
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002783
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002783
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00002783
|
| 896 | CCND3 | cyclin D3 |
C00002783
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00002783
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00002783
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00002783
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00002783
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002783
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00002783
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00002783
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002783
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002783
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00002783
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00002783
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00002783
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002783
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002783
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00002783
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002783
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002783
|
| 27035 | NOX1, GP91-2, MOX1, NOH-1, NOH1 | NADPH oxidase 1 |
C00002783
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002783
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00002783
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002783
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002783
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00002783
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00002783
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00002783
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00002783
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002783
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002783
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (10)
| OMIM | preferred title | UniProt |
|---|---|---|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #613496 | Immunodeficiency, common variable, 6; cvid6 |
P60033
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #602111 | Spondyloepimetaphyseal dysplasia, missouri type |
P45452
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (12)
| KEGG | name | UniProt |
|---|---|---|
| H00028 | Choriocarcinoma |
P03956
(related)
P08253 (related) |
| H00025 | Penile cancer |
P08253
(related)
P14780 (related) P35354 (related) |
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
P45452 (related) |
| H00017 | Esophageal cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00088 | Common variable immunodeficiency (CVID) |
P60033
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
Q13148
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|