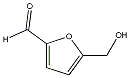
Metabolite
KNApSAcK Entry
| id | C00029547 |
|---|---|
| Name | 5-Hydroxymethylfuraldehyde / 5-Hydroxymethyl-2-furfural / 5-Hydroxymethyl-2-furaldehyde / 5-(Hydroxymethyl)-2-furaldehyde |
| CAS RN | 67-47-0 |
| Standard InChI | InChI=1S/C6H6O3/c7-3-5-1-2-6(4-8)9-5/h1-3,8H,4H2 |
| Standard InChI (Main Layer) | InChI=1S/C6H6O3/c7-3-5-1-2-6(4-8)9-5/h1-3,8H,4H2 |
Cluster
| Phytochemical cluster | |
|---|---|
| KCF-S cluster | No. 6025 |
Link
ChEMBL
| By standard InChI | CHEMBL185885 |
|---|---|
| By standard InChI Main Layer | CHEMBL185885 |
KEGG
| By LinkDB | C11101 |
|---|
CTD
| By CAS RN | C008046 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| asterids | 4 |
| Liliopsida | 3 |
Family
| family name | count |
|---|---|
| Cardiopteridaceae | 1 |
| Elaphomycetaceae | 1 |
| Eucommiaceae | 1 |
| Orchidaceae | 1 |
| Scrophulariaceae | 1 |
| Amaryllidaceae | 1 |
| Asparagaceae | 1 |
List (8)
* NCBI
Human Protein / Gene in interaction
4 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| P16473 | Thyrotropin receptor | Glycohormone receptor | CHEMBL185885 |
CHEMBL1614281
(1)
CHEMBL1614361
(1)
|
3 / 2 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | CHEMBL185885 |
CHEMBL1614458
(1)
|
0 / 0 |
| P04150 | Glucocorticoid receptor | NR3C1 | CHEMBL185885 |
CHEMBL1794456
(1)
|
0 / 1 |
| P51449 | Nuclear receptor ROR-gamma | Nuclear hormone receptor subfamily 1 group F member 3 | CHEMBL185885 |
CHEMBL2114842
(1)
|
0 / 0 |
CTD interaction (6)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| C008046 | 6817 |
SULT1A1
HAST1/HAST2 P-PST PST ST1A1 ST1A3 STP STP1 TSPST1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) | 5-hydroxymethylfurfural affects the activity of SULT1A1 protein |
affects activity
|
protein |
22563731
|
| C008046 | 6817 |
SULT1A1
HAST1/HAST2 P-PST PST ST1A1 ST1A3 STP STP1 TSPST1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) | SULT1A1 protein affects the metabolism of 5-hydroxymethylfurfural |
affects metabolic processing
|
protein |
22349055
|
| C008046 | 6817 |
SULT1A1
HAST1/HAST2 P-PST PST ST1A1 ST1A3 STP STP1 TSPST1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) | [SULT1A1 protein co-treated with SULT1A2 protein] results in increased susceptibility to 5-hydroxymethylfurfural |
affects cotreatment
/ increases response to substance |
protein |
22006426
|
| C008046 | 6817 |
SULT1A1
HAST1/HAST2 P-PST PST ST1A1 ST1A3 STP STP1 TSPST1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) | SULT1A1 protein results in increased activity of 5-hydroxymethylfurfural |
increases activity
|
protein |
22006426
|
| C008046 | 6799 |
SULT1A2
HAST4 P-PST ST1A2 STP2 TSPST2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (EC:2.8.2.1) | [SULT1A1 protein co-treated with SULT1A2 protein] results in increased susceptibility to 5-hydroxymethylfurfural |
affects cotreatment
/ increases response to substance |
protein |
22006426
|
| C008046 | 6799 |
SULT1A2
HAST4 P-PST ST1A2 STP2 TSPST2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (EC:2.8.2.1) | SULT1A2 protein results in increased activity of 5-hydroxymethylfurfural |
increases activity
|
protein |
22006426
|
Related Disease
Diseases related to proteins in ChEMBL interactions
Diseases related to CTD interactions
5 disease from interactions of metabolites
| MeSH disease | OMIM | compound | disease name | evidence type |
reference
pmid |
|---|---|---|---|---|---|
| D058739 | C008046 | Aberrant Crypt Foci |
marker/mechanism
|
19382817
22006426 |
|
| D018248 | C008046 | Adenoma, Liver Cell |
marker/mechanism
|
19382817
|
|
| D008113 | C008046 | Liver Neoplasms |
marker/mechanism
|
22563731
23038016 |
|
| D010212 | C008046 | Papilloma |
marker/mechanism
|
19382817
|
|
| D012878 | C008046 | Skin Neoplasms |
marker/mechanism
|
19382817
|