Dianthus superbus var.longicalycinus (MAXIM.) WILL
Species
KNApSAcK Entry
| Organism name | Dianthus superbus var.longicalycinus (MAXIM.) WILL |
|---|---|
| Genus | Dianthus |
| Family | Caryophyllaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Dianthus |
|---|---|
| Linked NCBI taxonomy ID | 3569 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Caryophyllaceae |
|---|---|
| ID | 3568 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | eudicotyledons |
|---|---|
| ID | 71240 |
Metabolite list (9)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00031435
|
Vaccaroside A
/ (+)-Vaccaroside A |
CHEMBL554779
|
No. 4 | No. 51 |

|
|||
|
C00031735
|
Dianoside G
|
C423334
|
No. 9 | No. 51 |

|
|||
|
C00031737
|
Dianthin D
|
CHEMBL500250
|
No. 290 |

|
||||
|
C00031739
|
Dianthin F
/ (-)-Dianthin F |
No. 745 |

|
|||||
|
C00031983
|
Longicalycinin A
|
No. 745 |

|
|||||
|
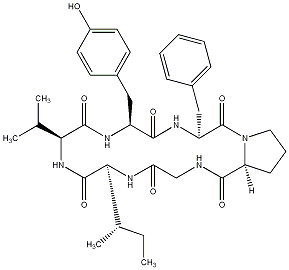
C00031736
|
Dianthin C
/ (-)-Dianthin C |
No. 745 |
|
|||||
|
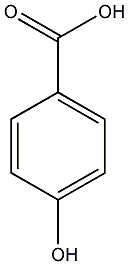
C00000856
|
4-Hydroxybenzoic acid
/ p-Hydroxybenzoic acid |
CHEMBL441343
|
C038193
|
21 / 7 / 16 | 2 / 1 | No. 817 | No. 81 |
|
|
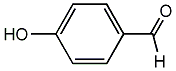
C00002657
|
p-Formylphenol
/ 4-Hydroxybenzaldehyde / p-Hydroxybenzaldehyde |
CHEMBL14193
|
C011483
|
3 / 2 / 2 | No. 2076 |
|
||
|
C00031738
|
Dianthin E
/ (-)-Dianthin E |
CHEMBL1957467
|
No. 2574 |

|
Human Protein / Gene in interactions
22 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00000856 C00002657 | 1 / 1 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00000856 C00002657 | 1 / 1 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00000856 | 1 / 2 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00000856 | 0 / 0 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00000856 | 0 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00000856 | 1 / 2 |
| Q9UQ49 | Sialidase-3 | Enzyme | C00000856 | 0 / 0 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00000856 | 0 / 0 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00000856 | 1 / 8 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00000856 | 0 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00000856 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00000856 | 0 / 0 |
| P14902 | Indoleamine 2,3-dioxygenase 1 | Enzyme | C00002657 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00000856 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00000856 | 0 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00000856 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00000856 | 0 / 1 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00000856 | 0 / 0 |
| P51580 | Thiopurine S-methyltransferase | Enzyme | C00000856 | 1 / 1 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00000856 | 0 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00000856 | 0 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00000856 | 1 / 1 |
2 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 3952 | LEP, LEPD, OB, OBS | leptin |
C00000856
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00000856
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (7)
| OMIM | preferred title | UniProt |
|---|---|---|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #211980 | Lung cancer |
P00533
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
| #610460 | Thiopurine s-methyltransferase deficiency |
P51580
|
KEGG DISEASE (16)
| KEGG | name | UniProt |
|---|---|---|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
|
| H00018 | Gastric cancer |
P00533
(related)
|
| H00022 | Bladder cancer |
P00533
(related)
|
| H00028 | Choriocarcinoma |
P00533
(related)
|
| H00030 | Cervical cancer |
P00533
(related)
|
| H00042 | Glioma |
P00533
(related)
P00533 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H00964 | Thiopurine S-methyltransferase deficiency (TPMT deficiency) |
P51580
(related)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|