Caesalpinia decapetala
Species
KNApSAcK Entry
| Organism name | Caesalpinia decapetala |
|---|---|
| Genus | Caesalpinia |
| Family | Fabaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Caesalpinia decapetala |
|---|---|
| Linked NCBI taxonomy ID | 191885 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Fabaceae |
|---|---|
| ID | 3803 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (6)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00000919
|
beta-Carotene
|
CHEMBL1293
CHEMBL1966639 |
D019207
|
7 / 12 / 11 | 104 / 19 | No. 26 | No. 59 |
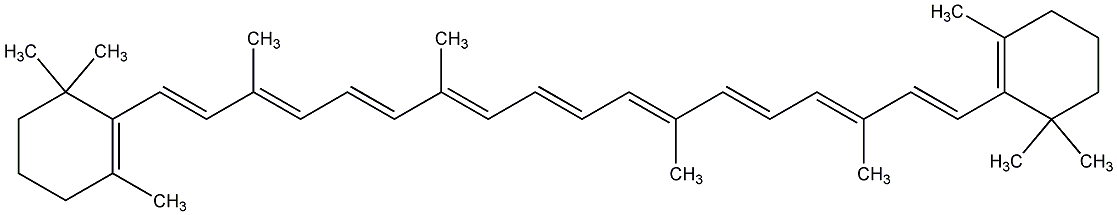
|
|
C00003800
|
7,4'-Dihydroxyflavone
|
CHEMBL294878
|
C085172
|
17 / 19 / 16 | No. 76 | No. 15 |
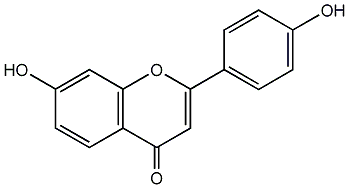
|
|
|
C00001260
|
Nonacosane
|
CHEMBL428955
|
C047577
|
No. 115 |
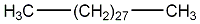
|
|||
|
C00010268
|
Brazilin
|
CHEMBL598951
CHEMBL1370456 |
C044362
|
32 / 25 / 25 | No. 1670 |

|
||
|
C00020152
|
4-O-Methylsappanol
|
CHEMBL477780
|
7 / 20 / 19 | No. 1670 |

|
|||
|
C00020451
|
Episappanol
|
CHEMBL477779
|
7 / 20 / 19 | No. 1670 |

|
Human Protein / Gene in interactions
55 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00000919 C00003800 C00010268 | 0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00003800 C00010268 | 3 / 3 |
| P10721 | Mast/stem cell growth factor receptor Kit | Pdgfr | C00020152 C00020451 | 4 / 3 |
| P21802 | Fibroblast growth factor receptor 2 | TK tyrosine-protein kinase TLK subfamily | C00020152 C00020451 | 9 / 3 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00003800 C00010268 | 0 / 1 |
| P11362 | Fibroblast growth factor receptor 1 | Fgfr | C00020152 C00020451 | 4 / 5 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00003800 C00010268 | 2 / 2 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00003800 C00010268 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00003800 C00010268 | 0 / 0 |
| P08581 | Hepatocyte growth factor receptor | TK tyrosine-protein kinase MET subfamily | C00020152 C00020451 | 2 / 3 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00020152 C00020451 | 1 / 8 |
| P35968 | Vascular endothelial growth factor receptor 2 | Vegfr | C00020152 C00020451 | 1 / 0 |
| O75496 | Geminin | Unclassified protein | C00000919 C00010268 | 0 / 0 |
| P12931 | Proto-oncogene tyrosine-protein kinase Src | Src | C00020152 C00020451 | 0 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00003800 | 4 / 1 |
| P39748 | Flap endonuclease 1 | Enzyme | C00010268 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00000919 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00010268 | 2 / 0 |
| Q9Y3R4 | Sialidase-2 | Enzyme | C00003800 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00010268 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00010268 | 1 / 1 |
| P11308 | Transcriptional regulator ERG | Unclassified protein | C00010268 | 1 / 2 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00010268 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00010268 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00010268 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00003800 | 2 / 2 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00003800 | 5 / 3 |
| O15296 | Arachidonate 15-lipoxygenase B | Enzyme | C00010268 | 0 / 0 |
| P06280 | Alpha-galactosidase A | Enzyme | C00003800 | 1 / 1 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00010268 | 2 / 3 |
| P05091 | Aldehyde dehydrogenase, mitochondrial | Oxidoreductase | C00003800 | 1 / 1 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00010268 | 3 / 1 |
| P14735 | Insulin-degrading enzyme | Enzyme | C00010268 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00000919 | 11 / 10 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00010268 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00010268 | 0 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00003800 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00010268 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00010268 | 4 / 3 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00010268 | 0 / 0 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | C00003800 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00010268 | 2 / 0 |
| P06239 | Tyrosine-protein kinase Lck | Src | C00003800 | 0 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00010268 | 0 / 0 |
| Q14145 | Kelch-like ECH-associated protein 1 | Unclassified protein | C00010268 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00010268 | 1 / 1 |
| O00255 | Menin | Unclassified protein | C00010268 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00010268 | 1 / 2 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00000919 | 0 / 1 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00000919 | 1 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00000919 | 0 / 0 |
| P27338 | Amine oxidase [flavin-containing] B | Oxidoreductase | C00003800 | 0 / 0 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00003800 | 1 / 1 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00010268 | 0 / 0 |
| Q14191 | Werner syndrome ATP-dependent helicase | Enzyme | C00010268 | 2 / 1 |
104 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 124 | ADH1A, ADH1 | alcohol dehydrogenase 1A (class I), alpha polypeptide (EC:1.1.1.1) |
C00000919
|
| 128 | ADH5, ADH-3, ADHX, FALDH, FDH, GSH-FDH, GSNOR | alcohol dehydrogenase 5 (class III), chi polypeptide (EC:1.1.1.1 1.1.1.284) |
C00000919
|
| 221 | ALDH3B1, ALDH4, ALDH7 | aldehyde dehydrogenase 3 family, member B1 (EC:1.2.1.5) |
C00000919
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00000919
|
| 287 | ANK2, ANK-2, LQT4, brank-2 | ankyrin 2, neuronal |
C00000919
|
| 288 | ANK3, ANKYRIN-G, MRT37 | ankyrin 3, node of Ranvier (ankyrin G) |
C00000919
|
| 427 | ASAH1, AC, ACDase, ASAH, PHP, PHP32, SMAPME | N-acylsphingosine amidohydrolase (acid ceramidase) 1 (EC:3.5.1.23) |
C00000919
|
| 51665 | ASB1, ASB-1 | ankyrin repeat and SOCS box containing 1 |
C00000919
|
| 140462 | ASB9 | ankyrin repeat and SOCS box containing 9 |
C00000919
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00000919
|
| 9530 | BAG4, BAG-4, SODD | BCL2-associated athanogene 4 |
C00000919
|
| 9529 | BAG5, BAG-5 | BCL2-associated athanogene 5 |
C00000919
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00000919
|
| 53630 | BCMO1, BCDO, BCDO1, BCMO, BCO, BCO1 | beta-carotene 15,15'-monooxygenase 1 (EC:1.14.99.36) |
C00000919
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00000919
|
| 999 | CDH1, Arc-1, CD324, CDHE, ECAD, LCAM, UVO | cadherin 1, type 1, E-cadherin (epithelial) |
C00000919
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00000919
|
| 9071 | CLDN10, CPETRL3, OSP-L | claudin 10 |
C00000919
|
| 51208 | CLDN18, SFTA5, SFTPJ | claudin 18 |
C00000919
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00000919
|
| 1282 | COL4A1, HANAC, ICH, POREN1, arresten | collagen, type IV, alpha 1 |
C00000919
|
| 1284 | COL4A2, ICH, POREN2 | collagen, type IV, alpha 2 |
C00000919
|
| 8738 | CRADD, MRT34, RAIDD | CASP2 and RIPK1 domain containing adaptor with death domain |
C00000919
|
| 1508 | CTSB, APPS, CPSB | cathepsin B (EC:3.4.22.1) |
C00000919
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00000919
|
| 6372 | CXCL6, CKA-3, GCP-2, GCP2, SCYB6 | chemokine (C-X-C motif) ligand 6 |
C00000919
|
| 4283 | CXCL9, CMK, Humig, MIG, SCYB9, crg-10 | chemokine (C-X-C motif) ligand 9 |
C00000919
|
| 7852 | CXCR4, CD184, D2S201E, FB22, HM89, HSY3RR, LAP3, LCR1, LESTR, NPY3R, NPYR, NPYRL, NPYY3R, WHIM | chemokine (C-X-C motif) receptor 4 |
C00000919
|
| 1592 | CYP26A1, CP26, CYP26, P450RAI, P450RAI1 | cytochrome P450, family 26, subfamily A, polypeptide 1 (EC:1.14.-.-) |
C00000919
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00000919
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00000919
|
| 64816 | CYP3A43 | cytochrome P450, family 3, subfamily A, polypeptide 43 (EC:1.14.14.1) |
C00000919
|
| 1746 | DLX2, TES-1, TES1 | distal-less homeobox 2 |
C00000919
|
| 1748 | DLX4, BP1, DLX7, DLX8, DLX9 | distal-less homeobox 4 |
C00000919
|
| 2068 | ERCC2, COFS2, EM9, TTD, XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 (EC:3.6.4.12) |
C00000919
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00000919
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00000919
|
| 23017 | FAIM2, LFG, LFG2, NGP35, NMP35, TMBIM2 | Fas apoptotic inhibitory molecule 2 |
C00000919
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00000919
|
| 3169 | FOXA1, HNF3A, TCF3A | forkhead box A1 |
C00000919
|
| 2627 | GATA6 | GATA binding protein 6 |
C00000919
|
| 2633 | GBP1 | guanylate binding protein 1, interferon-inducible |
C00000919
|
| 81025 | GJA9, CX58, CX59, GJA10 | gap junction protein, alpha 9, 59kDa |
C00000919
|
| 373156 | GSTK1, GST, GST_13-13, GST13, GST13-13, GSTK1-1, hGSTK1 | glutathione S-transferase kappa 1 (EC:2.5.1.18) |
C00000919
|
| 2946 | GSTM2, GST4, GSTM, GSTM2-2, GTHMUS | glutathione S-transferase mu 2 (muscle) (EC:2.5.1.18) |
C00000919
|
| 2948 | GSTM4, GSTM4-4, GTM4 | glutathione S-transferase mu 4 (EC:2.5.1.18) |
C00000919
|
| 2949 | GSTM5, GSTM5-5, GTM5 | glutathione S-transferase mu 5 (EC:2.5.1.18) |
C00000919
|
| 2954 | GSTZ1, GSTZ1-1, MAAI, MAI | glutathione S-transferase zeta 1 (EC:2.5.1.18 5.2.1.2) |
C00000919
|
| 3204 | HOXA7, ANTP, HOX1, HOX1.1, HOX1A | homeobox A7 |
C00000919
|
| 3216 | HOXB6, HOX2, HOX2B, HU-2, Hox-2.2 | homeobox B6 |
C00000919
|
| 3226 | HOXC10, HOX3I | homeobox C10 |
C00000919
|
| 3448 | IFNA14, IFN-alphaH, LEIF2H | interferon, alpha 14 |
C00000919
|
| 3449 | IFNA16, IFN-alphaO | interferon, alpha 16 |
C00000919
|
| 3451 | IFNA17, IFN-alphaI, IFNA, INFA, LEIF2C1 | interferon, alpha 17 |
C00000919
|
| 3442 | IFNA5, IFN-alpha-5, IFN-alphaG, INA5, INFA5, leIF_G | interferon, alpha 5 |
C00000919
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00000919
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00000919
|
| 3394 | IRF8, H-ICSBP, ICSBP, ICSBP1, IRF-8 | interferon regulatory factor 8 |
C00000919
|
| 50805 | IRX4, IRXA3 | iroquois homeobox 4 |
C00000919
|
| 3688 | ITGB1, CD29, FNRB, GPIIA, MDF2, MSK12, VLA-BETA, VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
C00000919
|
| 3691 | ITGB4, CD104 | integrin, beta 4 |
C00000919
|
| 3713 | IVL | involucrin |
C00000919
|
| 3875 | KRT18, CYK18, K18 | keratin 18 |
C00000919
|
| 3880 | KRT19, CK19, K19, K1CS | keratin 19 |
C00000919
|
| 3849 | KRT2, CK-2e, K2e, KRT2A, KRT2E, KRTE | keratin 2 |
C00000919
|
| 3855 | KRT7, CK7, K2C7, K7, SCL | keratin 7 |
C00000919
|
| 3856 | KRT8, CARD2, CK-8, CK8, CYK8, K2C8, K8, KO | keratin 8 |
C00000919
|
| 8022 | LHX3, CPHD3, LIM3, M2-LHX3 | LIM homeobox 3 |
C00000919
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00000919
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00000919
|
| 4153 | MBL2, COLEC1, HSMBPC, MBL, MBL2D, MBP, MBP-C, MBP1, MBPD | mannose-binding lectin (protein C) 2, soluble |
C00000919
|
| 4170 | MCL1, BCL2L3, EAT, MCL1-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT | myeloid cell leukemia sequence 1 (BCL2-related) |
C00000919
|
| 4212 | MEIS2, HsT18361, MRG1 | Meis homeobox 2 |
C00000919
|
| 4222 | MEOX1, KFS2, MOX1 | mesenchyme homeobox 1 |
C00000919
|
| 4255 | MGMT | O-6-methylguanine-DNA methyltransferase (EC:2.1.1.63) |
C00000919
|
| 3110 | MNX1, HB9, HLXB9, HOXHB9, SCRA1 | motor neuron and pancreas homeobox 1 |
C00000919
|
| 4439 | MSH5, G7, MUTSH5, NG23 | mutS homolog 5 |
C00000919
|
| 4586 | MUC5AC, MUC5, TBM, leB | mucin 5AC, oligomeric mucus/gel-forming |
C00000919
|
| 4842 | NOS1, IHPS1, N-NOS, NC-NOS, NOS, bNOS, nNOS | nitric oxide synthase 1 (neuronal) (EC:1.14.13.39) |
C00000919
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00000919
|
| 4973 | OLR1, CLEC8A, LOX1, LOXIN, SCARE1, SLOX1 | oxidized low density lipoprotein (lectin-like) receptor 1 |
C00000919
|
| 5098 | PCDHGC3, PC43, PCDH-GAMMA-C3, PCDH2 | protocadherin gamma subfamily C, 3 |
C00000919
|
| 5359 | PLSCR1, MMTRA1B | phospholipid scramblase 1 |
C00000919
|
| 57048 | PLSCR3 | phospholipid scramblase 3 |
C00000919
|
| 57088 | PLSCR4, TRA1 | phospholipid scramblase 4 |
C00000919
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00000919
|
| 5892 | RAD51D, BROVCA4, R51H3, RAD51L3, TRAD | RAD51 paralog D |
C00000919
|
| 5914 | RARA, NR1B1, RAR | retinoic acid receptor, alpha |
C00000919
|
| 5915 | RARB, HAP, NR1B2, RRB2 | retinoic acid receptor, beta |
C00000919
|
| 5918 | RARRES1, LXNL, TIG1 | retinoic acid receptor responder (tazarotene induced) 1 |
C00000919
|
| 5950 | RBP4, RDCCAS | retinol binding protein 4, plasma |
C00000919
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00000919
|
| 949 | SCARB1, CD36L1, CLA-1, CLA1, HDLQTL6, SR-BI, SRB1 | scavenger receptor class B, member 1 |
C00000919
|
| 4990 | SIX6, MCOPCT2, OPTX2, Six9 | SIX homeobox 6 |
C00000919
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00000919
|
| 9517 | SPTLC2, HSN1C, LCB2, LCB2A, NSAN1C, SPT2, hLCB2a | serine palmitoyltransferase, long chain base subunit 2 (EC:2.3.1.50) |
C00000919
|
| 6720 | SREBF1, SREBP-1c, SREBP1, bHLHd1 | sterol regulatory element binding transcription factor 1 |
C00000919
|
| 7052 | TGM2, G-ALPHA-h, GNAH, TG2, TGC | transglutaminase 2 (EC:2.3.2.13) |
C00000919
|
| 7047 | TGM4, TGP, hTGP | transglutaminase 4 (EC:2.3.2.13) |
C00000919
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00000919
|
| 3604 | TNFRSF9, 4-1BB, CD137, CDw137, ILA | tumor necrosis factor receptor superfamily, member 9 |
C00000919
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00000919
|
| 10190 | TXNDC9, APACD, PHLP3 | thioredoxin domain containing 9 |
C00000919
|
| 7517 | XRCC3, CMM6 | X-ray repair complementing defective repair in Chinese hamster cells 3 |
C00000919
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (71)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #610251 | Alcohol sensitivity, acute |
P05091
|
| #207410 | Antley-bixler syndrome without genital anomalies or disordered steroidogenesis; abs2 |
P21802
|
| #101200 | Apert syndrome |
P21802
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #123790 | Beare-stevenson cutis gyrata syndrome; bstvs |
P21802
|
| #614592 | Bent bone dysplasia syndrome; bbds |
P21802
|
| %606641 | Body mass index; bmi |
P37231
|
| #300615 | Brunner syndrome |
P21397
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #114500 | Colorectal cancer; crc |
P84022
Q14191 |
| #123500 | Crouzon syndrome |
P21802
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #612219 | Ewing sarcoma; es |
P11308
|
| #301500 | Fabry disease |
P06280
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #606764 | Gastrointestinal stromal tumor; gist |
P10721
|
| #137800 | Glioma susceptibility 1; glm1 |
P37231
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #602089 | Hemangioma, capillary infantile |
P35968
|
| #114550 | Hepatocellular carcinoma |
P08581
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #147950 | Hypogonadotropic hypogonadism 2 with or without anosmia; hh2 |
P11362
|
| #123150 | Jackson-weiss syndrome; jws |
P21802
|
| #149730 | Lacrimoauriculodentodigital syndrome; ladd |
P21802
|
| #601626 | Leukemia, acute myeloid; aml |
P10721
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #211980 | Lung cancer |
P00533
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #601665 | Obesity |
P37231
|
| #166250 | Osteoglophonic dysplasia; ogd |
P11362
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #101600 | Pfeiffer syndrome |
P11362
P21802 |
| #172700 | Pick disease of brain |
P10636
|
| #172800 | Piebald trait; pbt |
P10721
|
| #605074 | Renal cell carcinoma, papillary, 1; rccp1 |
P08581
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #609579 | Scaphocephaly, maxillary retrusion, and mental retardation |
P21802
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #273300 | Testicular germ cell tumor; tgct |
P10721
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #190440 | Trigonocephaly 1; trigno1 |
P11362
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #277700 | Werner syndrome; wrn |
Q14191
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (64)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
|
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
|
| H00018 | Gastric cancer |
P00533
(related)
P08581 (related) P21802 (related) |
| H00022 | Bladder cancer |
P00533
(related)
|
| H00028 | Choriocarcinoma |
P00533
(related)
|
| H00030 | Cervical cancer |
P00533
(related)
|
| H00042 | Glioma |
P00533
(related)
P00533 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) |
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
P37231 (related) |
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H01071 | Acute alcohol sensitivity |
P05091
(related)
|
| H00093 | Combined immunodeficiencies (CIDs) |
P06239
(related)
|
| H00125 | Fabry disease |
P06280
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H00021 | Renal cell carcinoma |
P08581
(related)
|
| H00046 | Cholangiocarcinoma |
P08581
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00003 | Acute myeloid leukemia (AML) |
P10721
(related)
P10721 (marker) |
| H00170 | Piebaldism |
P10721
(related)
|
| H00023 | Testicular cancer |
P10721
(marker)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00024 | Prostate cancer |
P11308
(related)
|
| H00035 | Ewing's sarcoma |
P11308
(related)
|
| H00255 | Hypogonadotropic hypogonadism |
P11362
(related)
|
| H00443 | Osteoglophonic dysplasia (OD) |
P11362
(related)
|
| H00458 | Craniosynostosis |
P11362
(related)
P21802 (related) |
| H00516 | Isolated orofacial clefts |
P11362
(related)
|
| H01207 | Trigonocephaly |
P11362
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H00642 | Lacrimo-auriculo-dento-digital syndrome (LADD) |
P21802
(related)
|
| H00032 | Thyroid cancer |
P37231
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q03164
(related)
Q03164 (marker) |
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00296 | Defects in RecQ helicases |
Q14191
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
19 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D000435 | Alcoholic Intoxication |
C00000919
|
| D001145 | Arrhythmias, Cardiac |
C00000919
|
| D053627 | Asthenozoospermia |
C00000919
|
| D002545 | Brain Ischemia |
C00000919
|
| D006528 | Carcinoma, Hepatocellular |
C00000919
|
| D002318 | Cardiovascular Diseases |
C00000919
|
| D003930 | Diabetic Retinopathy |
C00000919
|
| D056486 | Drug-Induced Liver Injury |
C00000919
|
| D006526 | Hepatitis C |
C00000919
|
| D015658 | HIV Infections |
C00000919
|
| D006986 | Hypervitaminosis A |
C00000919
|
| D008107 | Liver Diseases |
C00000919
|
| D008113 | Liver Neoplasms |
C00000919
|
| D008114 | Liver Neoplasms, Experimental |
C00000919
|
| D008175 | Lung Neoplasms |
C00000919
|
| D009369 | Neoplasms |
C00000919
|
| D009845 | Oligospermia |
C00000919
|
| D013272 | Stomach Diseases |
C00000919
|
| D013276 | Stomach Ulcer |
C00000919
|