Sedum sarmentosum
Species
KNApSAcK Entry
| Organism name | Sedum sarmentosum |
|---|---|
| Genus | Sedum |
| Family | Crassulaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Sedum sarmentosum |
|---|---|
| Linked NCBI taxonomy ID | 91146 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Crassulaceae |
|---|---|
| ID | 3781 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | eudicotyledons |
|---|---|
| ID | 71240 |
Metabolite list (44)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
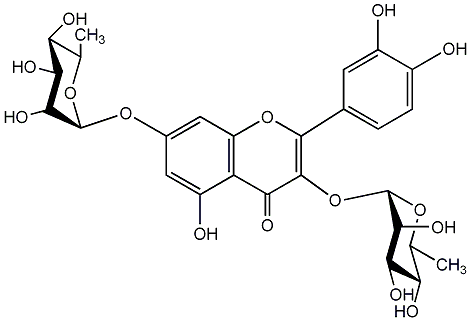
C00005432
|
Quercetin 3,7-dirhamnoside
/ Quercetin 3,7-di-O-alpha-L-rhamnopyranoside |
No. 1 | No. 15 |
|
||||
|
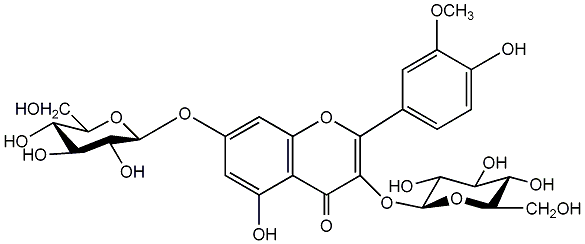
C00005556
|
Isorhamnetin 3,7-diglucoside
/ Isorhamnetin 3,7-di-O-beta-glucopyranoside |
No. 1 | No. 15 |
|
||||
|
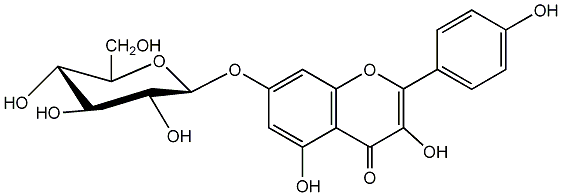
C00005149
|
Populnin
/ Kaempferol 7-glucoside |
CHEMBL469441
CHEMBL1159471 |
No. 2 | No. 15 |
|
|||
|
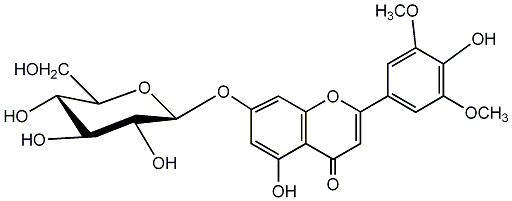
C00004444
|
Tricin 7-glucoside
/ Tricin 7-O-glucoside / Tricin 7-O-beta-D-glucopyranoside |
No. 2 | No. 15 |
|
||||
|
C00005530
|
Isorhamnetin 7-glucoside
/ Isorhamnetin 7-O-beta-D-glucopyranoside |
No. 2 | No. 15 |

|
||||
|
C00001017
|
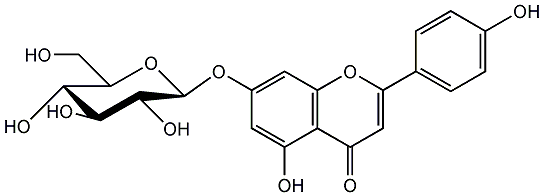
Cosmosiin
/ Apigenin 7-glucoside / (-)-Apigenin 7-glucoside / Apigenin 7-O-beta-D-glucopyranoside |
CHEMBL487995
CHEMBL487017 CHEMBL1591566 CHEMBL2165585 |
C057792
|
5 / 6 / 1 | No. 2 | No. 15 |
|
|
|
C00005715
|
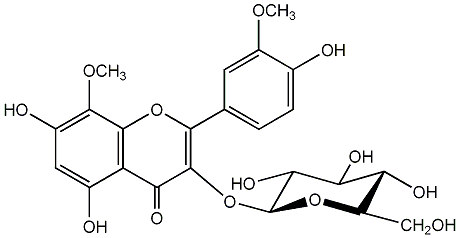
Limocitrin 3-glucoside
/ Limocitrin 3-O-beta-glucopyranoside / Limocitrin 3-O-beta-D-glucopyranoside |
No. 2 | No. 15 |
|
||||
|
C00005250
|
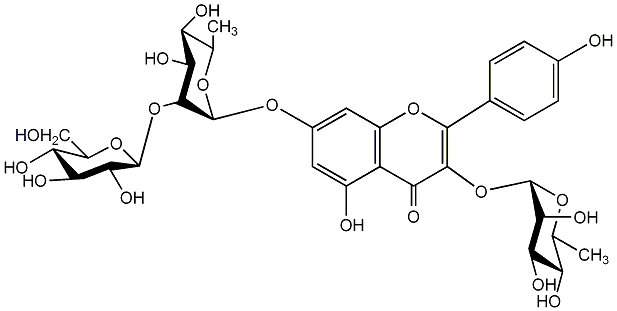
Grosvenorin
/ Grosvenorine / Kaempferol 3-rhamnoside-7-glucosyl-(1->2)-rhamnoside / 7-[(6-Deoxy-2-O-beta-D-glucopyranosyl-alpha-L-mannopyranosyl)oxy]-3-[(6-deoxy-alpha-L-mannopyranosyl)oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
No. 5 | No. 15 |
|
||||
|
C00040937
|
delta-Amyrin
|
No. 13 | No. 51 |

|
||||
|
C00041113
|
Sarmentolin
/ (+)-Sarmentolin |
No. 13 | No. 51 |

|
||||
|
C00030227
|
Eugenol glucoside
/ Eugenol beta-D-glucopyranoside |
CHEMBL463487
|
No. 45 | No. 72 |

|
|||
|
C00031450
|
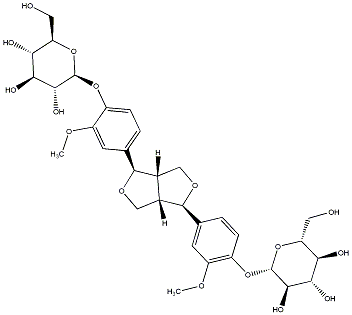
(-)-Pinoresinol 4,4'-di-O-beta-D-glucopyranoside
|
CHEMBL450911
CHEMBL573785 CHEMBL1712579 |
2 / 0 / 0 | No. 152 |
|
|||
|
C00031358
|
Sedumoside F1
/ (-)-Sedumoside F1 |
No. 153 |

|
|||||
|
C00029658
|
Alangioside A
/ Alangionoside A / Turpinionoside B / (-)-Alangioside A / (-)-Turpinionoside B |
CHEMBL1365711
CHEMBL1366071 |
15 / 3 / 7 | No. 153 |

|
|||
|
C00029499
|
3-Hydroxy-5,6-epoxy-beta-ionol 9-O-beta-D-glucopyranoside
|
CHEMBL1814430
|
No. 153 |

|
||||
|
C00031357
|
Sedumoside E3
/ (-)-Sedumoside E3 |
No. 167 |

|
|||||
|
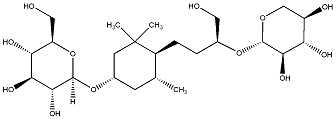
C00031351
|
Sedumoside A6
/ (-)-Sedumoside A6 |
No. 167 |
|
|||||
|
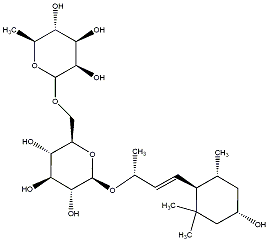
C00031359
|
Sedumoside F2
/ (-)-Sedumoside F2 |
No. 167 |
|
|||||
|
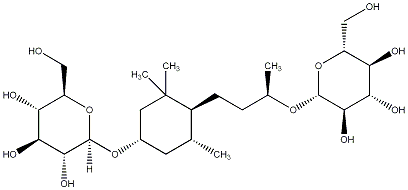
C00031053
|
Platanionoside D
/ (-)-Platanionoside D |
No. 167 |
|
|||||
|
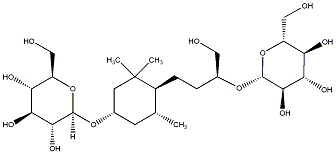
C00031350
|
Sedumoside A5
/ (-)-Sedumoside A5 |
No. 167 |
|
|||||
|
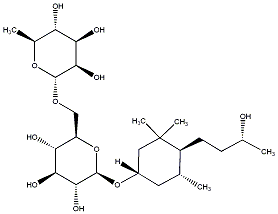
C00031355
|
Sedumoside E1
/ (-)-Sedumoside E1 |
No. 167 |
|
|||||
|
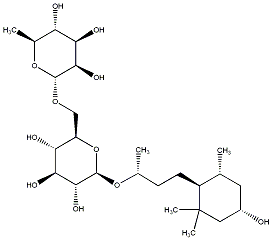
C00031356
|
Sedumoside E2
/ (-)-Sedumoside E2 |
No. 167 |
|
|||||
|
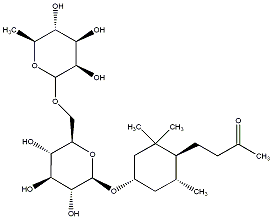
C00031360
|
Sedumoside G
/ (-)-Sedumoside G |
No. 167 |
|
|||||
|
C00032514
|
Woorenoside XI
/ (-)-Woorenoside XI |
No. 174 | No. 22 |

|
||||
|
C00031361
|
Sedumoside H
/ (-)-Sedumoside H |
No. 225 |

|
|||||
|
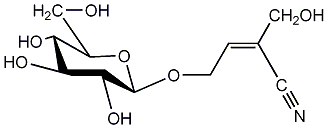
C00001455
|
Sarmentosin
|
C082842
|
No. 258 | No. 73 |
|
|||
|
C00031388
|
Staphylionoside D
/ (-)-Staphylionoside D |
No. 316 |

|
|||||
|
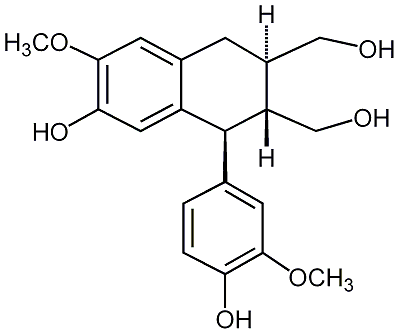
C00007204
|
Isolariciresinol
/ (+)-Isolariciresinol |
CHEMBL399512
CHEMBL1760593 |
C060284
|
2 / 1 / 1 | No. 406 |
|
||
|
C00031349
|
Sedumoside A3
/ (-)-Sedumoside A3 |
No. 431 |

|
|||||
|
C00031353
|
Sedumoside C
/ (-)-Sedumoside C |
No. 431 |

|
|||||
|
C00031354
|
Sedumoside D
/ (-)-Sedumoside D |
No. 431 |

|
|||||
|
C00031352
|
Sedumoside B
/ (-)-Sedumoside B |
No. 431 |

|
|||||
|
C00031362
|
Sedumoside I
/ (-)-Sedumoside I |
No. 431 |

|
|||||
|
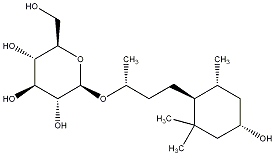
C00030802
|
Myrsinionoside D
|
No. 431 |
|
|||||
|
C00030801
|
Myrsinionoside A
/ (-)-Myrsinionoside A |
No. 431 |

|
|||||
|
C00029659
|
Alangionoside J
|
No. 431 |

|
|||||
|
C00031347
|
Sedumoside A1
/ (-)-Sedumoside A1 |
No. 431 |

|
|||||
|
C00031348
|
Sedumoside A2
/ (-)-Sedumoside A2 |
No. 431 |

|
|||||
|
C00029325
|
(3S,5R,6S,9R)-Megastigman-3,9-diol
|
No. 1981 |

|
|||||
|
C00031309
|
Sarmentol B
/ (+)-Sarmentol B / 9-epi-Sarmentol A |
No. 1981 |

|
|||||
|
C00031308
|
Sarmentol A
/ (-)-Sarmentol A |
No. 1981 |

|
|||||
|
C00031310
|
Sarmentol D
|
No. 1981 |

|
|||||
|
C00031307
|
Sarmentoic acid
/ (-)-Sarmentoic acid |
No. 1981 |

|
|||||
|
C00029415
|
1-Acetyl beta-Carboline
|
CHEMBL1682931
|
No. 2044 |

|
Human Protein / Gene in interactions
22 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00029658 C00031450 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00001017 C00029658 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00007204 | 1 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00029658 | 0 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00029658 | 1 / 2 |
| P39748 | Flap endonuclease 1 | Enzyme | C00029658 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00031450 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00029658 | 1 / 1 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00001017 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00001017 | 0 / 0 |
| Q9UNQ0 | ATP-binding cassette sub-family G member 2 | ATP binding cassette | C00001017 | 2 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00029658 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00007204 | 0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00029658 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00029658 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00029658 | 0 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00001017 | 4 / 1 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00029658 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00029658 | 0 / 0 |
| O00167 | Eyes absent homolog 2 | Enzyme | C00029658 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00029658 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00029658 | 1 / 4 |
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (10)
| OMIM | preferred title | UniProt |
|---|---|---|
| #614490 | Blood group, junior system; jr |
Q9UNQ0
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #138900 | Uric acid concentration, serum, quantitative trait locus 1; uaqtl1 |
Q9UNQ0
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (9)
| KEGG | name | UniProt |
|---|---|---|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|