phytochemical cluster No. 17 (140 metabolites)
Plant Species
Cumulative plant class count
| class name | count |
|---|---|
| rosids | 228 |
| asterids | 27 |
| Liliopsida | 14 |
| Embryophyta | 4 |
| eudicotyledons | 1 |
Cumulative family count
| class name | count |
|---|---|
| Fabaceae | 165 |
| Calophyllaceae | 52 |
| Rubiaceae | 18 |
| Orchidaceae | 12 |
| Clusiaceae | 8 |
| Asteraceae | 6 |
| Mniaceae | 3 |
| Cucurbitaceae | 2 |
| Hydrangeaceae | 2 |
| Thorectidae | 1 |
| Marchantiaceae | 1 |
| Aplysinidae | 1 |
| Amaranthaceae | 1 |
| Passifloraceae | 1 |
| Eriocaulaceae | 1 |
| Xanthorrhoeaceae | 1 |
KEGG BRITE br08003
Categories (2)
| br08003 Category | # of metabolite |
|---|---|
| Neoflavonoids | 4 |
| Coumestanes | 4 |
metabolites link (8)
| br08003 Category | KEGG ID | KNApSAcK ID |
|---|---|---|
| Neoflavonoids | C09158 | C00002455 |
| Neoflavonoids | C09275 | C00002482 |
| Coumestanes | C10205 | C00002514 |
| Neoflavonoids | C10414 | C00002519 |
| Neoflavonoids | C10504 | C00002548 |
| Coumestanes | C10523 | C00002566 |
| Coumestanes | C10529 | C00002572 |
| Coumestanes | C10541 | C00002585 |
Metabolite list (140)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
figure |
|---|---|---|---|---|---|---|---|
|
C00050256
|
Bavacoumestan B
|
No. 54 |
|
||||
|
C00038520
|
Apetalolide
|
No. 54 |

|
||||
|
C00002514
|
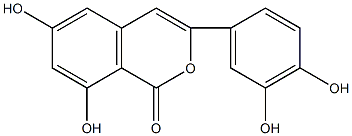
8-Methoxycoumestrol
/ 3'-Methoxycoumestrol / 7,12-Dihydroxy-11-methoxycoumestan |
No. 54 |
|
||||
|
C00002519
|
Trifoliol
/ 3,7-Dihydroxy-9-methoxycoumestan / 7,10-Dihydroxy-12-methoxycoumestan |
No. 54 |
|
||||
|
C00002548
|
Isotrifoliol
/ 3,9-Dihydroxy-1-methoxycoumestan |
No. 54 |

|
||||
|
C00035794
|
Mesuagin
/ Mammea A/AD cyclo D |
CHEMBL198546
|
No. 54 |

|
|||
|
C00034616
|
Mammea A/AB cyclo D
|
CHEMBL451993
|
No. 54 |

|
|||
|
C00002585
|
Wairol
/ 3-Hydroxy-7,9-dimethoxycoumestan / 7-Hydroxy-10,12-dimethoxycoumestan |
No. 54 |

|
||||
|
C00009757
|
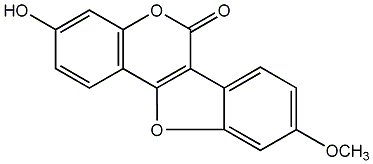
3-Hydroxy-8,9-dimethoxycoumestan
|
No. 54 |
|
||||
|
C00009758
|
4,2'-Epoxy-7,4'-dihydroxy-5,5'-dimethoxy-3-phenylcoumarin
|
No. 54 |

|
||||
|
C00009759
|
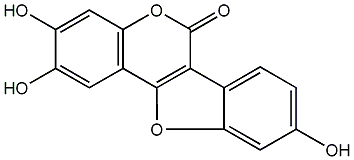
4,2'-Epoxy-5,4'-dihydroxy-7,5'-dimethoxy-3-phenylcoumarin
|
No. 54 |
|
||||
|
C00009761
|
4',5'-Dihydroxy-7-methoxy-5,2'-oxido-4-phenylcoumarin
|
No. 54 |

|
||||
|
C00009762
|
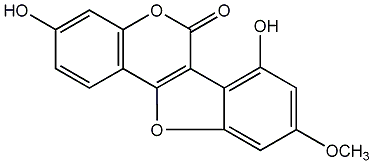
5'-Hydroxy-7,4'-dimethoxy-5,2'-oxido-4-phenylcoumarin
|
CHEMBL486220
|
No. 54 |
|
|||
|
C00009763
|
9-O-Methylcoumestrol
/ 4'-O-Methylcoumestrol / 3-Hydroxy-9-methoxycoumestan |
No. 54 |

|
||||
|
C00009764
|
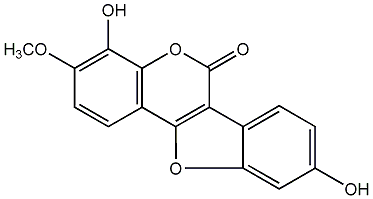
3,9-Dimethylcoumestan
/ Di-O-methylcoumestrol / Coumestrol dimethyl ether / 3,9-Dimethoxy-6-oxopterocarpene |
No. 54 |
|
||||
|
C00009765
|
7,10,12-Trihydroxycoumestan
/ Repensol3,7,9-Trihydroxycoumestan |
No. 54 |

|
||||
|
C00009767
|
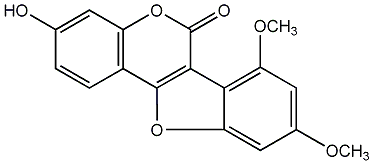
Aureol
/ 1,3,9-Trihydroxycoumestan |
No. 54 |
|
||||
|
C00009768
|
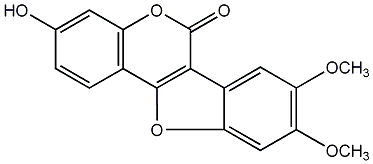
Lucernol
/ 2,3,9-Trihydroxycoumestan |
No. 54 |
|
||||
|
C00032364
|
MAB5
/ 5-Hydroxy-6'',6''-dimethyl-6-(2-methylbutyryl)-4-phenylpyrano[2'',3'':7,8]coumarin |
CHEMBL451993
|
No. 54 |

|
|||
|
C00030475
|
Mammeigin
/ 5-Hydroxy-6'',6''-dimethyl-6-(3-methylbutyryl)-4-phenylpyrano[2'',3'':7,8]coumarin |
CHEMBL195013
|
No. 54 |

|
|||
|
C00030474
|
Mammea A/AC cyclo D
|
CHEMBL455697
|
No. 54 |

|
|||
|
C00019955
|
Licoarylcoumarin
|
No. 54 |

|
||||
|
C00019597
|
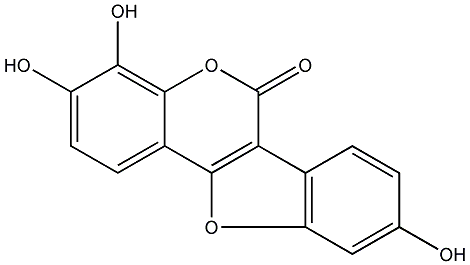
Sigmoidin K
/ 3,9-Dihydroxy-2,10-diprenylcoumestan |
CHEMBL1095262
|
C090620
|
No. 54 |
|
||
|
C00019289
|
Gancaonin W
/ 7,2',4'-Trihydroxy-5-methoxy-3'-prenyl-3-phenylcoumarin |
No. 54 |

|
||||
|
C00009781
|
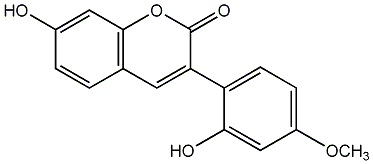
3',4'-Dihydroxy-5,7-dimethoxy-4-phenylcoumarin
|
No. 54 |
|
||||
|
C00009782
|
5,3'-Dihydroxy-7,4'-dimethoxy-4-phenylcoumarin
|
CHEMBL513547
|
No. 54 |

|
|||
|
C00019204
|
Glycycoumarin
|
CHEMBL1223642
|
1 / 0 / 0 | No. 54 |

|
||
|
C00019203
|
Lespecyrtin G1
|
CHEMBL564378
|
No. 54 |

|
|||
|
C00019197
|
Psoralidin
/ 3,9-Dihydroxy-2-prenylcoumestan |
C102768
|
No. 54 |

|
|||
|
C00019141
|
Phaseol
/ 3,9-Dihydroxy-4-prenylcoumestan |
No. 54 |
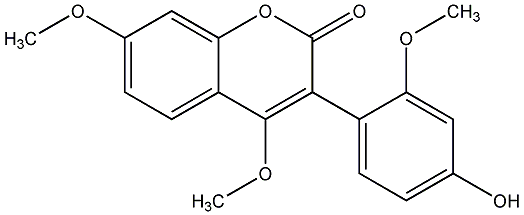
|
||||
|
C00019140
|
Isosojagol
|
CHEMBL1094311
|
No. 54 |
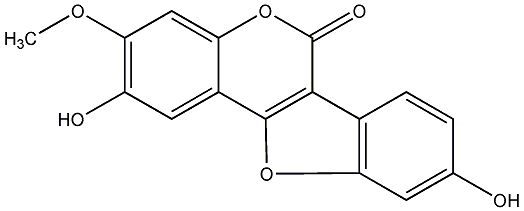
|
|||
|
C00010230
|
MAB3
/ 1,2-Dihydro-5-hydroxy-2-(1-hydroxy-1-methylethyl)-4-(2-methylbutyryl)-6-phenylfurano[2,3-h][1]benzopyran-8-one |
CHEMBL451991
CHEMBL481039 |
No. 54 |

|
|||
|
C00018990
|
1-O-Methylglycyrol
/ 5-O-Methylglycyrol / 9-Hydroxy-1,3-dimethoxy-2-prenylcoumestan |
CHEMBL522369
|
No. 54 |
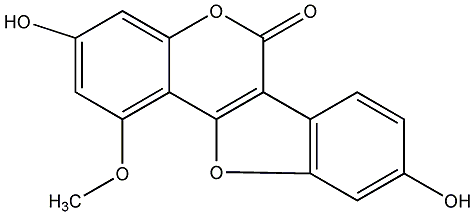
|
|||
|
C00015832
|
Glycyrol
/ Neoglycyrol / 1,9-Dihydroxy-3-methoxy-2-prenylcoumestan |
C071429
|
No. 54 |

|
|||
|
C00015829
|
Teysmanone B
|
No. 54 |

|
||||
|
C00015796
|
Calaustralin
|
No. 54 |
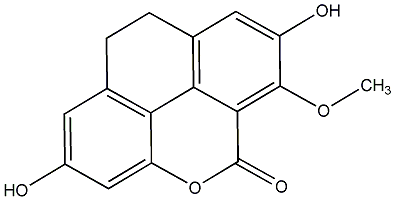
|
||||
|
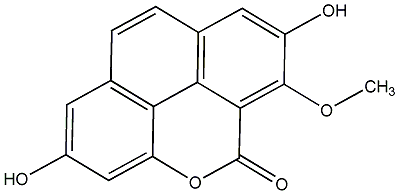
C00015792
|
Calocoumarin A
|
CHEMBL476875
|
No. 54 |
|
|||
|
C00015767
|
Brasimarin C
/ (+)-Brasimarin C |
CHEMBL476875
|
No. 54 |

|
|||
|
C00015755
|
Mammea A/AC
|
No. 54 |

|
||||
|
C00015274
|
Isoracemosol
|
No. 54 |

|
||||
|
C00010245
|
Mammeisin
/ Mammea A/AA / 5,7-Dihydroxy-8-(3-methyl-2-butenyl)-6-(3-methylbutyryl)-4-phenylcoumarin |
CHEMBL194485
|
C001878
|
No. 54 |

|
||
|
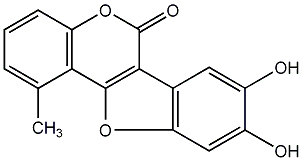
C00010060
|
Stevein
/ Stevenin |
No. 54 |
|
||||
|
C00010244
|
Mammea A/BA
/ 5,7-Dihydroxy-6-(3-methylbut-2-enyl)-8-(3-methylbutyryl)-4-phenylcoumarin |
CHEMBL511810
|
No. 54 |

|
|||
|
C00010193
|
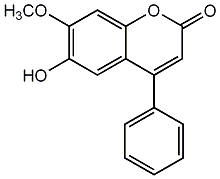
5,7-Dihydroxy-4'-methoxy-4-phenylcoumarin
|
No. 54 |

|
||||
|
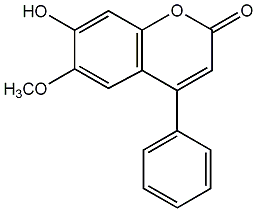
C00010194
|
5,3',4'-Trihydroxy-7-methoxy-4-phenylcoumarin
|
CHEMBL486219
|
No. 54 |
|
|||
|
C00010195
|
7',3',4'-Trihydroxy-5-methoxy-4-phenylcoumarin
|
No. 54 |

|
||||
|
C00010196
|
3'-Hydroxymelanettin
|
No. 54 |

|
||||
|
C00010198
|
7,2'-Dihydroxy-4',5'-methylenedioxy-3-phenylcoumarin
|
No. 54 |

|
||||
|
C00010199
|
Serratin
/ 5,7-Dihydroxy-4-phenylcoumarin |
CHEMBL600635
|
8 / 7 / 6 | No. 54 |

|
||
|
C00010200
|
Nordalbergin
/ 6,7-Dihydroxy-4-phenylcoumarin |
CHEMBL1255818
|
2 / 0 / 0 | No. 54 |

|
||
|
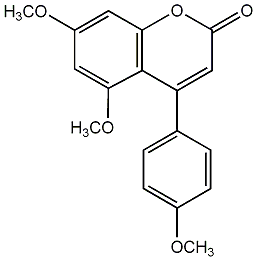
C00010201
|
4-Phenyl 5,7-dihydroxycoumarin
|
CHEMBL1233867
|
No. 54 |
|
|||
|
C00010202
|
Isodalbergin
/ 7-Hydroxy-6-methoxy-4-phenylcoumarin |
No. 54 |

|
||||
|
C00010203
|
Dalbergin
/ 6-Hydroxy-7-methoxy-4-phenylcoumarin |
CHEMBL1829658
|
No. 54 |

|
|||
|
C00010204
|
Methyldalbergin
/ 6,7-Dimethoxy-4-phenylcoumarin |
No. 54 |

|
||||
|
C00010205
|
Kuhlmannin
/ 6-Hydroxy-7,8-dimethoxy-4-phenylcoumarin |
CHEMBL1568181
|
2 / 0 / 1 | No. 54 |
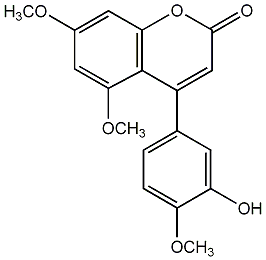
|
||
|
C00010207
|
Asphodelin A
|
CHEMBL268685
|
No. 54 |
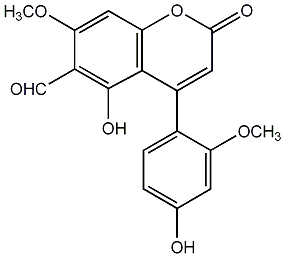
|
|||
|
C00010208
|
Thunberginol B
|
CHEMBL70501
|
C079056
|
20 / 16 / 45 | No. 54 |
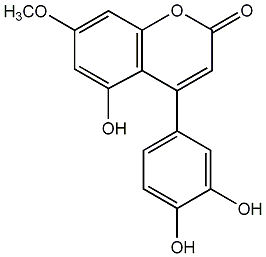
|
|
|
C00010209
|
Thunberginol A
/ 3-(3,4-dihydroxyphenyl)-8-hydroxyisocoumarin |
CHEMBL68810
|
C079055
|
14 / 5 / 4 | No. 54 |

|
|
|
C00010243
|
Mesuol
/ 5,7-Dihydroxy-8-(3-methylbut-2-enyl)-6-isobutyryl-4-phenylcoumarin |
CHEMBL197069
|
No. 54 |

|
|||
|
C00010242
|
Mammea A/BB
/ 5,7-Dihydroxy-6-(3-methylbut-2-enyl)-8-(2-methylbutyryl)-4-phenylcoumarin |
CHEMBL511832
CHEMBL1689186 |
No. 54 |

|
|||
|
C00010241
|
5,7-Dihydroxy-8-(3-methylbut-2-enyl)-6-(2-methylbutyryl)-4-phenylcoumarin
|
CHEMBL194782
CHEMBL1689185 |
No. 54 |

|
|||
|
C00010240
|
1,2-Dihydro-5-hydroxy-2-(1-hydroxy-1-methylethyl)-4-(3-methylbutyryl)-6-phenylfurano[2,3-h][1]benzopyran-8-one
|
CHEMBL196157
CHEMBL451992 |
No. 54 |

|
|||
|
C00010239
|
Mammea A/AC cyclo F
|
CHEMBL453305
CHEMBL478710 |
No. 54 |
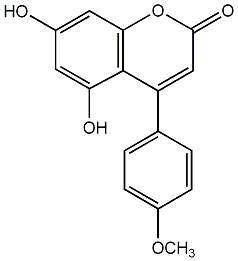
|
|||
|
C00010234
|
1,2-Dihydro-5-hydroxy-2-(1-hydroxy-1-methylethyl)-4-(butyryl)-6-phenylfurano[2,3-h][1]benzopyran-8-one
|
CHEMBL453305
CHEMBL478710 |
No. 54 |

|
|||
|
C00010233
|
Ochrocarpin E
|
CHEMBL486024
|
No. 54 |

|
|||
|
C00010232
|
1,2-Dihydro-5-hydroxy-2-(1-hydroxy-1-methylethyl)-4-(isobutyryl)-6-phenylfurano[2,3-h][1]benzopyran-8-one
|
CHEMBL198103
|
No. 54 |
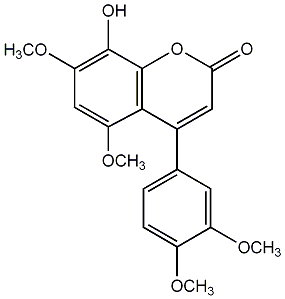
|
|||
|
C00010231
|
Brasimarin B
/ (+)-Brasimarin B |
CHEMBL476706
CHEMBL516283 |
No. 54 |

|
|||
|
C00010048
|
7,3'-Dihydroxy-5,4'-dimethoxy-6-formyl-4-phenylcoumarin
|
No. 54 |

|
||||
|
C00045863
|
Licopyranocoumarin
|
CHEMBL597425
|
C063348
|
1 / 0 | No. 266 |

|
|
|
C00010224
|
Ochrocarpin C
|
CHEMBL487415
|
No. 266 |
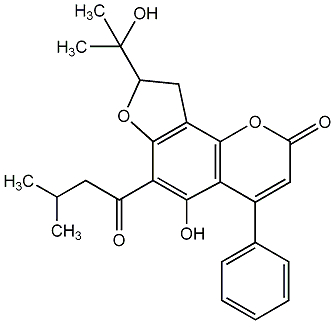
|
|||
|
C00010225
|
Ochrocarpin B
/ (-)-Ochrocarpin B |
CHEMBL460239
|
No. 266 |

|
|||
|
C00010226
|
Ochrocarpin D
/ (-)-Ochrocarpin D |
CHEMBL487416
|
No. 266 |

|
|||
|
C00010227
|
Disparacetylfuran A
|
No. 266 |

|
||||
|
C00010228
|
Disparfuran B
|
No. 266 |
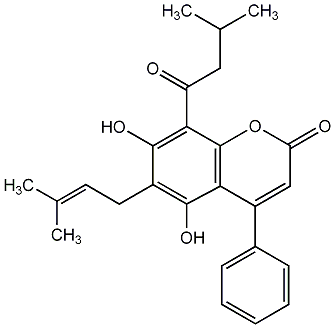
|
||||
|
C00010229
|
Isodisparfuran A
|
No. 266 |

|
||||
|
C00044611
|
Inophyllum A
/ (+)-Inophyllum A |
CHEMBL337029
CHEMBL132692 CHEMBL341394 CHEMBL422798 CHEMBL192847 CHEMBL466773 |
2 / 0 / 0 | No. 266 |

|
||
|
C00044571
|
Tomentolide A
|
No. 266 |

|
||||
|
C00010220
|
Paepalantine
|
CHEMBL1833982
|
2 / 0 / 0 | No. 266 |

|
||
|
C00010219
|
Toralactone
|
C005505
|
No. 266 |

|
|||
|
C00010217
|
Coeloginin
|
No. 266 |

|
||||
|
C00044570
|
Inophyllum C
|
CHEMBL334946
CHEMBL336955 |
No. 266 |

|
|||
|
C00041052
|
Teysmanone A
|
No. 266 |

|
||||
|
C00045862
|
Isoglycycoumarin
|
No. 266 |
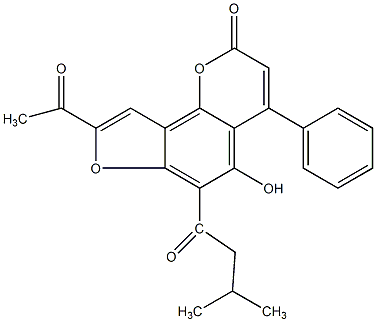
|
||||
|
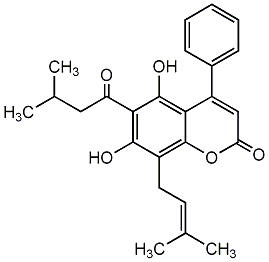
C00002482
|
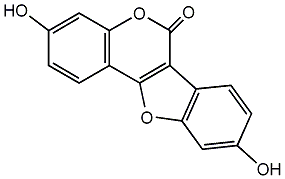
2-Methoxy-3,9-dihydroxycoumestone
|
No. 266 |
|
||||
|
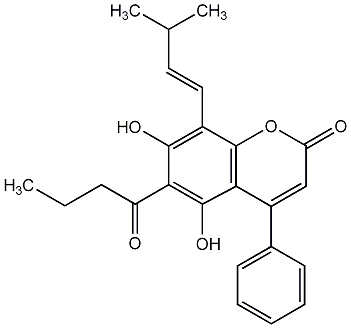
C00010211
|
Flaccidinin
|
No. 266 |
|
||||
|
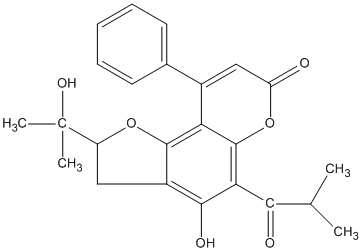
C00046219
|
Psoralidin oxide
|
No. 266 |
|
||||
|
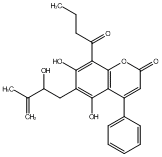
C00047462
|
Lespedezacoumestan
|
CHEMBL486292
|
No. 266 |
|
|||
|
C00046218
|
Bavacoumestan A
|
No. 266 |

|
||||
|
C00046046
|
Gancaonol A
/ 7-Hydroxy-5,2'-dimethoxy-6'',6''-dimethyl-4'',5''-dihydropyrano[2'',3'':4',3']-3-phenylcoumarin |
No. 266 |

|
||||
|
C00046217
|
Corylidin
|
No. 266 |

|
||||
|
C00049018
|
Licofuranocoumarin
/ 2,4'-Dihydroxy-5-methoxy-5''-(1-hydroxy-1-methylethyl)-4'',5''-dihydrofurano[2'',3'':7,6]-3-phenylcoumarin |
No. 266 |

|
||||
|
C00046215
|
Sojagol
/ Soyagol |
No. 266 |

|
||||
|
C00046216
|
Isoglycyrol
|
No. 266 |

|
||||
|
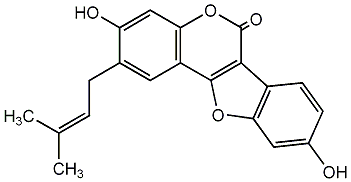
C00002566
|
Sativol
/ 4,9-Dihydroxy-3-methoxycoumestan / 8,12-Dihydroxy-7-methoxycoumestan |
No. 436 |
|
||||
|
C00018865
|
Mirificoumestan
/ 3,9-Dihydroxy-8-methoxy-7-prenylcoumestan |
No. 436 |

|
||||
|
C00018969
|
3,9-Dihydroxy-1-methoxy-8-prenylcoumestan
|
No. 436 |

|
||||
|
C00010055
|
3'-Hydroxy-5,7,8,4'-tetramethoxy-4-phenylcoumarin
|
No. 436 |

|
||||
|
C00010050
|
Exostemin
/ 8-Hydroxy-5,7,4'-trimethoxy-4-phenylcoumarin |
No. 436 |

|
||||
|
C00010043
|
Sisafolin
/ 5,4'-Dihydroxy-7,2'-dimethoxy-6-formyl-4-phenylcoumarin |
No. 436 |

|
||||
|
C00010051
|
3'-Hydroxy-5,7,4'-trimethoxy-4-phenylcoumarin
|
CHEMBL153529
|
3 / 2 / 0 | No. 436 |

|
||
|
C00010058
|
7,3'-Dihydroxy-4'-methoxy-4-phenylcoumarin
|
No. 436 |

|
||||
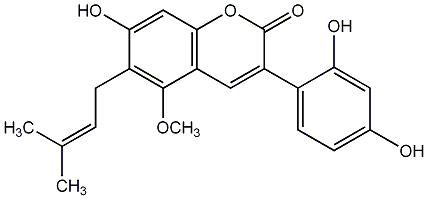
|
C00010040
|
Melannein
/ 6,3'-Dihydroxy-7,4'-dimethoxy-4-phenylcoumarin |
CHEMBL1801610
|
No. 436 |
|
|||
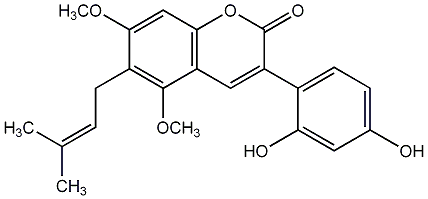
|
C00009785
|
7,3'-Dihydroxy-5,4'-dimethoxy-4-phenylcoumarin
|
No. 436 |
|
||||
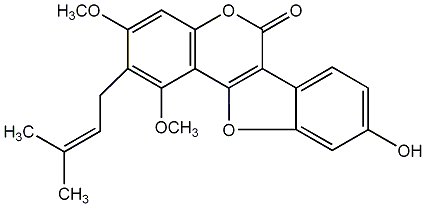
|
C00009780
|
Mutisifurocoumarin
/ 8,9-Dihydroxy-1-methylcoumestan |
C056049
|
No. 436 |
|
|||
|
C00019315
|
Glycyrin
/ 3-(2,4-Dihydroxyphenyl)-5,7-dimethoxy-6-prenylcoumarin |
CHEMBL132535
|
No. 436 |

|
|||
|
C00010059
|
Melannin
/ Melanettin |
No. 436 |

|
||||
|
C00019360
|
Lespedezol A6
/ 3,8,9-Trihydroxy-10-(3,7-dimethyl-2,6-octadienyl)coumestan |
No. 436 |

|
||||
|
C00010061
|
7,2'-Dihydroxy-4'-methoxy-3-phenylcoumarin
|
No. 436 |

|
||||
|
C00009777
|
Norwedelolactone
/ Demethylwedelolactone / Desmethylwedelolactone |
CHEMBL2089002
|
C051123
|
6 / 4 / 6 | No. 436 |

|
|
|
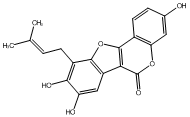
C00047282
|
Mirificoumestan hydrate
/ 3,9-Dihydroxy-8-methoxy-7-(3-hydroxy-3-methylbutyl)coumestan |
No. 436 |
|
||||
|
C00010223
|
Ochrocarpin A
/ (-)-Ochrocarpin A |
CHEMBL517870
|
No. 464 |

|
|||
|
C00044816
|
Inophyllolide chromanol
|
CHEMBL337029
CHEMBL132692 CHEMBL341394 CHEMBL422798 CHEMBL192847 CHEMBL466773 |
2 / 0 / 0 | No. 464 |

|
||
|
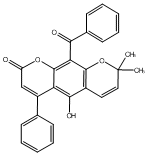
C00049017
|
Mirificoumestan glycol
|
No. 464 |
|
||||
|
C00019808
|
Licocoumarin A
/ 7,2'4'-Trihydroxy-8,3'-diprenyl-3-phenylcoumarin |
No. 464 |

|
||||
|
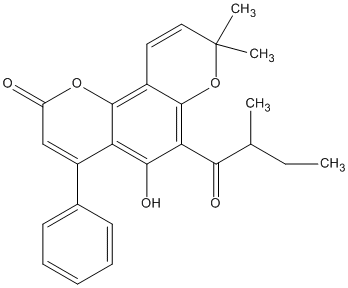
C00046114
|
Isopsoralidin
/ 1,2-Dideoxycorylidin |
No. 464 |
|
||||
|
C00035840
|
Brasimarin A
|
CHEMBL514685
|
No. 464 |

|
|||
|
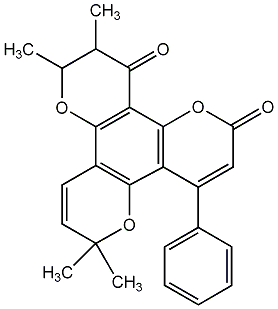
C00010213
|
Isoflaccidinin
|
No. 464 |
|
||||
|
C00010214
|
Ochrolide
|
No. 464 |

|
||||
|
C00041051
|
Calanone
|
CHEMBL518283
|
No. 464 |

|
|||
|
C00010215
|
Isooxoflaccidin
|
No. 464 |

|
||||
|
C00044569
|
Inophyllum E
/ cis-and-trans-Inophyllolide |
CHEMBL334946
CHEMBL336955 |
No. 464 |

|
|||
|
C00010216
|
Oxoflaccidin
|
No. 464 |

|
||||
|
C00010221
|
Herpetolide A
|
No. 464 |

|
||||
|
C00010222
|
Herpetolide B
|
No. 464 |

|
||||
|
C00044815
|
Inophyllum D
/ (+)-Inophyllum D |
CHEMBL337029
CHEMBL132692 CHEMBL341394 CHEMBL422798 CHEMBL192847 CHEMBL466773 |
2 / 0 / 0 | No. 464 |

|
||
|
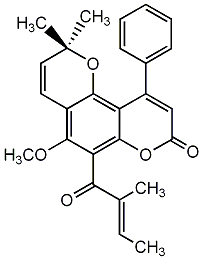
C00002455
|
Melimessanol A
/ 2,9-Dihydroxy-3-methoxycoumestan |
No. 464 |
|
||||
|
C00039617
|
Calophyllolide
|
C018859
|
No. 582 |

|
|||
|
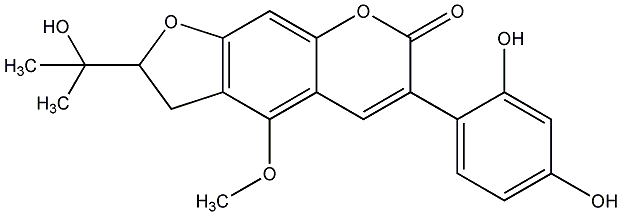
C00018989
|
Puerarostan
/ 3,9-Dihydroxy-4-methoxy-8-prenylcoumestan |
No. 582 |
|
||||
|
C00010057
|
5,7,4'-Trimethoxy-4-phenylcoumarin
|
No. 582 |

|
||||
|
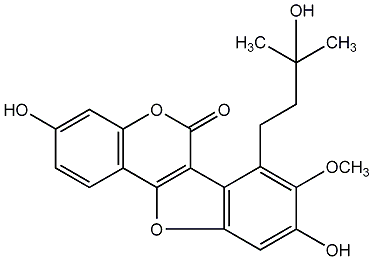
C00010056
|
8,3'-Dihydroxy-5,7,4'-trimethoxy-4-phenylcoumarin
|
No. 582 |
|
||||
|
C00010054
|
8-Hydroxy-5,7,3',4'-tetramethoxy-4-phenylcoumarin
|
No. 582 |

|
||||
|
C00010053
|
Melimessanol B
|
No. 582 |

|
||||
|
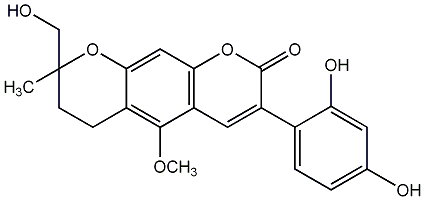
C00010042
|
8,3',4'-Trihydroxy-5,7-dimethoxy-4-phenylcoumarin
|
No. 582 |
|
||||
|
C00010041
|
4'-Hydroxy-5,7-dimethoxy-4-phenylcoumarin
|
CHEMBL153569
|
No. 582 |
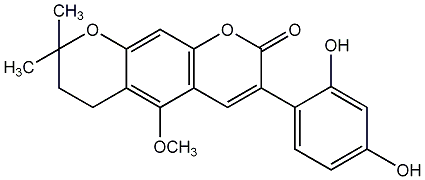
|
|||
|
C00009779
|
7,4',5'-Trihydroxy-5,2'-oxido-4-phenylcoumarin
|
No. 582 |

|
||||
|
C00009778
|
Wedelolactone
/ 1,8,9-Trihydroxy-3-methoxycoumestan / 5,11,12-Trihydroxy-7-methoxycoumestan |
CHEMBL97453
|
C051122
|
22 / 13 / 20 | No. 582 |
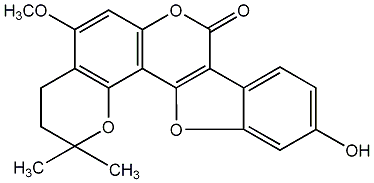
|
|
|
C00009776
|
Cumestrol
/ Coumestrol / 3,9-Dihydroxycoumestan |
CHEMBL30707
|
D003375
|
41 / 40 / 40 | 1795 / 2 | No. 582 |
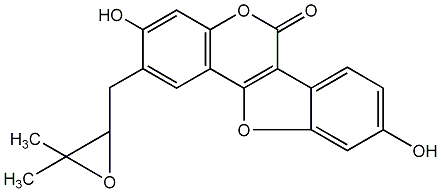
|
|
C00009775
|
4-Hydroxycoumestrol
/ 3,4,9-Trihydroxycoumestan |
No. 582 |

|
||||
|
C00002572
|
Pongacoumestan
/ 3,9-Dihydroxy-4-methoxycoumestan |
No. 582 |

|
||||
|
C00019321
|
Puerarol
/ 3,9-Dihydroxy-2-geranylcoumestan |
No. 582 |

|
Human Protein / Gene in interactions
82 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P06746 | DNA polymerase beta | Enzyme | C00002514 C00010214 C00044815 C00044816 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002514 C00002585 C00015274 C00050256 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00002514 C00010196 C00015274 C00050256 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00002514 C00015274 C00050256 | 1 / 1 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002514 C00015274 C00050256 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00002514 C00015274 C00050256 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002514 C00015274 C00050256 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002514 C00015274 C00050256 | 0 / 0 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00002514 C00015274 C00050256 | 0 / 0 |
| Q14191 | Werner syndrome ATP-dependent helicase | Enzyme | C00002514 C00015274 C00050256 | 2 / 1 |
| P09884 | DNA polymerase alpha catalytic subunit | Transferase | C00010214 C00044815 C00044816 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00002514 C00015274 C00050256 | 0 / 0 |
| P25963 | NF-kappa-B inhibitor alpha | Other cytosolic protein | C00002585 C00009765 | 1 / 2 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00015274 C00050256 | 0 / 0 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00002514 C00002585 | 3 / 1 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00002514 C00002585 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00002514 C00015274 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00002585 C00009765 | 0 / 0 |
| P27361 | Mitogen-activated protein kinase 3 | Erk | C00002585 C00009765 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00002514 C00002585 | 0 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00002514 C00002585 | 1 / 2 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00002585 C00050256 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00002514 C00002585 | 1 / 1 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00002514 C00002585 | 2 / 3 |
| Q05397 | Focal adhesion kinase 1 | Fak | C00002585 C00009765 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002514 C00010198 | 0 / 0 |
| P17405 | Sphingomyelin phosphodiesterase | Enzyme | C00002514 C00015274 | 2 / 2 |
| O00255 | Menin | Unclassified protein | C00002514 C00002585 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00002514 C00002585 | 1 / 2 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00002585 C00010196 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00002514 | 4 / 3 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00002514 | 0 / 0 |
| Q92731 | Estrogen receptor beta | NR3A2 | C00002514 | 0 / 1 |
| P15121 | Aldose reductase | Enzyme | C00010195 | 0 / 0 |
| O14920 | Inhibitor of nuclear factor kappa-B kinase subunit beta | Other serine/threonine protein kinase | C00002585 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00015274 | 0 / 0 |
| P11387 | DNA topoisomerase 1 | Isomerase | C00010205 | 0 / 0 |
| P09923 | Intestinal-type alkaline phosphatase | Enzyme | C00050256 | 0 / 0 |
| P05186 | Alkaline phosphatase, tissue-nonspecific isozyme | Enzyme | C00050256 | 3 / 1 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00050256 | 0 / 0 |
| P11308 | Transcriptional regulator ERG | Unclassified protein | C00050256 | 1 / 2 |
| Q9NPD5 | Solute carrier organic anion transporter family member 1B3 | Electrochemical transporter | C00002514 | 1 / 0 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00010196 | 1 / 0 |
| P51843 | Nuclear receptor subfamily 0 group B member 1 | Nuclear hormone receptor subfamily 0 group B member 1 | C00050256 | 2 / 2 |
| P51679 | C-C chemokine receptor type 4 | CC chemokine receptor | C00010196 | 0 / 0 |
| P06280 | Alpha-galactosidase A | Enzyme | C00002514 | 1 / 1 |
| O15111 | Inhibitor of nuclear factor kappa-B kinase subunit alpha | Other serine/threonine protein kinase | C00002585 | 1 / 1 |
| P10696 | Alkaline phosphatase, placental-like | Enzyme | C00050256 | 0 / 1 |
| P08253 | 72 kDa type IV collagenase | M10A | C00009765 | 1 / 3 |
| Q9UNQ0 | ATP-binding cassette sub-family G member 2 | ATP binding cassette | C00010205 | 2 / 0 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00002514 | 2 / 2 |
| O75828 | Carbonyl reductase [NADPH] 3 | Enzyme | C00002585 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00002514 | 1 / 1 |
| P49862 | Kallikrein-7 | S1A | C00034616 | 0 / 0 |
| Q00796 | Sorbitol dehydrogenase | Enzyme | C00010195 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00002514 | 0 / 0 |
| Q9Y337 | Kallikrein-5 | S1A | C00034616 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00010198 | 0 / 1 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00002514 | 0 / 0 |
| P37840 | Alpha-synuclein | Unclassified protein | C00010196 | 4 / 2 |
| P02545 | Prelamin-A/C | Unclassified protein | C00002514 | 11 / 10 |
| P16152 | Carbonyl reductase [NADPH] 1 | Enzyme | C00002585 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00009765 | 2 / 2 |
| P10275 | Androgen receptor | NR3C4 | C00002514 | 3 / 4 |
| P35869 | Aryl hydrocarbon receptor | Transcription Factor | C00002514 | 0 / 0 |
| Q9GZV3 | High affinity choline transporter 1 | Choline Na-symporter | C00010196 | 1 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00002514 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00002514 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00002514 | 1 / 1 |
| Q2M3G0 | ATP-binding cassette sub-family B member 5 | Unclassified protein | C00010205 | 0 / 0 |
| P56937 | 3-keto-steroid reductase | Enzyme | C00002514 | 0 / 0 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00002585 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00010196 | 1 / 1 |
| O94956 | Solute carrier organic anion transporter family member 2B1 | Unclassified protein | C00002514 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00002585 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00002585 | 1 / 4 |
| P63165 | Small ubiquitin-related modifier 1 | Unclassified protein | C00002514 | 1 / 1 |
| Q9Y6L6 | Solute carrier organic anion transporter family member 1B1 | Electrochemical transporter | C00002514 | 1 / 0 |
| P01215 | Glycoprotein hormones alpha chain | Unclassified protein | C00010196 | 0 / 3 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00050256 | 7 / 37 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00010040 | 0 / 0 |
| Q9HCT0 | Fibroblast growth factor 22 | Unclassified protein | C00050256 | 0 / 0 |
1796 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00010042
|
| 8086 | AAAS, AAA, AAASb, ADRACALA, ADRACALIN, ALADIN | achalasia, adrenocortical insufficiency, alacrimia |
C00002514
|
| 16 | AARS, CMT2N | alanyl-tRNA synthetase (EC:6.1.1.7) |
C00002514
|
| 18 | ABAT, GABA-AT, GABAT, NPD009 | 4-aminobutyrate aminotransferase (EC:2.6.1.19 2.6.1.22) |
C00002514
|
| 10057 | ABCC5, ABC33, EST277145, MOAT-C, MOATC, MRP5, SMRP, pABC11 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
C00002514
|
| 5825 | ABCD3, ABC43, PMP70, PXMP1, ZWS2 | ATP-binding cassette, sub-family D (ALD), member 3 |
C00002514
|
| 6059 | ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I | ATP-binding cassette, sub-family E (OABP), member 1 |
C00002514
|
| 23 | ABCF1, ABC27, ABC50 | ATP-binding cassette, sub-family F (GCN20), member 1 |
C00002514
|
| 9619 | ABCG1, ABC8, WHITE1 | ATP-binding cassette, sub-family G (WHITE), member 1 |
C00002514
|
| 83451 | ABHD11, WBSCR21 | abhydrolase domain containing 11 |
C00002514
|
| 26090 | ABHD12, ABHD12A, BEM46L2, C20orf22, PHARC, dJ965G21.2 | abhydrolase domain containing 12 (EC:3.1.1.23) |
C00002514
|
| 10006 | ABI1, ABI-1, ABLBP4, E3B1, NAP1BP, SSH3BP, SSH3BP1 | abl-interactor 1 |
C00002514
|
| 10449 | ACAA2, DSAEC | acetyl-CoA acyltransferase 2 (EC:2.3.1.16) |
C00002514
|
| 39 | ACAT2 | acetyl-CoA acetyltransferase 2 (EC:2.3.1.9) |
C00002514
|
| 64746 | ACBD3, GCP60, GOCAP1, GOLPH1, PAP7 | acyl-CoA binding domain containing 3 |
C00002514
|
| 57007 | ACKR3, CMKOR1, CXC-R7, CXCR-7, CXCR7, GPR159, RDC-1, RDC1 | atypical chemokine receptor 3 |
C00002514
|
| 47 | ACLY, ACL, ATPCL, CLATP | ATP citrate lyase (EC:2.3.3.8) |
C00002514
|
| 122970 | ACOT4, PTE-Ib, PTE1B, PTE2B | acyl-CoA thioesterase 4 (EC:3.1.2.2) |
C00002514
|
| 11332 | ACOT7, ACH1, ACT, BACH, CTE-II, LACH, LACH1, RP1-120G22.10, hBACH | acyl-CoA thioesterase 7 (EC:3.1.2.2) |
C00002514
|
| 2181 | ACSL3, ACS3, FACL3, PRO2194 | acyl-CoA synthetase long-chain family member 3 (EC:6.2.1.3) |
C00002514
|
| 79611 | ACSS3 | acyl-CoA synthetase short-chain family member 3 (EC:6.2.1.1) |
C00002514
|
| 90 | ACVR1, ACTRI, ACVR1A, ACVRLK2, ALK2, FOP, SKR1, TSRI | activin A receptor, type I (EC:2.7.11.30) |
C00002514
|
| 97 | ACYP1, ACYPE | acylphosphatase 1, erythrocyte (common) type (EC:3.6.1.7) |
C00002514
|
| 23536 | ADAT1, HADAT1 | adenosine deaminase, tRNA-specific 1 (EC:3.5.4.34) |
C00002514
|
| 120 | ADD3, ADDL | adducin 3 (gamma) |
C00002514
|
| 132 | ADK, AK | adenosine kinase (EC:2.7.1.20) |
C00002514
|
| 150 | ADRA2A, ADRA2, ADRA2R, ADRAR, ALPHA2AAR, ZNF32 | adrenoceptor alpha 2A |
C00002514
|
| 60312 | AFAP1, AFAP, AFAP-110, AFAP110 | actin filament associated protein 1 |
C00002514
|
| 27125 | AFF4, AF5Q31, MCEF | AF4/FMR2 family, member 4 |
C00002514
|
| 116987 | AGAP1, AGAP-1, CENTG2, GGAP1, cnt-g2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
C00002514
|
| 27161 | AGO2, EIF2C2, Q10 | argonaute RISC catalytic component 2 |
C00002514
|
| 56894 | AGPAT3, LPAAT-GAMMA1, LPAAT3 | 1-acylglycerol-3-phosphate O-acyltransferase 3 (EC:2.3.1.51) |
C00002514
|
| 137964 | AGPAT6, 1-AGPAT_6, LPAAT-zeta, LPAATZ, TSARG7 | 1-acylglycerol-3-phosphate O-acyltransferase 6 (EC:2.3.1.15) |
C00002514
|
| 10551 | AGR2, AG2, GOB-4, HAG-2, PDIA17, XAG-2 | anterior gradient 2 |
C00002514
|
| 155465 | AGR3, AG3, BCMP11, HAG3, PDIA18, hAG-3 | anterior gradient 3 |
C00002514
|
| 57085 | AGTRAP, ATRAP | angiotensin II receptor-associated protein |
C00002514
|
| 10768 | AHCYL1, DCAL, IRBIT, PRO0233, XPVKONA | adenosylhomocysteinase-like 1 (EC:3.3.1.1) |
C00002514
|
| 10598 | AHSA1, AHA1, C14orf3, p38 | AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
C00002514
|
| 84962 | AJUBA, JUB | ajuba LIM protein |
C00002514
|
| 11214 | AKAP13, AKAP-13, AKAP-Lbc, ARHGEF13, BRX, HA-3, Ht31, LBC, PRKA13, PROTO-LB, PROTO-LBC, c-lbc, p47 | A kinase (PRKA) anchor protein 13 |
C00002514
|
| 214 | ALCAM, CD166, MEMD | activated leukocyte cell adhesion molecule |
C00002514
|
| 126133 | ALDH16A1 | aldehyde dehydrogenase 16 family, member A1 |
C00002514
|
| 222 | ALDH3B2, ALDH8 | aldehyde dehydrogenase 3 family, member B2 (EC:1.2.1.5) |
C00002514
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00002514
|
| 29929 | ALG6, CDG1C | ALG6, alpha-1,3-glucosyltransferase (EC:2.4.1.267) |
C00002514
|
| 79053 | ALG8, CDG1H | ALG8, alpha-1,3-glucosyltransferase (EC:2.4.1.265) |
C00002514
|
| 84266 | ALKBH7, ABH7, SPATA11, UNQ6002 | alkB, alkylation repair homolog 7 (E. coli) |
C00002514
|
| 249 | ALPL, AP-TNAP, APTNAP, HOPS, TNAP, TNSALP | alkaline phosphatase, liver/bone/kidney (EC:3.1.3.1) |
C00002514
|
| 57679 | ALS2, ALS2CR6, ALSJ, IAHSP, PLSJ | amyotrophic lateral sclerosis 2 (juvenile) |
C00002514
|
| 702262 |
C00002514
|
||
| 10189 | ALYREF, ALY, ALY/REF, BEF, REF, THOC4 | Aly/REF export factor |
C00002514
|
| 262 | AMD1, ADOMETDC, AMD, SAMDC | adenosylmethionine decarboxylase 1 (EC:4.1.1.50) |
C00002514
|
| 267 | AMFR, GP78, RNF45 | autocrine motility factor receptor, E3 ubiquitin protein ligase (EC:6.3.2.19) |
C00002514
|
| 9949 | AMMECR1, AMMERC1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
C00002514
|
| 51421 | AMOTL2, LCCP | angiomotin like 2 |
C00002514
|
| 51433 | ANAPC5, APC5 | anaphase promoting complex subunit 5 |
C00002514
|
| 51434 | ANAPC7, APC7 | anaphase promoting complex subunit 7 |
C00002514
|
| 253650 | ANKRD18A | ankyrin repeat domain 18A |
C00002514
|
| 84250 | ANKRD32, BRCTD1, BRCTx | ankyrin repeat domain 32 |
C00002514
|
| 54443 | ANLN, Scraps, scra | anillin, actin binding protein |
C00002514
|
| 196527 | ANO6, BDPLT7, SCTS, TMEM16F | anoctamin 6 |
C00002514
|
| 8125 | ANP32A, C15orf1, HPPCn, I1PP2A, LANP, MAPM, PHAP1, PHAPI, PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
C00002514
|
| 81611 | ANP32E, LANP-L, LANPL | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
C00002514
|
| 302 | ANXA2, ANX2, ANX2L4, CAL1H, LIP2, LPC2, LPC2D, P36, PAP-IV | annexin A2 |
C00002514
|
| 304 | ANXA2P2, ANX2L2, ANX2P2, LPC2B | annexin A2 pseudogene 2 |
C00002514
|
| 306 | ANXA3, ANX3 | annexin A3 (EC:3.1.4.43) |
C00002514
|
| 8416 | ANXA9, ANX31 | annexin A9 |
C00002514
|
| 162 | AP1B1, ADTB1, AP105A, BAM22, CLAPB2 | adaptor-related protein complex 1, beta 1 subunit |
C00002514
|
| 8907 | AP1M1, AP47, CLAPM2, CLTNM, MU-1A | adaptor-related protein complex 1, mu 1 subunit |
C00002514
|
| 1175 | AP2S1, AP17, AP17-DELTA, CLAPS2, HHC3 | adaptor-related protein complex 2, sigma 1 subunit |
C00002514
|
| 323 | APBB2, FE65L, FE65L1 | amyloid beta (A4) precursor protein-binding, family B, member 2 |
C00002514
|
| 378708 | APITD1, CENP-S, CENPS, FAAP16, MHF1 | apoptosis-inducing, TAF9-like domain 1 |
C00002514
|
| 9582 | APOBEC3B, A3B, APOBEC1L, ARCD3, ARP4, DJ742C19.2, PHRBNL, bK150C2.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
C00002514
|
| 347 | APOD | apolipoprotein D |
C00002514
|
| 351 | APP, AAA, ABETA, ABPP, AD1, APPI, CTFgamma, CVAP, PN-II, PN2 | amyloid beta (A4) precursor protein |
C00002514
|
| 360 | AQP3, AQP-3, GIL | aquaporin 3 (Gill blood group) |
C00002514
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00002514
|
| 10564 | ARFGEF2, BIG2, PVNH2, dJ1164I10.1 | ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
C00002514
|
| 84986 | ARHGAP19 | Rho GTPase activating protein 19 |
C00002514
|
| 80728 | ARHGAP39, CrGAP, Vilse | Rho GTPase activating protein 39 |
C00002514
|
| 396 | ARHGDIA, GDIA1, NPHS8, RHOGDI, RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha |
C00002514
|
| 26084 | ARHGEF26, CSGEF, HMFN1864, SGEF | Rho guanine nucleotide exchange factor (GEF) 26 |
C00002514
|
| 389337 | ARHGEF37 | Rho guanine nucleotide exchange factor (GEF) 37 |
C00002514
|
| 84904 | ARHGEF39, C9orf100, RP11-331F9.7 | Rho guanine nucleotide exchange factor (GEF) 39 |
C00002514
|
| 51742 | ARID4B, BCAA, BRCAA1, RBBP1L1, RBP1L1, SAP180 | AT rich interactive domain 4B (RBP1-like) |
C00002514
|
| 403 | ARL3, ARFL3 | ADP-ribosylation factor-like 3 |
C00002514
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00002514
|
| 57561 | ARRDC3, TLIMP | arrestin domain containing 3 |
C00002514
|
| 35539 |
C00002514
|
||
| 414 | ARSD, ASD | arylsulfatase D (EC:3.1.6.1) |
C00002514
|
| 170302 | ARX, CT121, EIEE1, ISSX, MRX29, MRX32, MRX33, MRX36, MRX38, MRX43, MRX54, MRX76, MRX87, MRXS1, PRTS | aristaless related homeobox |
C00002514
|
| 55723 | ASF1B, CIA-II | anti-silencing function 1B histone chaperone |
C00002514
|
| 259266 | ASPM, ASP, Calmbp1, MCPH5 | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
C00002514
|
| 29028 | ATAD2, ANCCA, PRO2000 | ATPase family, AAA domain containing 2 (EC:3.6.1.3) |
C00002514
|
| 1386 | ATF2, CRE-BP1, CREB2, HB16, TREB7 | activating transcription factor 2 (EC:2.3.1.48) |
C00002514
|
| 467 | ATF3 | activating transcription factor 3 |
C00002514
|
| 22809 | ATF5, ATFX, HMFN0395 | activating transcription factor 5 |
C00002514
|
| 22863 | ATG14, ATG14L, BARKOR, KIAA0831 | autophagy related 14 |
C00002514
|
| 481 | ATP1B1, ATP1B | ATPase, Na+/K+ transporting, beta 1 polypeptide |
C00002514
|
| 489 | ATP2A3, SERCA3 | ATPase, Ca++ transporting, ubiquitous (EC:3.6.3.8) |
C00002514
|
| 490 | ATP2B1, PMCA1, PMCA1kb | ATPase, Ca++ transporting, plasma membrane 1 (EC:3.6.3.8) |
C00002514
|
| 27032 | ATP2C1, ATP2C1A, BCPM, HHD, PMR1, SPCA1, hSPCA1 | ATPase, Ca++ transporting, type 2C, member 1 (EC:3.6.3.8) |
C00002514
|
| 516 | ATP5G1, ATP5A, ATP5G | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
C00002514
|
| 27109 | ATP5S, ATPW, FB, HSU79253 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) (EC:3.6.1.14) |
C00002514
|
| 8992 | ATP6V0E1, ATP6H, ATP6V0E, M9.2, Vma21, Vma21p | ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (EC:3.6.3.14) |
C00002514
|
| 5205 | ATP8B1, ATPIC, BRIC, FIC1, ICP1, PFIC, PFIC1 | ATPase, aminophospholipid transporter, class I, type 8B, member 1 (EC:3.6.3.1) |
C00002514
|
| 57198 | ATP8B2, ATPID | ATPase, aminophospholipid transporter, class I, type 8B, member 2 (EC:3.6.3.1) |
C00002514
|
| 10079 | ATP9A, ATPIIA | ATPase, class II, type 9A (EC:3.6.3.1) |
C00002514
|
| 6310 | ATXN1, ATX1, D6S504E, SCA1 | ataxin 1 |
C00002514
|
| 6790 | AURKA, AIK, ARK1, AURA, AURORA2, BTAK, PPP1R47, STK15, STK6, STK7 | aurora kinase A (EC:2.7.11.1) |
C00002514
|
| 9212 | AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, PPP1R48, STK12, STK5, aurkb-sv1, aurkb-sv2 | aurora kinase B (EC:2.7.11.1) |
C00002514
|
| 26053 | AUTS2, FBRSL2 | autism susceptibility candidate 2 |
C00002514
|
| 33419 |
C00002514
|
||
| 567 | B2M | beta-2-microglobulin |
C00002514
|
| 8706 | B3GALNT1, B3GALT3, GLCT3, GLOB, Gb4Cer, P, P1, beta3Gal-T3, galT3 | beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) (EC:2.4.1.79) |
C00002514
|
| 84002 | B3GNT5, B3GN-T5, beta3Gn-T5 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 (EC:2.4.1.206) |
C00002514
|
| 23621 | BACE1, ASP2, BACE, HSPC104 | beta-site APP-cleaving enzyme 1 (EC:3.4.23.46) |
C00002514
|
| 25825 | BACE2, AEPLC, ALP56, ASP1, ASP21, BAE2, CEAP1, DRAP | beta-site APP-cleaving enzyme 2 (EC:3.4.23.45) |
C00002514
|
| 9532 | BAG2, BAG-2, dJ417I1.2 | BCL2-associated athanogene 2 |
C00002514
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00002514
|
| 9529 | BAG5, BAG-5 | BCL2-associated athanogene 5 |
C00002514
|
| 8815 | BANF1, BAF, BCRP1, D14S1460, NGPS | barrier to autointegration factor 1 |
C00002514
|
| 580 | BARD1 | BRCA1 associated RING domain 1 |
C00002514
|
| 393917 |
C00002514
|
||
| 100155979 |
C00002514
|
||
| 11176 | BAZ2A, TIP5, WALp3 | bromodomain adjacent to zinc finger domain, 2A |
C00002514
|
| 582 | BBS1, BBS2L2 | Bardet-Biedl syndrome 1 |
C00002514
|
| 8412 | BCAR3, NSP2, SH2D3B | breast cancer anti-estrogen resistance 3 |
C00002514
|
| 54828 | BCAS3, GAOB1, MAAB | breast carcinoma amplified sequence 3 |
C00002514
|
| 594 | BCKDHB, E1B, dJ279A18.1 | branched chain keto acid dehydrogenase E1, beta polypeptide (EC:1.2.4.4) |
C00002514
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00002514
|
| 83596 | BCL2L12 | BCL2-like 12 (proline rich) |
C00002514
|
| 602 | BCL3, BCL4, D19S37 | B-cell CLL/lymphoma 3 |
C00002514
|
| 607 | BCL9, LGS | B-cell CLL/lymphoma 9 |
C00002514
|
| 283149 | BCL9L, BCL9-2, DLNB11 | B-cell CLL/lymphoma 9-like |
C00002514
|
| 9774 | BCLAF1, BTF, bK211L9.1 | BCL2-associated transcription factor 1 |
C00002514
|
| 617 | BCS1L, BCS, BCS1, BJS, FLNMS, GRACILE, Hs.6719, MC3DN1, PTD, h-BCS | BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
C00002514
|
| 84707 | BEX2, BEX1, DJ79P11.1 | brain expressed X-linked 2 |
C00002514
|
| 100172339 |
C00002514
|
||
| 638 | BIK, BIP1, BP4, NBK | BCL2-interacting killer (apoptosis-inducing) |
C00002514
|
| 55909 | BIN3 | bridging integrator 3 |
C00002514
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00002514
|
| 641 | BLM, BS, RECQ2, RECQL2, RECQL3 | Bloom syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00002514
|
| 29760 | BLNK, AGM4, BASH, BLNK-S, LY57, SLP-65, SLP65, bca | B-cell linker |
C00002514
|
| 26258 | BLOC1S6, BLOS6, HPS9, PA, PALLID, PLDN | biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
C00002514
|
| 645 | BLVRB, BVRB, FLR, SDR43U1 | biliverdin reductase B (flavin reductase (NADPH)) (EC:1.3.1.24 1.5.1.30) |
C00002514
|
| 8548 | BLZF1, GOLGIN-45, JEM-1, JEM-1s, JEM1 | basic leucine zipper nuclear factor 1 |
C00002514
|
| 90427 | BMF | Bcl2 modifying factor |
C00002514
|
| 653 | BMP5 | bone morphogenetic protein 5 |
C00002514
|
| 655 | BMP7, OP-1 | bone morphogenetic protein 7 |
C00002514
|
| 659 | BMPR2, BMPR-II, BMPR3, BMR2, BRK-3, PPH1, T-ALK | bone morphogenetic protein receptor, type II (serine/threonine kinase) (EC:2.7.11.30) |
C00002514
|
| 665 | BNIP3L, BNIP3a, NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like |
C00002514
|
| 79866 | BORA, C13orf34 | bora, aurora kinase A activator |
C00002514
|
| 221927 | BRAT1, BAAT1, C7orf27, RMFSL | BRCA1-associated ATM activator 1 |
C00002514
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00002514
|
| 675 | BRCA2, BRCC2, BROVCA2, FACD, FAD, FAD1, FANCB, FANCD, FANCD1, GLM3, PNCA2 | breast cancer 2, early onset |
C00002514
|
| 8019 | BRD3, ORFX, RING3L | bromodomain containing 3 |
C00002514
|
| 25798 | BRI3, I3 | brain protein I3 |
C00002514
|
| 140707 | BRI3BP, BNAS1, HCCR-1, HCCR-2, HCCRBP-1, KG19 | BRI3 binding protein |
C00002514
|
| 54014 | BRWD1, C21orf107, N143, WDR9 | bromodomain and WD repeat domain containing 1 |
C00002514
|
| 55108 | BSDC1 | BSD domain containing 1 |
C00002514
|
| 682 | BSG, 5F7, CD147, EMMPRIN, M6, OK, TCSF | basigin |
C00002514
|
| 55727 | BTBD7, FUP1 | BTB (POZ) domain containing 7 |
C00002514
|
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative |
C00002514
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00002514
|
| 699 | BUB1, BUB1A, BUB1L, hBUB1 | BUB1 mitotic checkpoint serine/threonine kinase (EC:2.7.11.1) |
C00002514
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00002514
|
| 100528825 |
C00002514
|
||
| 220042 | C11orf82, noxin | chromosome 11 open reading frame 82 |
C00002514
|
| 790955 | C11orf83, CCDS41658.1, UNQ655 | chromosome 11 open reading frame 83 |
C00002514
|
| 57103 | C12orf5, FR2BP, TIGAR | chromosome 12 open reading frame 5 (EC:3.1.3.46) |
C00002514
|
| 283643 | C14orf80 | chromosome 14 open reading frame 80 |
C00002514
|
| 89927 | C16orf45 | chromosome 16 open reading frame 45 |
C00002514
|
| 29105 | C16orf80, EVORF, GTL3, fSAP23 | chromosome 16 open reading frame 80 |
C00002514
|
| 256302 | C17orf103, Gtlf3b | chromosome 17 open reading frame 103 |
C00002514
|
| 497661 | C18orf32 | chromosome 18 open reading frame 32 |
C00002514
|
| 84798 | C19orf48 | chromosome 19 open reading frame 48 |
C00002514
|
| 55732 | C1orf112, RP1-97P20.1 | chromosome 1 open reading frame 112 |
C00002514
|
| 79762 | C1orf115, RP11-322F10.4 | chromosome 1 open reading frame 115 |
C00002514
|
| 199920 | C1orf168 | chromosome 1 open reading frame 168 |
C00002514
|
| 149563 | C1orf64, ERRF, RP11-5P18.4 | chromosome 1 open reading frame 64 |
C00002514
|
| 112770 | C1orf85, NCU-G1 | chromosome 1 open reading frame 85 |
C00002514
|
| 205327 | C2orf69 | chromosome 2 open reading frame 69 |
C00002514
|
| 57415 | C3orf14 | chromosome 3 open reading frame 14 |
C00002514
|
| 79669 | C3orf52, TTMP | chromosome 3 open reading frame 52 |
C00002514
|
| 401152 | C4orf3 | chromosome 4 open reading frame 3 |
C00002514
|
| 201725 | C4orf46 | chromosome 4 open reading frame 46 |
C00002514
|
| 727 | C5, C5a, C5b, CPAMD4 | complement component 5 |
C00002514
|
| 375444 | C5orf34 | chromosome 5 open reading frame 34 |
C00002514
|
| 63920 | C5orf54, Buster3, ZBED8 | chromosome 5 open reading frame 54 |
C00002514
|
| 78996 | C7orf49, MRI | chromosome 7 open reading frame 49 |
C00002514
|
| 254778 | C8orf46 | chromosome 8 open reading frame 46 |
C00002514
|
| 286257 | C9orf142 | chromosome 9 open reading frame 142 |
C00002514
|
| 401546 | C9orf152, bA470J20.2 | chromosome 9 open reading frame 152 |
C00002514
|
| 55071 | C9orf40 | chromosome 9 open reading frame 40 |
C00002514
|
| 157983 | C9orf66, RP11-59O6.1 | chromosome 9 open reading frame 66 |
C00002514
|
| 771 | CA12, CAXII, HsT18816 | carbonic anhydrase XII (EC:4.2.1.1) |
C00002514
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00002514
|
| 27101 | CACYBP, GIG5, RP1-102G20.6, S100A6BP, SIP | calcyclin binding protein |
C00002514
|
| 799 | CALCR, CRT, CT-R, CTR, CTR1 | calcitonin receptor |
C00002514
|
| 808 | CALM3, PHKD, PHKD3 | calmodulin 3 (phosphorylase kinase, delta) (EC:2.7.11.19) |
C00002514
|
| 813 | CALU | calumenin |
C00002514
|
| 57118 | CAMK1D, CKLiK, CaM-K1, CaMKID | calcium/calmodulin-dependent protein kinase ID (EC:2.7.11.17) |
C00002514
|
| 817 | CAMK2D, CAMKD | calcium/calmodulin-dependent protein kinase II delta (EC:2.7.11.17) |
C00002514
|
| 55450 | CAMK2N1, PRO1489, RP11-401M16.1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
C00002514
|
| 124583 | CANT1, DBQD, SCAN-1, SCAN1, SHAPY | calcium activated nucleotidase 1 (EC:3.6.1.6) |
C00002514
|
| 22900 | CARD8, CARDINAL, DACAR, DAKAR, NDPP, NDPP1, TUCAN | caspase recruitment domain family, member 8 |
C00002514
|
| 23589 | CARHSP1, CRHSP-24, CSDC1 | calcium regulated heat stable protein 1, 24kDa |
C00002514
|
| 57082 | CASC5, AF15Q14, CT29, D40, KNL1, PPP1R55, Spc7, hKNL-1, hSpc105 | cancer susceptibility candidate 5 |
C00002514
|
| 9994 | CASP8AP2, CED-4, FLASH, RIP25 | caspase 8 associated protein 2 |
C00002514
|
| 831 | CAST, BS-17 | calpastatin |
C00002514
|
| 857 | CAV1, BSCL3, CGL3, MSTP085, PPH3, VIP21, CAV | caveolin 1, caveolae protein, 22kDa |
C00002514
|
| 865 | CBFB, PEBP2B | core-binding factor, beta subunit |
C00002514
|
| 868 | CBLB, Cbl-b, RNF56 | Cbl proto-oncogene B, E3 ubiquitin protein ligase |
C00002514
|
| 873 | CBR1, CBR, SDR21C1, hCBR1 | carbonyl reductase 1 (EC:1.1.1.189 1.1.1.197 1.1.1.184) |
C00002514
|
| 8535 | CBX4, NBP16, PC2 | chromobox homolog 4 |
C00002514
|
| 23468 | CBX5, HP1, HP1A | chromobox homolog 5 |
C00002514
|
| 112869 | CCDC101, SGF29, STAF36 | coiled-coil domain containing 101 |
C00002514
|
| 55013 | CCDC109B, MCUb | coiled-coil domain containing 109B |
C00002514
|
| 150275 | CCDC117, dJ366L4.1 | coiled-coil domain containing 117 |
C00002514
|
| 64770 | CCDC14 | coiled-coil domain containing 14 |
C00002514
|
| 154467 | CCDC167, C6orf129, HSPC265 | coiled-coil domain containing 167 |
C00002514
|
| 374969 | CCDC23 | coiled-coil domain containing 23 |
C00002514
|
| 91057 | CCDC34, NY-REN-41, RAMA3 | coiled-coil domain containing 34 |
C00002514
|
| 57003 | CCDC47, MSTP041 | coiled-coil domain containing 47 |
C00002514
|
| 100286684 |
C00002514
|
||
| 79714 | CCDC51 | coiled-coil domain containing 51 |
C00002514
|
| 131076 | CCDC58 | coiled-coil domain containing 58 |
C00002514
|
| 440193 | CCDC88C, DAPLE, HKRP2, KIAA1509 | coiled-coil domain containing 88C |
C00002514
|
| 461205 |
C00002514
|
||
| 890 | CCNA2, CCN1, CCNA | cyclin A2 |
C00002514
|
| 891 | CCNB1, CCNB | cyclin B1 |
C00002514
|
| 9133 | CCNB2, HsT17299 | cyclin B2 |
C00002514
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002514
|
| 9134 | CCNE2, CYCE2 | cyclin E2 |
C00002514
|
| 899 | CCNF, FBX1, FBXO1 | cyclin F |
C00002514
|
| 901 | CCNG2 | cyclin G2 |
C00002514
|
| 10983 | CCNI, CYC1, CYI | cyclin I |
C00002514
|
| 9236 | CCPG1, CPR8 | cell cycle progression 1 |
C00002514
|
| 100133941 | CD24, CD24A | CD24 molecule |
C00002514
|
| 51293 | CD320, 8D6, 8D6A, TCBLR | CD320 molecule |
C00002514
|
| 960 | CD44, CDW44, CSPG8, ECMR-III, HCELL, HUTCH-I, IN, LHR, MC56, MDU2, MDU3, MIC4, Pgp1 | CD44 molecule (Indian blood group) |
C00002514
|
| 1604 | CD55, CR, CROM, DAF, TC | CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
C00002514
|
| 966 | CD59, 16.3A5, 1F5, EJ16, EJ30, EL32, G344, HRF-20, HRF20, MAC-IP, MACIF, MEM43, MIC11, MIN1, MIN2, MIN3, MIRL, MSK21, p18-20 | CD59 molecule, complement regulatory protein |
C00002514
|
| 8555 | CDC14B, CDC14B3, Cdc14B1, Cdc14B2, hCDC14B | cell division cycle 14B (EC:3.1.3.16 3.1.3.48) |
C00002514
|
| 993 | CDC25A, CDC25A2 | cell division cycle 25A (EC:3.1.3.48) |
C00002514
|
| 995 | CDC25C, CDC25, PPP1R60 | cell division cycle 25C (EC:3.1.3.48) |
C00002514
|
| 996 | CDC27, ANAPC3, APC3, CDC27Hs, D0S1430E, D17S978E, HNUC, NUC2 | cell division cycle 27 |
C00002514
|
| 9578 | CDC42BPB, MRCKB | CDC42 binding protein kinase beta (DMPK-like) (EC:2.7.11.1) |
C00002514
|
| 31052 |
C00002514
|
||
| 988 | CDC5L, CDC5, CDC5-LIKE, CEF1, PCDC5RP, dJ319D22.1 | cell division cycle 5-like |
C00002514
|
| 990 | CDC6, CDC18L, HsCDC18, HsCDC6 | cell division cycle 6 |
C00002514
|
| 8317 | CDC7, CDC7L1, HsCDC7, Hsk1, huCDC7 | cell division cycle 7 (EC:2.7.11.1) |
C00002514
|
| 157313 | CDCA2, Repo-Man | cell division cycle associated 2 |
C00002514
|
| 83461 | CDCA3, GRCC8, TOME-1 | cell division cycle associated 3 |
C00002514
|
| 55038 | CDCA4, HEPP, SEI-3/HEPP | cell division cycle associated 4 |
C00002514
|
| 113130 | CDCA5, SORORIN | cell division cycle associated 5 |
C00002514
|
| 55536 | CDCA7L, JPO2, R1, RAM2 | cell division cycle associated 7-like |
C00002514
|
| 55143 | CDCA8, BOR, BOREALIN, DasraB, MESRGP | cell division cycle associated 8 |
C00002514
|
| 1010 | CDH12, CDHB | cadherin 12, type 2 (N-cadherin 2) |
C00002514
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00002514
|
| 8621 | CDK13, CDC2L, CDC2L5, CHED, hCDK13 | cyclin-dependent kinase 13 (EC:2.7.11.23 2.7.11.22) |
C00002514
|
| 10263 | CDK2AP2, DOC-1R, p14 | cyclin-dependent kinase 2 associated protein 2 |
C00002514
|
| 55755 | CDK5RAP2, C48, Cep215, MCPH3 | CDK5 regulatory subunit associated protein 2 |
C00002514
|
| 6792 | CDKL5, EIEE2, ISSX, STK9 | cyclin-dependent kinase-like 5 (EC:2.7.11.22) |
C00002514
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002514
|
| 1028 | CDKN1C, BWCR, BWS, KIP2, WBS, p57, p57Kip2 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
C00002514
|
| 55602 | CDKN2AIP, CARF | CDKN2A interacting protein |
C00002514
|
| 1030 | CDKN2B, CDK4I, INK4B, MTS2, P15, TP15, p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
C00002514
|
| 1031 | CDKN2C, INK4C, p18, p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
C00002514
|
| 1033 | CDKN3, CDI1, CIP2, KAP, KAP1 | cyclin-dependent kinase inhibitor 3 (EC:3.1.3.16 3.1.3.48) |
C00002514
|
| 1040 | CDS1, CDS | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 (EC:2.7.7.41) |
C00002514
|
| 81620 | CDT1, DUP, RIS2 | chromatin licensing and DNA replication factor 1 |
C00002514
|
| 55573 | CDV3, H41 | CDV3 homolog (mouse) |
C00002514
|
| 9425 | CDYL, CDYL1 | chromodomain protein, Y-like (EC:2.3.1.48) |
C00002514
|
| 1052 | CEBPD, C/EBP-delta, CELF, CRP3, NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta |
C00002514
|
| 1952 | CELSR2, CDHF10, EGFL2, Flamingo1, MEGF3 | cadherin, EGF LAG seven-pass G-type receptor 2 |
C00002514
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00002514
|
| 1062 | CENPE, CENP-E, KIF10, PPP1R61 | centromere protein E, 312kDa |
C00002514
|
| 1063 | CENPF, CENF, PRO1779, hcp-1 | centromere protein F, 350/400kDa |
C00002514
|
| 64946 | CENPH | centromere protein H |
C00002514
|
| 2491 | CENPI, CENP-I, FSHPRH1, LRPR1, Mis6 | centromere protein I |
C00002514
|
| 55835 | CENPJ, BM032, CENP-J, CPAP, LAP, LIP1, MCPH6, SASS4, SCKL4, Sas-4 | centromere protein J |
C00002514
|
| 91687 | CENPL, C1orf155, CENP-L, RP3-383J4.1, dJ383J4.3 | centromere protein L |
C00002514
|
| 79019 | CENPM, C22orf18, CENP-M, PANE1, bK250D10.2 | centromere protein M |
C00002514
|
| 55839 | CENPN, BM039, C16orf60, CENP-N, ICEN32 | centromere protein N |
C00002514
|
| 79172 | CENPO, CENP-O, ICEN-36, MCM21R | centromere protein O |
C00002514
|
| 55166 | CENPQ, C6orf139, CENP-Q | centromere protein Q |
C00002514
|
| 387103 | CENPW, C6orf173, CENP-W, CUG2 | centromere protein W |
C00002514
|
| 145508 | CEP128, C14orf145, C14orf61, LEDP/132 | centrosomal protein 128kDa |
C00002514
|
| 22995 | CEP152, MCPH4, MCPH9, SCKL5 | centrosomal protein 152kDa |
C00002514
|
| 55125 | CEP192, PPP1R62 | centrosomal protein 192kDa |
C00002514
|
| 100196261 |
C00002514
|
||
| 55165 | CEP55, C10orf3, CT111, URCC6 | centrosomal protein 55kDa |
C00002514
|
| 9702 | CEP57, MVA2, PIG8, TSP57 | centrosomal protein 57kDa |
C00002514
|
| 285753 | CEP57L1, C6orf182, bA487F23.2, cep57R | centrosomal protein 57kDa-like 1 |
C00002514
|
| 84131 | CEP78, C9orf81, IP63 | centrosomal protein 78kDa |
C00002514
|
| 64793 | CEP85, CCDC21 | centrosomal protein 85kDa |
C00002514
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00002514
|
| 57530 | CGN | cingulin |
C00002514
|
| 494143 | CHAC2 | ChaC, cation transport regulator homolog 2 (E. coli) |
C00002514
|
| 10036 | CHAF1A, CAF-1, CAF1, CAF1B, CAF1P150, P150 | chromatin assembly factor 1, subunit A (p150) |
C00002514
|
| 8208 | CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSPH7, MPP7 | chromatin assembly factor 1, subunit B (p60) |
C00002514
|
| 1106 | CHD2, EEOC | chromodomain helicase DNA binding protein 2 (EC:3.6.4.12) |
C00002514
|
| 84181 | CHD6, CHD-6, CHD5, RIGB | chromodomain helicase DNA binding protein 6 (EC:3.6.4.12) |
C00002514
|
| 1111 | CHEK1, CHK1 | checkpoint kinase 1 (EC:2.7.11.1) |
C00002514
|
| 11200 | CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53, hCds1 | checkpoint kinase 2 (EC:2.7.11.1) |
C00002514
|
| 555843 |
C00002514
|
||
| 1123 | CHN1, ARHGAP2, CHN, DURS2, NC, RHOGAP2 | chimerin 1 |
C00002514
|
| 56994 | CHPT1, CPT, CPT1 | choline phosphotransferase 1 (EC:2.7.8.2) |
C00002514
|
| 1138 | CHRNA5, LNCR2 | cholinergic receptor, nicotinic, alpha 5 (neuronal) |
C00002514
|
| 55847 | CISD1, C10orf70, ZCD1, mitoNEET | CDGSH iron sulfur domain 1 |
C00002514
|
| 11113 | CIT, CRIK, STK21 | citron (rho-interacting, serine/threonine kinase 21) (EC:2.7.11.1) |
C00002514
|
| 10370 | CITED2, ASD8, MRG-1, MRG1, P35SRJ, VSD2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
C00002514
|
| 26586 | CKAP2, LB1, TMAP, se20-10 | cytoskeleton associated protein 2 |
C00002514
|
| 150468 | CKAP2L | cytoskeleton associated protein 2-like |
C00002514
|
| 10970 | CKAP4, CLIMP-63, ERGIC-63, p63 | cytoskeleton-associated protein 4 |
C00002514
|
| 9793 | CKAP5, CHTOG, MSPS, TOG, TOGp, ch-TOG | cytoskeleton associated protein 5 |
C00002514
|
| 51192 | CKLF, C32, CKLF1, CKLF2, CKLF3, CKLF4, UCK-1 | chemokine-like factor |
C00002514
|
| 1163 | CKS1B, CKS1, PNAS-16, PNAS-18, ckshs1 | CDC28 protein kinase regulatory subunit 1B |
C00002514
|
| 1164 | CKS2, CKSHS2 | CDC28 protein kinase regulatory subunit 2 |
C00002514
|
| 23155 | CLCC1, MCLC | chloride channel CLIC-like 1 |
C00002514
|
| 9076 | CLDN1, CLD1, ILVASC, SEMP1 | claudin 1 |
C00002514
|
| 1364 | CLDN4, CPE-R, CPER, CPETR, CPETR1, WBSCR8, hCPE-R | claudin 4 |
C00002514
|
| 6249 | CLIP1, CLIP, CLIP-170, CLIP170, CYLN1, RSN | CAP-GLY domain containing linker protein 1 |
C00002514
|
| 54982 | CLN6, CLN4A, HsT18960, nclf | ceroid-lipofuscinosis, neuronal 6, late infantile, variant |
C00002514
|
| 2055 | CLN8, C8orf61, EPMR | ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) |
C00002514
|
| 1191 | CLU, APO-J, APOJ, CLI, CLU1, CLU2, KUB1, NA1/NA2, SGP-2, SGP2, SP-40, TRPM-2, TRPM2 | clusterin |
C00002514
|
| 56942 | CMC2, 2310061C15Rik, C16orf61 | C-x(9)-C motif containing 2 |
C00002514
|
| 84319 | CMSS1, C3orf26 | cms1 ribosomal small subunit homolog (yeast) |
C00002514
|
| 152189 | CMTM8, CKLFSF8, CKLFSF8-V2 | CKLF-like MARVEL transmembrane domain containing 8 |
C00002514
|
| 55744 | COA1, C7orf44, MITRAC15 | cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
C00002514
|
| 23242 | COBL | cordon-bleu WH2 repeat protein |
C00002514
|
| 1690 | COCH, COCH-5B2, COCH5B2, DFNA9, DFNA31 | cochlin |
C00002514
|
| 25839 | COG4, CDG2J, COD1 | component of oligomeric golgi complex 4 |
C00002514
|
| 10466 | COG5, CDG2I, GOLTC1, GTC90 | component of oligomeric golgi complex 5 |
C00002514
|
| 57511 | COG6, CDG2L, COD2, SHNS | component of oligomeric golgi complex 6 |
C00002514
|
| 81578 | COL21A1, COLA1L, dJ682J15.1, dJ708F5.1 | collagen, type XXI, alpha 1 |
C00002514
|
| 54939 | COMMD4 | COMM domain containing 4 |
C00002514
|
| 1312 | COMT | catechol-O-methyltransferase (EC:2.1.1.6) |
C00002514
|
| 9318 | COPS2, ALIEN, CSN2, SGN2, TRIP15 | COP9 signalosome subunit 2 |
C00002514
|
| 27235 | COQ2, CL640, COQ10D1, MSA1 | coenzyme Q2 4-hydroxybenzoate polyprenyltransferase (EC:2.5.1.39) |
C00002514
|
| 10229 | COQ7, CAT5, CLK-1, CLK1 | coenzyme Q7 homolog, ubiquinone (yeast) |
C00002514
|
| 57175 | CORO1B, CORONIN-2 | coronin, actin binding protein, 1B |
C00002514
|
| 23603 | CORO1C, HCRNN4 | coronin, actin binding protein, 1C |
C00002514
|
| 7464 | CORO2A, CLIPINB, IR10, WDR2 | coronin, actin binding protein, 2A |
C00002514
|
| 33136 |
C00002514
|
||
| 1363 | CPE | carboxypeptidase E (EC:3.4.17.10) |
C00002514
|
| 132864 | CPEB2, CPE-BP2, CPEB-2, hCPEB-2 | cytoplasmic polyadenylation element binding protein 2 |
C00002514
|
| 51692 | CPSF3, CPSF-73, CPSF73 | cleavage and polyadenylation specific factor 3, 73kDa |
C00002514
|
| 148327 | CREB3L4, AIBZIP, ATCE1, CREB3, CREB4, JAL, hJAL | cAMP responsive element binding protein 3-like 4 |
C00002514
|
| 1397 | CRIP2, CRIP, CRP2, ESP1 | cysteine-rich protein 2 |
C00002514
|
| 1407 | CRY1, PHLL1 | cryptochrome 1 (photolyase-like) |
C00002514
|
| 1434 | CSE1L, CAS, CSE1, XPO2 | CSE1 chromosome segregation 1-like (yeast) |
C00002514
|
| 1452 | CSNK1A1, CK1, CK1a, CKIa, HLCDGP1, PRO2975 | casein kinase 1, alpha 1 (EC:2.7.11.1) |
C00002514
|
| 53944 | CSNK1G1, CK1gamma1 | casein kinase 1, gamma 1 (EC:2.7.11.1) |
C00002514
|
| 1456 | CSNK1G3, CKI-gamma_3, CSNK1G3L | casein kinase 1, gamma 3 (EC:2.7.11.1) |
C00002514
|
| 1459 | CSNK2A2, CK2A2, CSNK2A1 | casein kinase 2, alpha prime polypeptide (EC:2.7.11.1) |
C00002514
|
| 10675 | CSPG5, NGC | chondroitin sulfate proteoglycan 5 (neuroglycan C) |
C00002514
|
| 81566 | CSRNP2, C12orf2, C12orf22, FAM130A1, PPP1R72, TAIP-12 | cysteine-serine-rich nuclear protein 2 |
C00002514
|
| 1475 | CSTA, AREI, STF1, STFA | cystatin A (stefin A) |
C00002514
|
| 1477 | CSTF1, CstF-50, CstFp50 | cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
C00002514
|
| 1478 | CSTF2, CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
C00002514
|
| 1487 | CTBP1, BARS | C-terminal binding protein 1 |
C00002514
|
| 51496 | CTDSPL2, HSPC129 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 (EC:3.1.3.-) |
C00002514
|
| 8727 | CTNNAL1, CLLP, alpha-CATU | catenin (cadherin-associated protein), alpha-like 1 |
C00002514
|
| 1500 | CTNND1, CAS, CTNND, P120CAS, P120CTN, p120, p120(CAS), p120(CTN) | catenin (cadherin-associated protein), delta 1 |
C00002514
|
| 1501 | CTNND2, GT24, NPRAP | catenin (cadherin-associated protein), delta 2 |
C00002514
|
| 51797 |
C00002514
|
||
| 9646 | CTR9, SH2BP1, TSBP, p150, p150TSP | CTR9, Paf1/RNA polymerase II complex component |
C00002514
|
| 1508 | CTSB, APPS, CPSB | cathepsin B (EC:3.4.22.1) |
C00002514
|
| 1075 | CTSC, CPPI, DPP-I, DPP1, DPPI, HMS, JP, JPD, PALS, PDON1, PLS | cathepsin C (EC:3.4.14.1) |
C00002514
|
| 1512 | CTSH, ACC-4, ACC-5, CPSB, minichain | cathepsin H (EC:3.4.22.16) |
C00002514
|
| 1515 | CTSV, CATL2, CTSL2, CTSU | cathepsin V (EC:3.4.22.43) |
C00002514
|
| 2017 | CTTN, EMS1 | cortactin |
C00002514
|
| 55917 | CTTNBP2NL | CTTNBP2 N-terminal like |
C00002514
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00002514
|
| 256643 | CXorf23 | chromosome X open reading frame 23 |
C00002514
|
| 51523 | CXXC5, CF5, RINF, WID | CXXC finger protein 5 |
C00002514
|
| 80777 | CYB5B, CYB5-M, CYPB5M, OMB5 | cytochrome b5 type B (outer mitochondrial membrane) |
C00002514
|
| 51706 | CYB5R1, B5R.1, B5R1, B5R2, NQO3A2, humb5R2 | cytochrome b5 reductase 1 (EC:1.6.2.2) |
C00002514
|
| 26999 | CYFIP2, PIR121 | cytoplasmic FMR1 interacting protein 2 |
C00002514
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00002514
|
| 120227 | CYP2R1 | cytochrome P450, family 2, subfamily R, polypeptide 1 (EC:1.14.13.159) |
C00002514
|
| 1595 | CYP51A1, CP51, CYP51, CYPL1, LDM, P450-14DM, P450L1 | cytochrome P450, family 51, subfamily A, polypeptide 1 (EC:1.14.13.70) |
C00002514
|
| 3491 | CYR61, CCN1, GIG1, IGFBP10 | cysteine-rich, angiogenic inducer, 61 |
C00002514
|
| 9266 | CYTH2, ARNO, CTS18, CTS18.1, PSCD2, PSCD2L, SEC7L, Sec7p-L, Sec7p-like | cytohesin 2 |
C00002514
|
| 23002 | DAAM1 | dishevelled associated activator of morphogenesis 1 |
C00002514
|
| 26007 | DAK, NET45 | dihydroxyacetone kinase 2 homolog (S. cerevisiae) (EC:2.7.1.29 4.6.1.15 2.7.1.28) |
C00002514
|
| 57291 | DANCR, AGU2, ANCR, KIAA0114, SNHG13 | differentiation antagonizing non-protein coding RNA |
C00002514
|
| 55157 | DARS2, ASPRS, LBSL, MT-ASPRS, RP3-383J4.2 | aspartyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.12) |
C00002514
|
| 10926 | DBF4, ASK, CHIF, DBF4A, ZDBF1 | DBF4 homolog (S. cerevisiae) |
C00002514
|
| 1627 | DBN1, D0S117E | drebrin 1 |
C00002514
|
| 90379 | DCAF15, C19orf72 | DDB1 and CUL4 associated factor 15 |
C00002514
|
| 26094 | DCAF4, WDR21, WDR21A | DDB1 and CUL4 associated factor 4 |
C00002514
|
| 8816 | DCAF5, BCRG2, BCRP2, D14S1461E, WDR22 | DDB1 and CUL4 associated factor 5 |
C00002514
|
| 100528464 |
C00002514
|
||
| 1633 | DCK | deoxycytidine kinase (EC:2.7.1.74) |
C00002514
|
| 9201 | DCLK1, CL1, CLICK1, DCAMKL1, DCDC3A, DCLK | doublecortin-like kinase 1 (EC:2.7.11.1) |
C00002514
|
| 9937 | DCLRE1A, PSO2, SNM1, SNM1A | DNA cross-link repair 1A |
C00002514
|
| 64858 | DCLRE1B, APOLLO, SNM1B, SNMIB | DNA cross-link repair 1B |
C00002514
|
| 28960 | DCPS, DCS1, HINT-5, HINT5, HSL1 | decapping enzyme, scavenger (EC:3.6.1.59) |
C00002514
|
| 79077 | DCTPP1, RS21C6, XTP3TPA | dCTP pyrophosphatase 1 (EC:3.6.1.12) |
C00002514
|
| 23564 | DDAH2, DDAH, DDAHII, G6a, NG30 | dimethylarginine dimethylaminohydrolase 2 (EC:3.5.3.18) |
C00002514
|
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00002514
|
| 326709 |
C00002514
|
||
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00002514
|
| 1663 | DDX11, CHL1, CHLR1, KRG2, WABS | DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 (EC:3.6.4.13) |
C00002514
|
| 68278 |
C00002514
|
||
| 54606 | DDX56, DDX21, DDX26, NOH61 | DEAD (Asp-Glu-Ala-Asp) box helicase 56 (EC:3.6.4.13) |
C00002514
|
| 7913 | DEK, D6S231E | DEK oncogene |
C00002514
|
| 57706 | DENND1A, FAM31A, KIAA1608, RP11-230L22.3 | DENN/MADD domain containing 1A |
C00002514
|
| 163486 | DENND1B, C1ORF18, C1orf218, FAM31B | DENN/MADD domain containing 1B |
C00002514
|
| 79958 | DENND1C, FAM31C | DENN/MADD domain containing 1C |
C00002514
|
| 8562 | DENR, DRP, DRP1, SMAP-3 | density-regulated protein |
C00002514
|
| 55635 | DEPDC1, DEP.8, DEPDC1-V2, DEPDC1A, SDP35 | DEP domain containing 1 |
C00002514
|
| 55789 | DEPDC1B, BRCC3, XTP1 | DEP domain containing 1B |
C00002514
|
| 420357 |
C00002514
|
||
| 51071 | DERA, DEOC | deoxyribose-phosphate aldolase (putative) (EC:4.1.2.4) |
C00002514
|
| 51029 | DESI2, C1orf121, DESI, DESI1, DeSI-2, FAM152A, PNAS-4, PPPDE1 | desumoylating isopeptidase 2 |
C00002514
|
| 1717 | DHCR7, SLOS | 7-dehydrocholesterol reductase (EC:1.3.1.21) |
C00002514
|
| 1719 | DHFR, DHFRP1, DYR | dihydrofolate reductase (EC:1.5.1.3) |
C00002514
|
| 10202 | DHRS2, HEP27, SDR25C1 | dehydrogenase/reductase (SDR family) member 2 |
C00002514
|
| 51635 | DHRS7, SDR34C1, retDSR4, retSDR4 | dehydrogenase/reductase (SDR family) member 7 |
C00002514
|
| 55526 | DHTKD1, AMOXAD, CMT2Q | dehydrogenase E1 and transketolase domain containing 1 (EC:1.2.4.2) |
C00002514
|
| 81624 | DIAPH3, AN, AUNA1, DIA2, DRF3, NSDAN, diap3, mDia2 | diaphanous-related formin 3 |
C00002514
|
| 23405 | DICER1, DCR1, Dicer, HERNA, MNG1, RMSE2 | dicer 1, ribonuclease type III (EC:3.1.26.-) |
C00002514
|
| 22982 | DIP2C, KIAA0934 | DIP2 disco-interacting protein 2 homolog C (Drosophila) |
C00002514
|
| 85458 | DIXDC1, CCD1 | DIX domain containing 1 |
C00002514
|
| 1736 | DKC1, CBF5, DKC, DKCX, NAP57, NOLA4, XAP101 | dyskeratosis congenita 1, dyskerin |
C00002514
|
| 1737 | DLAT, DLTA, PDC-E2, PDCE2 | dihydrolipoamide S-acetyltransferase (EC:2.3.1.12) |
C00002514
|
| 79469 | DLEU2L, BCMSUNL | deleted in lymphocytic leukemia 2-like |
C00002514
|
| 9787 | DLGAP5, DLG7, HURP | discs, large (Drosophila) homolog-associated protein 5 |
C00002514
|
| 1746 | DLX2, TES-1, TES1 | distal-less homeobox 2 |
C00002514
|
| 1657 | DMXL1 | Dmx-like 1 |
C00002514
|
| 1763 | DNA2, DNA2L, PEOA6, hDNA2 | DNA replication helicase/nuclease 2 (EC:3.6.4.12) |
C00002514
|
| 10294 | DNAJA2, CPR3, DJ3, DJA2, DNAJ, DNJ3, HIRIP4, PRO3015, RDJ2 | DnaJ (Hsp40) homolog, subfamily A, member 2 |
C00002514
|
| 4189 | DNAJB9, ERdj4, MDG-1, MDG1, MST049, MSTP049 | DnaJ (Hsp40) homolog, subfamily B, member 9 |
C00002514
|
| 64215 | DNAJC1, DNAJL1, ERdj1, HTJ1, MTJ1 | DnaJ (Hsp40) homolog, subfamily C, member 1 |
C00002514
|
| 23341 | DNAJC16 | DnaJ (Hsp40) homolog, subfamily C, member 16 |
C00002514
|
| 23234 | DNAJC9, HDJC9, JDD1, SB73 | DnaJ (Hsp40) homolog, subfamily C, member 9 |
C00002514
|
| 1777 | DNASE2, DNASE2A, DNL, DNL2 | deoxyribonuclease II, lysosomal (EC:3.1.22.1) |
C00002514
|
| 10059 | DNM1L, DLP1, DRP1, DVLP, DYMPLE, EMPF, HDYNIV | dynamin 1-like (EC:3.6.5.5) |
C00002514
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00002514
|
| 81704 | DOCK8, MRD2, ZIR8 | dedicator of cytokinesis 8 |
C00002514
|
| 29980 | DONSON, B17, C21orf60 | downstream neighbor of SON |
C00002514
|
| 9980 | DOPEY2, 21orf5, C21orf5 | dopey family member 2 |
C00002514
|
| 1798 | DPAGT1, ALG7, CDG-Ij, CDG1J, CMSTA2, D11S366, DGPT, DPAGT, DPAGT2, G1PT, GPT, UAGT, UGAT | dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) (EC:2.7.8.15) |
C00002514
|
| 8818 | DPM2, CDG1U | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (EC:2.4.1.83) |
C00002514
|
| 23333 | DPY19L1 | dpy-19-like 1 (C. elegans) |
C00002514
|
| 1824 | DSC2, ARVD11, CDHF2, DG2, DGII/III, DSC3 | desmocollin 2 |
C00002514
|
| 380353 |
C00002514
|
||
| 79980 | DSN1, C20orf172, KNL3, MIS13, dJ469A13.2, hKNL-3 | DSN1, MIS12 kinetochore complex component |
C00002514
|
| 1837 | DTNA, D18S892E, DRP3, DTN, DTN-A, LVNC1 | dystrobrevin, alpha |
C00002514
|
| 1843 | DUSP1, CL100, HVH1, MKP-1, MKP1, PTPN10 | dual specificity phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00002514
|
| 11221 | DUSP10, MKP-5, MKP5 | dual specificity phosphatase 10 (EC:3.1.3.16 3.1.3.48) |
C00002514
|
| 56940 | DUSP22, JKAP, JSP-1, JSP1, LMW-DSP2, LMWDSP2, MKP-x, MKPX, VHX | dual specificity phosphatase 22 (EC:3.1.3.16 3.1.3.48) |
C00002514
|
| 1854 | DUT, dUTPase | deoxyuridine triphosphatase (EC:3.6.1.23) |
C00002514
|
| 51143 | DYNC1LI1, DNCLI1, LIC1 | dynein, cytoplasmic 1, light intermediate chain 1 |
C00002514
|
| 6990 | DYNLT3, RP3, TCTE1L, TCTEX1L | dynein, light chain, Tctex-type 3 |
C00002514
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002514
|
| 1870 | E2F2, E2F-2 | E2F transcription factor 2 |
C00002514
|
| 1875 | E2F5, E2F-5 | E2F transcription factor 5, p130-binding |
C00002514
|
| 144455 | E2F7 | E2F transcription factor 7 |
C00002514
|
| 79733 | E2F8, E2F-8 | E2F transcription factor 8 |
C00002514
|
| 10682 | EBP, CDPX2, CHO2, CPX, CPXD | emopamil binding protein (sterol isomerase) (EC:5.3.3.5) |
C00002514
|
| 1894 | ECT2, ARHGEF31 | epithelial cell transforming sequence 2 oncogene |
C00002514
|
| 80267 | EDEM3, C1orf22 | ER degradation enhancer, mannosidase alpha-like 3 (EC:3.2.1.113) |
C00002514
|
| 10085 | EDIL3, DEL1 | EGF-like repeats and discoidin I-like domains 3 |
C00002514
|
| 1906 | EDN1, ET1, HDLCQ7, PPET1 | endothelin 1 |
C00002514
|
| 8726 | EED, HEED, WAIT1 | embryonic ectoderm development |
C00002514
|
| 1915 | EEF1A1, CCS-3, CCS3, EE1A1, EEF-1, EEF1A, EF-Tu, EF1A, GRAF-1EF, HNGC:16303, LENG7, PTI1, eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 |
C00002514
|
| 2202 | EFEMP1, DHRD, DRAD, FBLN3, FBNL, FIBL-3, MLVT, MTLV, S1-5 | EGF containing fibulin-like extracellular matrix protein 1 |
C00002514
|
| 80303 | EFHD1, MST133, MSTP133, SWS2 | EF-hand domain family, member D1 |
C00002514
|
| 1942 | EFNA1, B61, ECKLG, EFL1, EPLG1, LERK-1, LERK1, TNFAIP4 | ephrin-A1 |
C00002514
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00002514
|
| 1949 | EFNB3, EFL6, EPLG8, LERK8 | ephrin-B3 |
C00002514
|
| 9343 | EFTUD2, MFDGA, MFDM, SNRNP116, Snrp116, Snu114, U5-116KD | elongation factor Tu GTP binding domain containing 2 |
C00002514
|
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00002514
|
| 1960 | EGR3, EGR-3, PILOT | early growth response 3 |
C00002514
|
| 79813 | EHMT1, EUHMTASE1, Eu-HMTase1, FP13812, GLP, GLP1, KMT1D, bA188C12.1 | euchromatic histone-lysine N-methyltransferase 1 (EC:2.1.1.43) |
C00002514
|
| 9451 | EIF2AK3, PEK, PERK, WRS | eukaryotic translation initiation factor 2-alpha kinase 3 (EC:2.7.11.1) |
C00002514
|
| 1965 | EIF2S1, EIF-2, EIF-2A, EIF-2alpha, EIF2, EIF2A | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
C00002514
|
| 1979 | EIF4EBP2, 4EBP2, PHASII | eukaryotic translation initiation factor 4E binding protein 2 |
C00002514
|
| 8672 | EIF4G3, eIF-4G_3, eIF4G_3, eIF4GII | eukaryotic translation initiation factor 4 gamma, 3 |
C00002514
|
| 2001 | ELF5, ESE2 | E74-like factor 5 (ets domain transcription factor) |
C00002514
|
| 2002 | ELK1 | ELK1, member of ETS oncogene family |
C00002514
|
| 2004 | ELK3, ERP, NET, SAP2 | ELK3, ETS-domain protein (SRF accessory protein 2) |
C00002514
|
| 63916 | ELMO2, CED-12, CED12, ELMO-2 | engulfment and cell motility 2 |
C00002514
|
| 54898 | ELOVL2, SSC2 | ELOVL fatty acid elongase 2 (EC:2.3.1.199) |
C00002514
|
| 60481 | ELOVL5, HELO1, dJ483K16.1 | ELOVL fatty acid elongase 5 (EC:2.3.1.199) |
C00002514
|
| 79071 | ELOVL6, FACE, FAE, LCE | ELOVL fatty acid elongase 6 (EC:2.3.1.199) |
C00002514
|
| 79993 | ELOVL7 | ELOVL fatty acid elongase 7 (EC:2.3.1.199) |
C00002514
|
| 23587 | ELP5, C17orf81, DERP6, MST071, MSTP071 | elongator acetyltransferase complex subunit 5 |
C00002514
|
| 54859 | ELP6, C3orf75, TMEM103 | elongator acetyltransferase complex subunit 6 |
C00002514
|
| 51016 | EMC9, C14orf122, FAM158A | ER membrane protein complex subunit 9 |
C00002514
|
| 2010 | EMD, EDMD, LEMD5, STA | emerin |
C00002514
|
| 146956 | EME1, MMS4L, SLX2A | essential meiotic structure-specific endonuclease 1 |
C00002514
|
| 8507 | ENC1, CCL28, ENC-1, KLHL35, KLHL37, NRPB, PIG10, TP53I10 | ectodermal-neural cortex 1 (with BTB domain) |
C00002514
|
| 23052 | ENDOD1 | endonuclease domain containing 1 |
C00002514
|
| 55556 | ENOSF1, HSRTSBETA, RTS, TYMSAS | enolase superfamily member 1 |
C00002514
|
| 954 | ENTPD2, CD39L1, NTPDase-2 | ectonucleoside triphosphate diphosphohydrolase 2 (EC:3.6.1.-) |
C00002514
|
| 9583 | ENTPD4, LALP70, LAP70, LYSAL1, NTPDase-4, UDPase | ectonucleoside triphosphate diphosphohydrolase 4 (EC:3.6.1.6) |
C00002514
|
| 2037 | EPB41L2, 4.1-G, 4.1G | erythrocyte membrane protein band 4.1-like 2 |
C00002514
|
| 57669 | EPB41L5, BE37, YMO1 | erythrocyte membrane protein band 4.1 like 5 |
C00002514
|
| 2043 | EPHA4, HEK8, SEK, TYRO1 | EPH receptor A4 (EC:2.7.10.1) |
C00002514
|
| 58513 | EPS15L1, EPS15R | epidermal growth factor receptor pathway substrate 15-like 1 |
C00002514
|
| 2064 | ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU, NGL, TKR1 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (EC:2.7.10.1) |
C00002514
|
| 2065 | ERBB3, ErbB-3, HER3, LCCS2, MDA-BF-1, c-erbB-3, c-erbB3, erbB3-S, p180-ErbB3, p45-sErbB3, p85-sErbB3 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 (EC:2.7.10.1) |
C00002514
|
| 2066 | ERBB4, ALS19, HER4, p180erbB4 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 (EC:2.7.10.1) |
C00002514
|
| 54821 | ERCC6L, PICH, RAD26L | excision repair cross-complementing rodent repair deficiency, complementation group 6-like (EC:3.6.4.12) |
C00002514
|
| 112479 | ERI2, EXOD1 | ERI1 exoribonuclease family member 2 (EC:3.1.-.-) |
C00002514
|
| 54206 | ERRFI1, GENE-33, MIG-6, MIG6, RALT | ERBB receptor feedback inhibitor 1 |
C00002514
|
| 157570 | ESCO2, 2410004I17Rik, EFO2, RBS | establishment of sister chromatid cohesion N-acetyltransferase 2 (EC:2.3.1.-) |
C00002514
|
| 9700 | ESPL1, ESP1, SEPA | extra spindle pole bodies homolog 1 (S. cerevisiae) (EC:3.4.22.49) |
C00002514
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00002514
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00002514
|
| 57488 | ESYT2, CHR2SYT, E-Syt2, FAM62B | extended synaptotagmin-like protein 2 |
C00002514
|
| 55500 | ETNK1, EKI, EKI_1, EKI1, Nbla10396 | ethanolamine kinase 1 (EC:2.7.1.82) |
C00002514
|
| 7813 | EVI5, NB4S | ecotropic viral integration site 5 |
C00002514
|
| 9156 | EXO1, HEX1, hExoI | exonuclease 1 |
C00002514
|
| 55770 | EXOC2, SEC5, SEC5L1, Sec5p | exocyst complex component 2 |
C00002514
|
| 23404 | EXOSC2, RRP4, Rrp4p, hRrp4p, p7 | exosome component 2 |
C00002514
|
| 51010 | EXOSC3, PCH1B, RP11-3J10.8, RRP40, Rrp40p, bA3J10.7, hRrp-40, p10 | exosome component 3 |
C00002514
|
| 11340 | EXOSC8, CIP3, EAP2, OIP2, RP11-421P11.3, RRP43, Rrp43p, bA421P11.3, p9 | exosome component 8 (EC:3.1.13.-) |
C00002514
|
| 5393 | EXOSC9, PM/Scl-75, PMSCL1, RRP45, Rrp45p, p5, p6 | exosome component 9 (EC:3.1.13.-) |
C00002514
|
| 2131 | EXT1, EXT, LGCR, LGS, TRPS2, TTV | exostosin glycosyltransferase 1 (EC:2.4.1.225 2.4.1.224) |
C00002514
|
| 2146 | EZH2, ENX-1, ENX1, EZH1, EZH2b, KMT6, KMT6A, WVS, WVS2 | enhancer of zeste homolog 2 (Drosophila) (EC:2.1.1.43) |
C00002514
|
| 7430 | EZR, CVIL, CVL, VIL2 | ezrin |
C00002514
|
| 2149 | F2R, CF2R, HTR, PAR-1, PAR1, TR | coagulation factor II (thrombin) receptor |
C00002514
|
| 2150 | F2RL1, GPR11, PAR2 | coagulation factor II (thrombin) receptor-like 1 |
C00002514
|
| 2171 | FABP5, E-FABP, EFABP, KFABP, PA-FABP, PAFABP | fatty acid binding protein 5 (psoriasis-associated) |
C00002514
|
| 3992 | FADS1, D5D, FADS6, FADSD5, LLCDL1, TU12 | fatty acid desaturase 1 (EC:1.14.19.-) |
C00002514
|
| 2184 | FAH | fumarylacetoacetate hydrolase (fumarylacetoacetase) (EC:3.7.1.2) |
C00002514
|
| 9214 | FAIM3, FCMR, TOSO | Fas apoptotic inhibitory molecule 3 |
C00002514
|
| 83641 | FAM107B, C10orf45 | family with sequence similarity 107, member B |
C00002514
|
| 90362 | FAM110B, C8orf72 | family with sequence similarity 110, member B |
C00002514
|
| 642273 | FAM110C | family with sequence similarity 110, member C |
C00002514
|
| 63901 | FAM111A, GCLEB, KCS2 | family with sequence similarity 111, member A |
C00002514
|
| 374393 | FAM111B, CANP | family with sequence similarity 111, member B |
C00002514
|
| 159090 | FAM122B, SPACIA2 | family with sequence similarity 122B |
C00002514
|
| 285172 | FAM126B, HYCC2 | family with sequence similarity 126, member B |
C00002514
|
| 317662 | FAM149B1, KIAA0974 | family with sequence similarity 149, member B1 |
C00002514
|
| 145483 | FAM161B, C14orf44, c14_5547 | family with sequence similarity 161, member B |
C00002514
|
| 26049 | FAM169A, SLAP75 | family with sequence similarity 169, member A |
C00002514
|
| 165215 | FAM171B, KIAA1946 | family with sequence similarity 171, member B |
C00002514
|
| 400451 | FAM174B | family with sequence similarity 174, member B |
C00002514
|
| 55719 | FAM178A, C10orf6 | family with sequence similarity 178, member A |
C00002514
|
| 9413 | FAM189A2, C9orf61, RP11-548B3.1, X123 | family with sequence similarity 189, member A2 |
C00002514
|
| 116151 | FAM210B, 5A3, C20orf108, dJ1167H4.1 | family with sequence similarity 210, member B |
C00002514
|
| 56204 | FAM214A, KIAA1370 | family with sequence similarity 214, member A |
C00002514
|
| 29902 | FAM216A, C12orf24, HSU79274 | family with sequence similarity 216, member A |
C00002514
|
| 55731 | FAM222B, C17orf63 | family with sequence similarity 222, member B |
C00002514
|
| 131583 | FAM43A | family with sequence similarity 43, member A |
C00002514
|
| 54855 | FAM46C | family with sequence similarity 46, member C |
C00002514
|
| 51571 | FAM49B, L1 | family with sequence similarity 49, member B |
C00002514
|
| 79850 | FAM57A, CT120 | family with sequence similarity 57, member A |
C00002514
|
| 54478 | FAM64A, CATS, RCS1 | family with sequence similarity 64, member A |
C00002514
|
| 2175 | FANCA, FA, FA-H, FA1, FAA, FACA, FAH, FANCH | Fanconi anemia, complementation group A |
C00002514
|
| 2177 | FANCD2, FA-D2, FA4, FACD, FAD, FAD2, FANCD | Fanconi anemia, complementation group D2 |
C00002514
|
| 2189 | FANCG, FAG, XRCC9 | Fanconi anemia, complementation group G |
C00002514
|
| 55215 | FANCI, KIAA1794 | Fanconi anemia, complementation group I |
C00002514
|
| 55120 | FANCL, FAAP43, PHF9, POG | Fanconi anemia, complementation group L |
C00002514
|
| 10056 | FARSB, FARSLB, FRSB, PheHB, PheRS | phenylalanyl-tRNA synthetase, beta subunit (EC:6.1.1.20) |
C00002514
|
| 54751 | FBLIM1, CAL, FBLP-1, FBLP1 | filamin binding LIM protein 1 |
C00002514
|
| 26234 | FBXL5, FBL4, FBL5, FLR1 | F-box and leucine-rich repeat protein 5 |
C00002514
|
| 81545 | FBXO38, Fbx38, MOKA | F-box protein 38 |
C00002514
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00002514
|
| 51077 | FCF1, Bka, C14orf111, UTP24 | FCF1 rRNA-processing protein |
C00002514
|
| 2222 | FDFT1, DGPT, ERG9, SQS, SS | farnesyl-diphosphate farnesyltransferase 1 (EC:2.5.1.21) |
C00002514
|
| 2224 | FDPS, FPPS, FPS | farnesyl diphosphate synthase (EC:2.5.1.1 2.5.1.10) |
C00002514
|
| 2237 | FEN1, FEN-1, MF1, RAD2 | flap structure-specific endonuclease 1 |
C00002514
|
| 2260 | FGFR1, BFGFR, CD331, CEK, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 | fibroblast growth factor receptor 1 (EC:2.7.10.1) |
C00002514
|
| 2271 | FH, HLRCC, LRCC, MCL, MCUL1 | fumarate hydratase (EC:4.2.1.2) |
C00002514
|
| 2273 | FHL1, FHL-1, FHL1A, FHL1B, FLH1A, KYOT, SLIM, SLIM-1, SLIM1, SLIMMER, XMPMA | four and a half LIM domains 1 |
C00002514
|
| 2274 | FHL2, AAG11, DRAL, FHL-2, SLIM-3, SLIM3 | four and a half LIM domains 2 |
C00002514
|
| 9158 | FIBP, FGFIBP, FIBP-1 | fibroblast growth factor (acidic) intracellular binding protein |
C00002514
|
| 63979 | FIGNL1 | fidgetin-like 1 |
C00002514
|
| 11259 | FILIP1L, DOC-1, DOC1, GIP130, GIP90 | filamin A interacting protein 1-like |
C00002514
|
| 51303 | FKBP11, FKBP19 | FK506 binding protein 11, 19 kDa (EC:5.2.1.8) |
C00002514
|
| 2288 | FKBP4, FKBP51, FKBP52, FKBP59, HBI, Hsp56, PPIase, p52 | FK506 binding protein 4, 59kDa (EC:5.2.1.8) |
C00002514
|
| 2289 | FKBP5, AIG6, FKBP51, FKBP54, P54, PPIase, Ptg-10 | FK506 binding protein 5 (EC:5.2.1.8) |
C00002514
|
| 63943 | FKBPL, DIR1, NG7, WISP39 | FK506 binding protein like |
C00002514
|
| 201163 | FLCN, BHD, FLCL | folliculin |
C00002514
|
| 114793 | FMNL2, FHOD2 | formin-like 2 |
C00002514
|
| 23048 | FNBP1, FBP17 | formin binding protein 1 |
C00002514
|
| 54874 | FNBP1L, C1orf39, TOCA1 | formin binding protein 1-like |
C00002514
|
| 64778 | FNDC3B, FAD104, PRO4979, YVTM2421 | fibronectin type III domain containing 3B |
C00002514
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00002514
|
| 2297 | FOXD1, FKHL8, FREAC4 | forkhead box D1 |
C00002514
|
| 2305 | FOXM1, FKHL16, FOXM1B, HFH-11, HFH11, HNF-3, INS-1, MPHOSPH2, MPP-2, MPP2, PIG29, TGT3, TRIDENT | forkhead box M1 |
C00002514
|
| 494532 |
C00002514
|
||
| 93986 | FOXP2, CAGH44, SPCH1, TNRC10 | forkhead box P2 |
C00002514
|
| 80020 | FOXRED2, ERFAD | FAD-dependent oxidoreductase domain containing 2 |
C00002514
|
| 257019 | FRMD3, 4.1O, EPB41L4O, EPB41LO, P410 | FERM domain containing 3 |
C00002514
|
| 122786 | FRMD6, C14orf31, EX1, Willin, c14_5320 | FERM domain containing 6 |
C00002514
|
| 6624 | FSCN1, FAN1, HSN, SNL, p55 | fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
C00002514
|
| 24140 | FTSJ1, CDLIV, MRX44, MRX9, SPB1, TRMT7 | FtsJ RNA methyltransferase homolog 1 (E. coli) (EC:2.1.1.205) |
C00002514
|
| 29960 | FTSJ2, FJH1, MRM2 | FtsJ RNA methyltransferase homolog 2 (E. coli) (EC:2.1.1.-) |
C00002514
|
| 2517 | FUCA1, FUCA | fucosidase, alpha-L- 1, tissue (EC:3.2.1.51) |
C00002514
|
| 79443 | FYCO1, CATC2, CTRCT18, RUFY3, ZFYVE7 | FYVE and coiled-coil domain containing 1 |
C00002514
|
| 2535 | FZD2, Fz2, fz-2, fzE2, hFz2 | frizzled family receptor 2 |
C00002514
|
| 55632 | G2E3, KIAA1333, PHF7B | G2/M-phase specific E3 ubiquitin protein ligase |
C00002514
|
| 9908 | G3BP2 | GTPase activating protein (SH3 domain) binding protein 2 |
C00002514
|
| 126626 | GABPB2 | GA binding protein transcription factor, beta subunit 2 |
C00002514
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00002514
|
| 90480 | GADD45GIP1, CKBBP2, CRIF1, MRP-L59, PLINP, PLINP-1, PRG6, Plinp1 | growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
C00002514
|
| 51083 | GAL, GAL-GMAP, GALN, GLNN, GMAP | galanin/GMAP prepropeptide |
C00002514
|
| 2588 | GALNS, GALNAC6S, GAS, GalN6S, MPS4A | galactosamine (N-acetyl)-6-sulfate sulfatase (EC:3.1.6.4) |
C00002514
|
| 2589 | GALNT1, GALNAC-T1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (EC:2.4.1.41) |
C00002514
|
| 55568 | GALNT10, GALNACT10, PPGALNACT10, PPGANTASE10 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) (EC:2.4.1.41) |
C00002514
|
| 51809 | GALNT7, GALNAC-T7, GalNAcT7 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (EC:2.4.1.41) |
C00002514
|
| 23193 | GANAB, G2AN, GLUII | glucosidase, alpha; neutral AB (EC:3.2.1.84) |
C00002514
|
| 2618 | GART, AIRS, GARS, GARTF, PAIS, PGFT, PRGS | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase (EC:2.1.2.2 6.3.3.1 6.3.4.13) |
C00002514
|
| 283431 | GAS2L3, G2L3 | growth arrest-specific 2 like 3 |
C00002514
|
| 2624 | GATA2, DCML, MONOMAC, NFE1B | GATA binding protein 2 |
C00002514
|
| 2625 | GATA3, HDR, HDRS | GATA binding protein 3 |
C00002514
|
| 57798 | GATAD1, CMD2B, ODAG, RG083M05.2 | GATA zinc finger domain containing 1 |
C00002514
|
| 2634 | GBP2 | guanylate binding protein 2, interferon-inducible |
C00002514
|
| 2644 | GCHFR, GFRP, HsT16933, P35 | GTP cyclohydrolase I feedback regulator |
C00002514
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00002514
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00002514
|
| 79153 | GDPD3 | glycerophosphodiester phosphodiesterase domain containing 3 |
C00002514
|
| 2669 | GEM, KIR | GTP binding protein overexpressed in skeletal muscle |
C00002514
|
| 8487 | GEMIN2, SIP1, SIP1-delta | gem (nuclear organelle) associated protein 2 |
C00002514
|
| 50628 | GEMIN4, HC56, HCAP1, HHRF-1, p97 | gem (nuclear organelle) associated protein 4 |
C00002514
|
| 79833 | GEMIN6 | gem (nuclear organelle) associated protein 6 |
C00002514
|
| 2674 | GFRA1, GDNFR, GDNFRA, GFR-ALPHA-1, RET1L, RETL1, TRNR1 | GDNF family receptor alpha 1 |
C00002514
|
| 23062 | GGA2, VEAR | golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
C00002514
|
| 79017 | GGCT, C7orf24, CRF21, GCTG, GGC | gamma-glutamylcyclotransferase (EC:2.3.2.4) |
C00002514
|
| 8836 | GGH, GH | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (EC:3.4.19.9) |
C00002514
|
| 9837 | GINS1, PSF1 | GINS complex subunit 1 (Psf1 homolog) |
C00002514
|
| 51659 | GINS2, HSPC037, PSF2, Pfs2 | GINS complex subunit 2 (Psf2 homolog) |
C00002514
|
| 64785 | GINS3, PSF3 | GINS complex subunit 3 (Psf3 homolog) |
C00002514
|
| 84296 | GINS4, SLD5 | GINS complex subunit 4 (Sld5 homolog) |
C00002514
|
| 2717 | GLA, GALA | galactosidase, alpha (EC:3.2.1.22) |
C00002514
|
| 2733 | GLE1, GLE1L, LCCS, LCCS1, hGLE1 | GLE1 RNA export mediator |
C00002514
|
| 11146 | GLMN, FAP, FAP48, FAP68, FKBPAP, GLML, GVM, VMGLOM | glomulin, FKBP associated protein |
C00002514
|
| 2744 | GLS, AAD20, GAC, GAM, GLS1, KGA | glutaminase (EC:3.5.1.2) |
C00002514
|
| 51053 | GMNN, Gem | geminin, DNA replication inhibitor |
C00002514
|
| 8833 | GMPS | guanine monphosphate synthase (EC:6.3.5.2) |
C00002514
|
| 2770 | GNAI1, Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
C00002514
|
| 2776 | GNAQ, CMC1, G-ALPHA-q, GAQ, SWS | guanine nucleotide binding protein (G protein), q polypeptide |
C00002514
|
| 54552 | GNL3L | guanine nucleotide binding protein-like 3 (nucleolar)-like |
C00002514
|
| 2799 | GNS, G6S | glucosamine (N-acetyl)-6-sulfatase (EC:3.1.6.14) |
C00002514
|
| 2801 | GOLGA2, GM130 | golgin A2 |
C00002514
|
| 2805 | GOT1, ASTQTL1, GIG18, cAspAT, cCAT | glutamic-oxaloacetic transaminase 1, soluble (EC:2.6.1.1 2.6.1.3) |
C00002514
|
| 8733 | GPAA1, GAA1, hGAA1 | glycosylphosphatidylinositol anchor attachment 1 |
C00002514
|
| 253635 | GPATCH11, CCDC75, CENP-Y, CENPY | G patch domain containing 11 |
C00002514
|
| 55668 | GPATCH2L, C14orf118 | G patch domain containing 2-like |
C00002514
|
| 54865 | GPATCH4, GPATC4 | G patch domain containing 4 |
C00002514
|
| 2820 | GPD2, GDH2, GPDM, mGPDH | glycerol-3-phosphate dehydrogenase 2 (mitochondrial) (EC:1.1.5.3) |
C00002514
|
| 2852 | GPER1, CEPR, CMKRL2, DRY12, FEG-1, GPCR-Br, GPER, GPR30, LERGU, LERGU2, LyGPR | G protein-coupled estrogen receptor 1 |
C00002514
|
| 7107 | GPR137B, TM7SF1 | G protein-coupled receptor 137B |
C00002514
|
| 26996 | GPR160, GPCR1, GPCR150 | G protein-coupled receptor 160 |
C00002514
|
| 9052 | GPRC5A, GPCR5A, RAI3, RAIG1 | G protein-coupled receptor, family C, group 5, member A |
C00002514
|
| 55890 | GPRC5C, RAIG-3, RAIG3 | G protein-coupled receptor, family C, group 5, member C |
C00002514
|
| 29899 | GPSM2, CMCS, DFNB82, LGN, PINS | G-protein signaling modulator 2 |
C00002514
|
| 380153 |
C00002514
|
||
| 493869 | GPX8, EPLA847, GPx-8, GSHPx-8, UNQ847 | glutathione peroxidase 8 (putative) (EC:1.11.1.9) |
C00002514
|
| 57655 | GRAMD1A, KIAA1533 | GRAM domain containing 1A |
C00002514
|
| 2887 | GRB10, GRB-IR, Grb-10, IRBP, MEG1, RSS | growth factor receptor-bound protein 10 |
C00002514
|
| 9687 | GREB1 | growth regulation by estrogen in breast cancer 1 |
C00002514
|
| 57822 | GRHL3, SOM, TFCP2L4 | grainyhead-like 3 (Drosophila) |
C00002514
|
| 2870 | GRK6, GPRK6 | G protein-coupled receptor kinase 6 (EC:2.7.11.16) |
C00002514
|
| 2896 | GRN, CLN11, GEP, GP88, PCDGF, PEPI, PGRN | granulin |
C00002514
|
| 80273 | GRPEL1, HMGE | GrpE-like 1, mitochondrial (E. coli) |
C00002514
|
| 83903 | GSG2, HASPIN | germ cell associated 2 (haspin) (EC:2.7.11.1) |
C00002514
|
| 2968 | GTF2H4, P52, TFB2, TFIIH | general transcription factor IIH, polypeptide 4, 52kDa |
C00002514
|
| 9569 | GTF2IRD1, BEN, CREAM1, GTF3, MUSTRD1, RBAP2, WBS, WBSCR11, WBSCR12, hMusTRD1alpha1 | GTF2I repeat domain containing 1 |
C00002514
|
| 51512 | GTSE1, B99 | G-2 and S-phase expressed 1 |
C00002514
|
| 51454 | GULP1, CED-6, CED6, GULP | GULP, engulfment adaptor PTB domain containing 1 |
C00002514
|
| 94239 | H2AFV, H2A.Z-2, H2AV | H2A histone family, member V |
C00002514
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00002514
|
| 3015 | H2AFZ, H2A.Z-1, H2A.z, H2A/z, H2AZ | H2A histone family, member Z |
C00002514
|
| 3021 | H3F3B, H3.3B | H3 histone, family 3B (H3.3B) |
C00002514
|
| 3033 | HADH, HAD, HADH1, HADHSC, HCDH, HHF4, MSCHAD, SCHAD | hydroxyacyl-CoA dehydrogenase (EC:1.1.1.35) |
C00002514
|
| 8520 | HAT1, KAT1 | histone acetyltransferase 1 (EC:2.3.1.48) |
C00002514
|
| 54930 | HAUS4, C14orf94 | HAUS augmin-like complex, subunit 4 |
C00002514
|
| 55559 | HAUS7, UCHL5IP, UIP1 | HAUS augmin-like complex, subunit 7 |
C00002514
|
| 93323 | HAUS8, DGT4, HICE1, NY-SAR-48 | HAUS augmin-like complex, subunit 8 |
C00002514
|
| 26959 | HBP1 | HMG-box transcription factor 1 |
C00002514
|
| 50865 | HEBP1, HBP, HEBP | heme binding protein 1 |
C00002514
|
| 79654 | HECTD3, RP11-69J16.1 | HECT domain containing E3 ubiquitin protein ligase 3 |
C00002514
|
| 3070 | HELLS, LSH, PASG, SMARCA6 | helicase, lymphoid-specific |
C00002514
|
| 85441 | HELZ2, PDIP-1, PRIC285 | helicase with zinc finger 2, transcriptional coactivator |
C00002514
|
| 8925 | HERC1, p532, p619 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
C00002514
|
| 64224 | HERPUD2 | HERPUD family member 2 |
C00002514
|
| 3280 | HES1, HES-1, HHL, HRY, bHLHb39 | hes family bHLH transcription factor 1 |
C00002514
|
| 204851 | HIPK1, Myak, Nbak2 | homeodomain interacting protein kinase 1 (EC:2.7.11.1) |
C00002514
|
| 28996 | HIPK2, PRO0593 | homeodomain interacting protein kinase 2 (EC:2.7.11.1) |
C00002514
|
| 8479 | HIRIP3 | HIRA interacting protein 3 |
C00002514
|
| 3006 | HIST1H1C, H1.2, H1C, H1F2, H1s-1 | histone cluster 1, H1c |
C00002514
|
| 8334 | HIST1H2AC, H2A/l, H2AFL, dJ221C16.4 | histone cluster 1, H2ac |
C00002514
|
| 3012 | HIST1H2AE, H2A.1, H2A.2, H2A/a, H2AFA | histone cluster 1, H2ae |
C00002514
|
| 8969 | HIST1H2AG, H2A.1b, H2A/p, H2AFP, H2AG, pH2A/f | histone cluster 1, H2ag |
C00002514
|
| 8329 | HIST1H2AI, H2A/c, H2AFC | histone cluster 1, H2ai |
C00002514
|
| 8330 | HIST1H2AK, H2A/d, H2AFD | histone cluster 1, H2ak |
C00002514
|
| 8347 | HIST1H2BC, H2B.1, H2B/l, H2BFL, dJ221C16.3 | histone cluster 1, H2bc |
C00002514
|
| 3017 | HIST1H2BD, H2B.1B, H2B/b, H2BFB, HIRIP2, dJ221C16.6 | histone cluster 1, H2bd |
C00002514
|
| 8344 | HIST1H2BE, H2B.h, H2B/h, H2BFH, dJ221C16.8 | histone cluster 1, H2be |
C00002514
|
| 8343 | HIST1H2BF, H2B/g, H2BFG | histone cluster 1, H2bf |
C00002514
|
| 8339 | HIST1H2BG, H2B.1A, H2B/a, H2BFA, dJ221C16.8 | histone cluster 1, H2bg |
C00002514
|
| 8345 | HIST1H2BH, H2B/j, H2BFJ | histone cluster 1, H2bh |
C00002514
|
| 8346 | HIST1H2BI, H2B/k, H2BFK | histone cluster 1, H2bi |
C00002514
|
| 85236 | HIST1H2BK, H2B/S, H2BFAiii, H2BFT, H2BK | histone cluster 1, H2bk |
C00002514
|
| 8351 | HIST1H3D, H3/b, H3FB | histone cluster 1, H3d |
C00002514
|
| 8357 | HIST1H3H, H3/k, H3F1K, H3FK | histone cluster 1, H3h |
C00002514
|
| 8365 | HIST1H4H, H4/h, H4FH | histone cluster 1, H4h |
C00002514
|
| 8349 | HIST2H2BE, GL105, H2B, H2B.1, H2BFQ, H2BGL105, H2BQ | histone cluster 2, H2be |
C00002514
|
| 92815 | HIST3H2A | histone cluster 3, H2a |
C00002514
|
| 3096 | HIVEP1, CIRIP, CRYBP1, MBP-1, PRDII-BF1, Schnurri-1, ZAS1, ZNF40, ZNF40A | human immunodeficiency virus type I enhancer binding protein 1 |
C00002514
|
| 55355 | HJURP, FAKTS, URLC9, hFLEG1 | Holliday junction recognition protein |
C00002514
|
| 3159 | HMGA1, HMG-R, HMGA1A, HMGIY | high mobility group AT-hook 1 |
C00002514
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00002514
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00002514
|
| 9324 | HMGN3, PNAS-25, TRIP7 | high mobility group nucleosomal binding domain 3 |
C00002514
|
| 3161 | HMMR, CD168, IHABP, RHAMM | hyaluronan-mediated motility receptor (RHAMM) |
C00002514
|
| 3187 | HNRNPH1, HNRPH, HNRPH1, hnRNPH | heterogeneous nuclear ribonucleoprotein H1 (H) |
C00002514
|
| 3189 | HNRNPH3, 2H9, HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) |
C00002514
|
| 3192 | HNRNPU, HNRPU, SAF-A, U21.1, hnRNP_U | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
C00002514
|
| 100195139 |
C00002514
|
||
| 3229 | HOXC13, ECTD9, HOX3, HOX3G | homeobox C13 |
C00002514
|
| 3223 | HOXC6, CP25, HHO.C8, HOX3, HOX3C | homeobox C6 |
C00002514
|
| 3251 | HPRT1, HGPRT, HPRT | hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8) |
C00002514
|
| 90161 | HS6ST2 | heparan sulfate 6-O-sulfotransferase 2 (EC:2.8.2.-) |
C00002514
|
| 8630 | HSD17B6, HSE, RODH, SDR9C6 | hydroxysteroid (17-beta) dehydrogenase 6 (EC:1.1.1.62 1.1.1.239 1.1.1.105) |
C00002514
|
| 7923 | HSD17B8, D6S2245E, FABG, FABGL, H2-KE6, HKE6, KE6, RING2, SDR30C1, dJ1033B10.9 | hydroxysteroid (17-beta) dehydrogenase 8 (EC:1.1.1.62 1.1.1.239) |
C00002514
|
| 7184 | HSP90B1, ECGP, GP96, GRP94, TRA1 | heat shock protein 90kDa beta (Grp94), member 1 |
C00002514
|
| 22824 | HSPA4L, APG-1, HSPH3, Osp94 | heat shock 70kDa protein 4-like |
C00002514
|
| 3313 | HSPA9, CSA, GRP-75, GRP75, HSPA9B, MOT, MOT2, MTHSP75, PBP74 | heat shock 70kDa protein 9 (mortalin) |
C00002514
|
| 219844 | HYLS1, HLS | hydrolethalus syndrome 1 |
C00002514
|
| 25764 | HYPK, C15orf63 | huntingtin interacting protein K |
C00002514
|
| 3382 | ICA1, ICA69, ICAp69 | islet cell autoantigen 1, 69kDa |
C00002514
|
| 23463 | ICMT, HSTE14, MST098, MSTP098, PCCMT, PCMT, PPMT | isoprenylcysteine carboxyl methyltransferase (EC:2.1.1.100) |
C00002514
|
| 3399 | ID3, HEIR-1, bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
C00002514
|
| 3422 | IDI1, IPP1, IPPI1 | isopentenyl-diphosphate delta isomerase 1 (EC:5.3.3.2) |
C00002514
|
| 3423 | IDS, MPS2, SIDS | iduronate 2-sulfatase (EC:3.1.6.13) |
C00002514
|
| 9592 | IER2, ETR101 | immediate early response 2 |
C00002514
|
| 8870 | IER3, DIF-2, DIF2, GLY96, IEX-1, IEX-1L, IEX1, PRG1 | immediate early response 3 |
C00002514
|
| 51278 | IER5, SBBI48 | immediate early response 5 |
C00002514
|
| 389792 | IER5L, bA247A12.2 | immediate early response 5-like |
C00002514
|
| 10964 | IFI44L, C1orf29 | interferon-induced protein 44-like |
C00002514
|
| 3433 | IFIT2, G10P2, GARG-39, IFI-54, IFI-54K, IFI54, IFIT-2, ISG-54_K, ISG-54K, ISG54, P54, cig42 | interferon-induced protein with tetratricopeptide repeats 2 |
C00002514
|
| 7866 | IFRD2, IFNRP, SKMc15, SM15 | interferon-related developmental regulator 2 |
C00002514
|
| 55764 | IFT122, CED, CED1, SPG, WDR10, WDR10p, WDR140 | intraflagellar transport 122 homolog (Chlamydomonas) |
C00002514
|
| 90410 | IFT20 | intraflagellar transport 20 homolog (Chlamydomonas) |
C00002514
|
| 112752 | IFT43, C14orf179, CED3 | intraflagellar transport 43 homolog (Chlamydomonas) |
C00002514
|
| 3485 | IGFBP2, IBP2, IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa |
C00002514
|
| 3487 | IGFBP4, BP-4, HT29-IGFBP, IBP4, IGFBP-4 | insulin-like growth factor binding protein 4 |
C00002514
|
| 3488 | IGFBP5, IBP5 | insulin-like growth factor binding protein 5 |
C00002514
|
| 79713 | IGFLR1, TMEM149, U2AF1L4 | IGF-like family receptor 1 |
C00002514
|
| 445438 |
C00002514
|
||
| 55540 | IL17RB, CRL4, EVI27, IL17BR, IL17RH1 | interleukin 17 receptor B |
C00002514
|
| 3554 | IL1R1, CD121A, D2S1473, IL-1R-alpha, IL1R, IL1RA, P80 | interleukin 1 receptor, type I |
C00002514
|
| 3558 | IL2, IL-2, TCGF, lymphokine | interleukin 2 |
C00002514
|
| 50604 | IL20, IL-20, IL10D, ZCYTO10 | interleukin 20 |
C00002514
|
| 3572 | IL6ST, CD130, CDW130, GP130, IL-6RB | interleukin 6 signal transducer (gp130, oncostatin M receptor) |
C00002514
|
| 3608 | ILF2, NF45, PRO3063 | interleukin enhancer binding factor 2 |
C00002514
|
| 10994 | ILVBL, 209L8, AHAS, ILV2H | ilvB (bacterial acetolactate synthase)-like (EC:2.2.1.-) |
C00002514
|
| 196294 | IMMP1L, IMP1, IMP1-LIKE | IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
C00002514
|
| 92856 | IMP4, BXDC4 | IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
C00002514
|
| 3613 | IMPA2 | inositol(myo)-1(or 4)-monophosphatase 2 (EC:3.1.3.25) |
C00002514
|
| 10207 | INADL, Cipp, PATJ, hINADL | InaD-like (Drosophila) |
C00002514
|
| 3619 | INCENP | inner centromere protein antigens 135/155kDa |
C00002514
|
| 3625 | INHBB | inhibin, beta B |
C00002514
|
| 283899 | INO80E, CCDC95 | INO80 complex subunit E |
C00002514
|
| 8821 | INPP4B | inositol polyphosphate-4-phosphatase, type II, 105kDa (EC:3.1.3.66) |
C00002514
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00002514
|
| 25896 | INTS7, C1orf73, INT7 | integrator complex subunit 7 |
C00002514
|
| 51447 | IP6K2, IHPK2, PIUS | inositol hexakisphosphate kinase 2 (EC:2.7.4.21) |
C00002514
|
| 51194 | IPO11, RanBP11 | importin 11 |
C00002514
|
| 55705 | IPO9, Imp9 | importin 9 |
C00002514
|
| 124152 | IQCK | IQ motif containing K |
C00002514
|
| 128239 | IQGAP3 | IQ motif containing GTPase activating protein 3 |
C00002514
|
| 359948 | IRF2BP2 | interferon regulatory factor 2 binding protein 2 |
C00002514
|
| 3664 | IRF6, LPS, OFC6, PIT, PPS, PPS1, VWS, VWS1 | interferon regulatory factor 6 |
C00002514
|
| 10379 | IRF9, IRF-9, ISGF3, ISGF3G, p48 | interferon regulatory factor 9 |
C00002514
|
| 3667 | IRS1, HIRS-1 | insulin receptor substrate 1 |
C00002514
|
| 8660 | IRS2, IRS-2 | insulin receptor substrate 2 |
C00002514
|
| 51477 | ISYNA1, INO1, INOS, IPS, IPS_1, IPS-1 | inositol-3-phosphate synthase 1 (EC:5.5.1.4) |
C00002514
|
| 3673 | ITGA2, BR, CD49B, GPIa, HPA-5, VLA-2, VLAA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
C00002514
|
| 3682 | ITGAE, CD103, HUMINAE | integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
C00002514
|
| 23421 | ITGB3BP, CENP-R, CENPR, HSU37139, NRIF3, TAP20 | integrin beta 3 binding protein (beta3-endonexin) |
C00002514
|
| 3708 | ITPR1, INSP3R1, IP3R, IP3R1, SCA15, SCA16, SCA29 | inositol 1,4,5-trisphosphate receptor, type 1 |
C00002514
|
| 10625 | IVNS1ABP, FLARA3, HSPC068, KLHL39, ND1, NS-1, NS1-BP, NS1BP | influenza virus NS1A binding protein |
C00002514
|
| 182 | JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1 | jagged 1 |
C00002514
|
| 133746 | JMY, WHDC1L3 | junction mediating and regulatory protein, p53 cofactor |
C00002514
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00002514
|
| 3726 | JUNB, AP-1 | jun B proto-oncogene |
C00002514
|
| 3728 | JUP, ARVD12, CTNNG, DP3, DPIII, PDGB, PKGB | junction plakoglobin |
C00002514
|
| 7994 | KAT6A, MOZ, MYST3, RUNXBP2, ZC2HC6A, ZNF220 | K(lysine) acetyltransferase 6A (EC:2.3.1.48) |
C00002514
|
| 84056 | KATNAL1 | katanin p60 subunit A-like 1 (EC:3.6.4.3) |
C00002514
|
| 8645 | KCNK5, K2p5.1, TASK-2, TASK2 | potassium channel, subfamily K, member 5 |
C00002514
|
| 54793 | KCTD9, BTBD27 | potassium channel tetramerization domain containing 9 |
C00002514
|
| 23028 | KDM1A, AOF2, BHC110, KDM1, LSD1 | lysine (K)-specific demethylase 1A |
C00002514
|
| 221656 | KDM1B, AOF1, C6orf193, LSD2, bA204B7.3, dJ298J15.2 | lysine (K)-specific demethylase 1B |
C00002514
|
| 55818 | KDM3A, JHDM2A, JHMD2A, JMJD1, JMJD1A, TSGA | lysine (K)-specific demethylase 3A |
C00002514
|
| 5927 | KDM5A, RBBP-2, RBBP2, RBP2, JARID1A | lysine (K)-specific demethylase 5A |
C00002514
|
| 10765 | KDM5B, CT31, JARID1B, PLU-1, PLU1, PUT1, RBBP2H1A | lysine (K)-specific demethylase 5B |
C00002514
|
| 9703 | BCOX, BCOX1, CT101 | KIAA0100 |
C00002514
|
| 9768 | NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | KIAA0101 |
C00002514
|
| 9764 | KIAA0513 |
C00002514
|
|
| 23247 | KIAA0556 |
C00002514
|
|
| 9786 | Talpid3 | KIAA0586 |
C00002514
|
| 23379 | KIAA0947 |
C00002514
|
|
| 23325 | SWIP | KIAA1033 |
C00002514
|
| 57189 | LCHN, PRO2561 | KIAA1147 |
C00002514
|
| 57179 | p60MONOX | KIAA1191 |
C00002514
|
| 57482 | KIAA1211 |
C00002514
|
|
| 57221 | A7322, ARFGEF3, BIG3, C6orf92, PPP1R33, dJ171N11.1 | KIAA1244 |
C00002514
|
| 57535 | EIG121 | KIAA1324 |
C00002514
|
| 57650 | CIP2A, p90 | KIAA1524 |
C00002514
|
| 57498 | KIDINS220, ARMS | kinase D-interacting substrate, 220kDa |
C00002514
|
| 3832 | KIF11, EG5, HKSP, KNSL1, MCLMR, TRIP5 | kinesin family member 11 |
C00002514
|
| 9928 | KIF14 | kinesin family member 14 |
C00002514
|
| 56992 | KIF15, HKLP2, KNSL7, NY-BR-62 | kinesin family member 15 |
C00002514
|
| 81930 | KIF18A, MS-KIF18A | kinesin family member 18A |
C00002514
|
| 146909 | KIF18B | kinesin family member 18B |
C00002514
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00002514
|
| 11004 | KIF2C, KNSL6, MCAK | kinesin family member 2C |
C00002514
|
| 3800 | KIF5C, CDCBM2, KINN, NKHC, NKHC-2, NKHC2 | kinesin family member 5C |
C00002514
|
| 3833 | KIFC1, HSET, KNSL2 | kinesin family member C1 |
C00002514
|
| 11278 | KLF12, AP-2rep, AP2REP | Kruppel-like factor 12 |
C00002514
|
| 9314 | KLF4, EZF, GKLF | Kruppel-like factor 4 (gut) |
C00002514
|
| 1316 | KLF6, BCD1, CBA1, COPEB, CPBP, GBF, PAC1, ST12, ZF9 | Kruppel-like factor 6 |
C00002514
|
| 687 | KLF9, BTEB, BTEB1 | Kruppel-like factor 9 |
C00002514
|
| 23008 | KLHDC10, slim | kelch domain containing 10 |
C00002514
|
| 116138 | KLHDC3, PEAS, RP1-20C7.3, dJ20C7.3 | kelch domain containing 3 |
C00002514
|
| 54800 | KLHL24, DRE1, KRIP6 | kelch-like family member 24 |
C00002514
|
| 51088 | KLHL5 | kelch-like family member 5 |
C00002514
|
| 346689 | KLRG2, CLEC15B | killer cell lectin-like receptor subfamily G, member 2 |
C00002514
|
| 8085 | KMT2D, AAD10, ALR, CAGL114, KABUK1, KMS, MLL2, MLL4, TNRC21 | lysine (K)-specific methyltransferase 2D (EC:2.1.1.43) |
C00002514
|
| 55904 | KMT2E, HDCMC04P, MLL5 | lysine (K)-specific methyltransferase 2E (EC:2.1.1.43) |
C00002514
|
| 90417 | KNSTRN, C15orf23, SKAP, TRAF4AF1 | kinetochore-localized astrin/SPAG5 binding protein |
C00002514
|
| 9735 | KNTC1, ROD | kinetochore associated 1 |
C00002514
|
| 779317 |
C00002514
|
||
| 3839 | KPNA3, IPOA4, SRP1, SRP1gamma, SRP4, hSRP1 | karyopherin alpha 3 (importin alpha 4) |
C00002514
|
| 3840 | KPNA4, IPOA3, QIP1, SRP3 | karyopherin alpha 4 (importin alpha 3) |
C00002514
|
| 3845 | KRAS, C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RASK2 | Kirsten rat sarcoma viral oncogene homolog |
C00002514
|
| 51315 | KRCC1, CHBP2 | lysine-rich coiled-coil 1 |
C00002514
|
| 144501 | KRT80, KB20 | keratin 80 |
C00002514
|
| 3895 | KTN1, CG1, KNT, MU-RMS-40.19 | kinectin 1 (kinesin receptor) |
C00002514
|
| 3897 | L1CAM, CAML1, CD171, HSAS, HSAS1, MASA, MIC5, N-CAM-L1, N-CAML1, NCAM-L1, S10, SPG1 | L1 cell adhesion molecule |
C00002514
|
| 79944 | L2HGDH, C14orf160 | L-2-hydroxyglutarate dehydrogenase (EC:1.1.99.2) |
C00002514
|
| 114294 | LACTB, G24, MRPL56 | lactamase, beta |
C00002514
|
| 8270 | LAGE3, CVG5, DXS9879E, DXS9951E, ESO3, ITBA2 | L antigen family, member 3 |
C00002514
|
| 3916 | LAMP1, CD107a, LAMPA, LGP120 | lysosomal-associated membrane protein 1 |
C00002514
|
| 55323 | LARP6, ACHN | La ribonucleoprotein domain family, member 6 |
C00002514
|
| 3930 | LBR, DHCR14B, LMN2R, PHA, TDRD18 | lamin B receptor |
C00002514
|
| 9836 | LCMT2, PPM2, TYW4 | leucine carboxyl methyltransferase 2 |
C00002514
|
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00002514
|
| 3953 | LEPR, CD295, LEP-R, LEPRD, OB-R, OBR | leptin receptor |
C00002514
|
| 3956 | LGALS1, GAL1, GBP | lectin, galactoside-binding, soluble, 1 |
C00002514
|
| 3964 | LGALS8, Gal-8, PCTA-1, PCTA1, Po66-CBP | lectin, galactoside-binding, soluble, 8 |
C00002514
|
| 9355 | LHX2, LH2, hLhx2 | LIM homeobox 2 |
C00002514
|
| 3978 | LIG1 | ligase I, DNA, ATP-dependent (EC:6.5.1.1) |
C00002514
|
| 3981 | LIG4 | ligase IV, DNA, ATP-dependent (EC:6.5.1.1) |
C00002514
|
| 51474 | LIMA1, EPLIN, SREBP3 | LIM domain and actin binding 1 |
C00002514
|
| 91750 | LIN52, C14orf46, c14_5549 | lin-52 homolog (C. elegans) |
C00002514
|
| 8825 | LIN7A, LIN-7A, LIN7, MALS-1, TIP-33, VELI1 | lin-7 homolog A (C. elegans) |
C00002514
|
| 55327 | LIN7C, LIN-7-C, LIN-7C, MALS-3, MALS3, VELI3 | lin-7 homolog C (C. elegans) |
C00002514
|
| 145978 | LINC00052, NCRNA00052, TMEM83 | long intergenic non-protein coding RNA 52 |
C00002514
|
| 29995 | LMCD1 | LIM and cysteine-rich domains 1 |
C00002514
|
| 4001 | LMNB1, ADLD, LMN, LMN2, LMNB | lamin B1 |
C00002514
|
| 84823 | LMNB2, LAMB2, LMN2 | lamin B2 |
C00002514
|
| 9361 | LONP1, LON, LONP, LonHS, PIM1, PRSS15, hLON | lon peptidase 1, mitochondrial |
C00002514
|
| 26018 | LRIG1, LIG-1, LIG1 | leucine-rich repeats and immunoglobulin-like domains 1 |
C00002514
|
| 26020 | LRP10, LRP9, MST087 | low density lipoprotein receptor-related protein 10 |
C00002514
|
| 7804 | LRP8, APOER2, HSZ75190, LRP-8, MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
C00002514
|
| 122769 | LRR1, 4-1BBLRR, LRR-1, PPIL5 | leucine rich repeat protein 1 |
C00002514
|
| 80131 | LRRC8E | leucine rich repeat containing 8 family, member E |
C00002514
|
| 57819 | LSM2, C6orf28, G7B, YBL026W, snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00002514
|
| 25804 | LSM4, GRP, YER112W | LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00002514
|
| 51690 | LSM7, YNL147W | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00002514
|
| 55692 | LUC7L, LUC7B1, Luc7, SR+89, hLuc7B1 | LUC7-like (S. cerevisiae) |
C00002514
|
| 55646 | LYAR, ZC2HC2, ZLYAR | Ly1 antibody reactive |
C00002514
|
| 4067 | LYN, JTK8, p53Lyn, p56Lyn | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (EC:2.7.10.2) |
C00002514
|
| 130574 | LYPD6 | LY6/PLAUR domain containing 6 |
C00002514
|
| 4085 | MAD2L1, HSMAD2, MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) |
C00002514
|
| 10459 | MAD2L2, MAD2B, POLZ2, REV7 | MAD2 mitotic arrest deficient-like 2 (yeast) |
C00002514
|
| 23764 | MAFF, U-MAF, hMafF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
C00002514
|
| 10916 | MAGED2, 11B6, BCG-1, BCG1, HCA10, MAGE-D2 | melanoma antigen family D, 2 |
C00002514
|
| 9223 | MAGI1, AIP-3, AIP3, BAIAP1, BAP-1, BAP1, MAGI-1, TNRC19, WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 |
C00002514
|
| 55110 | MAGOHB, MGN2, mago, magoh | mago-nashi homolog B (Drosophila) |
C00002514
|
| 378938 | MALAT1, HCN, LINC00047, MALAT-1, NCRNA00047, NEAT2, PRO2853 | metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
C00002514
|
| 4124 | MAN2A1, GOLIM7, MANA2, MANII | mannosidase, alpha, class 2A, member 1 (EC:3.2.1.114) |
C00002514
|
| 79694 | MANEA, ENDO, hEndo | mannosidase, endo-alpha (EC:3.2.1.130) |
C00002514
|
| 4131 | MAP1B, FUTSCH, MAP5 | microtubule-associated protein 1B |
C00002514
|
| 81631 | MAP1LC3B, ATG8F, LC3B, MAP1A/1BLC3, MAP1LC3B-a | microtubule-associated protein 1 light chain 3 beta |
C00002514
|
| 5606 | MAP2K3, MAPKK3, MEK3, MKK3, PRKMK3, SAPKK-2, SAPKK2 | mitogen-activated protein kinase kinase 3 (EC:2.7.12.2) |
C00002514
|
| 9448 | MAP4K4, FLH21957, HGK, MEKKK4, NIK | mitogen-activated protein kinase kinase kinase kinase 4 (EC:2.7.11.1) |
C00002514
|
| 64844 | MARCH7, AXO, AXOT, MARCH-VII, RNF177 | membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
C00002514
|
| 4082 | MARCKS, 80K-L, MACS, PKCSL, PRKCSL | myristoylated alanine-rich protein kinase C substrate |
C00002514
|
| 375449 | MAST4 | microtubule associated serine/threonine kinase family member 4 (EC:2.7.11.1) |
C00002514
|
| 84930 | MASTL, GREATWALL, GW, GWL, MAST-L, RP11-85G18.2, THC2, hGWL | microtubule associated serine/threonine kinase-like (EC:2.7.11.1) |
C00002514
|
| 4148 | MATN3, DIPOA, EDM5, HOA, OADIP, OS2 | matrilin 3 |
C00002514
|
| 4149 | MAX, bHLHd4 | MYC associated factor X |
C00002514
|
| 4150 | MAZ, PUR1, Pur-1, SAF-1, SAF-2, SAF-3, ZF87, ZNF801, Zif87 | MYC-associated zinc finger protein (purine-binding transcription factor) |
C00002514
|
| 151963 | MB21D2, C3orf59 | Mab-21 domain containing 2 |
C00002514
|
| 4162 | MCAM, CD146, MUC18 | melanoma cell adhesion molecule |
C00002514
|
| 55388 | MCM10, CNA43, DNA43 | minichromosome maintenance complex component 10 |
C00002514
|
| 4171 | MCM2, BM28, CCNL1, CDCL1, D3S3194, MITOTIN, cdc19 | minichromosome maintenance complex component 2 (EC:3.6.4.12) |
C00002514
|
| 4172 | MCM3, HCC5, P1-MCM3, P1.h, RLFB | minichromosome maintenance complex component 3 (EC:3.6.4.12) |
C00002514
|
| 4173 | MCM4, CDC21, CDC54, NKCD, NKGCD, P1-CDC21, hCdc21 | minichromosome maintenance complex component 4 (EC:3.6.4.12) |
C00002514
|
| 4174 | MCM5, CDC46, P1-CDC46 | minichromosome maintenance complex component 5 (EC:3.6.4.12) |
C00002514
|
| 4175 | MCM6, MCG40308, Mis5, P105MCM | minichromosome maintenance complex component 6 (EC:3.6.4.12) |
C00002514
|
| 4176 | MCM7, CDC47, MCM2, P1.1-MCM3, P1CDC47, P85MCM, PNAS146 | minichromosome maintenance complex component 7 (EC:3.6.4.12) |
C00002514
|
| 84515 | MCM8, C20orf154, dJ967N21.5 | minichromosome maintenance complex component 8 (EC:3.6.4.12) |
C00002514
|
| 9656 | MDC1, NFBD1 | mediator of DNA-damage checkpoint 1 |
C00002514
|
| 4201 | MEA1, HYS, MEA | male-enhanced antigen 1 |
C00002514
|
| 64769 | MEAF6, C1orf149, CENP-28, EAF6, NY-SAR-91 | MYST/Esa1-associated factor 6 |
C00002514
|
| 4205 | MEF2A, ADCAD1, RSRFC4, RSRFC9, mef2 | myocyte enhancer factor 2A |
C00002514
|
| 4213 | MEIS3P1, MEIS3, MEIS4, MRG2 | Meis homeobox 3 pseudogene 1 |
C00002514
|
| 9833 | MELK, HPK38 | maternal embryonic leucine zipper kinase (EC:2.7.11.1 2.7.10.2) |
C00002514
|
| 56257 | MEPCE, BCDIN3 | methylphosphate capping enzyme |
C00002514
|
| 59274 | MESDC1 | mesoderm development candidate 1 |
C00002514
|
| 10988 | METAP2, MAP2, MNPEP, p67, p67eIF2 | methionyl aminopeptidase 2 (EC:3.4.11.18) |
C00002514
|
| 4234 | METTL1, C12orf1, TRM8, TRMT8, YDL201w | methyltransferase like 1 (EC:2.1.1.33) |
C00002514
|
| 25840 | METTL7A, AAM-B | methyltransferase like 7A |
C00002514
|
| 4236 | MFAP1 | microfibrillar-associated protein 1 |
C00002514
|
| 23269 | MGA, MAD5, MXD5 | MGA, MAX dimerization protein |
C00002514
|
| 11282 | MGAT4B, GNT-IV, GNT-IVB | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (EC:2.4.1.145) |
C00002514
|
| 4249 | MGAT5, GNT-V, GNT-VA | mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase (EC:2.4.1.155) |
C00002514
|
| 10724 | MGEA5, MEA5, NCOAT, OGA | meningioma expressed antigen 5 (hyaluronidase) (EC:2.3.1.48 3.2.1.169) |
C00002514
|
| 11343 | MGLL, HU-K5, HUK5, MAGL, MGL | monoglyceride lipase (EC:3.1.1.23) |
C00002514
|
| 4256 | MGP, MGLAP, NTI | matrix Gla protein |
C00002514
|
| 85377 | MICALL1, MICAL-L1, MIRAB13 | MICAL-like 1 |
C00002514
|
| 79778 | MICALL2, JRAB, MICAL-L2 | MICAL-like 2 |
C00002514
|
| 4277 | MICB, PERB11.2 | MHC class I polypeptide-related sequence B |
C00002514
|
| 84864 | MINA, MDIG, MINA53, NO52, ROX | MYC induced nuclear antigen |
C00002514
|
| 406991 | MIR21, MIRN21, hsa-mir-21, miR-21, miRNA21 | microRNA 21 |
C00002514
|
| 55320 | MIS18BP1, C14orf106, HSA242977, KNL2, M18BP1 | MIS18 binding protein 1 |
C00002514
|
| 4288 | MKI67, KIA, MIB-1 | marker of proliferation Ki-67 |
C00002514
|
| 2872 | MKNK2, GPRK7, MNK2 | MAP kinase interacting serine/threonine kinase 2 (EC:2.7.11.1) |
C00002514
|
| 4291 | MLF1 | myeloid leukemia factor 1 |
C00002514
|
| 79682 | CENPU, CENP50, CENPU50, KLIP1, MLF1IP, PBIP1 | centromere protein U |
C00002514
|
| 4292 | MLH1, COCA2, FCC2, HNPCC, HNPCC2, hMLH1 | mutL homolog 1 |
C00002514
|
| 197259 | MLKL | mixed lineage kinase domain-like |
C00002514
|
| 326625 | MMAB, ATR, cblB, cob | methylmalonic aciduria (cobalamin deficiency) cblB type (EC:2.5.1.17) |
C00002514
|
| 4325 | MMP16, C8orf57, MMP-X2, MT-MMP2, MT-MMP3, MT3-MMP | matrix metallopeptidase 16 (membrane-inserted) (EC:3.4.24.-) |
C00002514
|
| 55329 | MNS1, SPATA40 | meiosis-specific nuclear structural 1 |
C00002514
|
| 64112 | MOAP1, MAP-1, PNMA4 | modulator of apoptosis 1 |
C00002514
|
| 9643 | MORF4L2, MORFL2, MRGX | mortality factor 4 like 2 |
C00002514
|
| 56180 | MOSPD1, DJ473B4 | motile sperm domain containing 1 |
C00002514
|
| 10198 | MPHOSPH9, MPP-9, MPP9 | M-phase phosphoprotein 9 |
C00002514
|
| 744 | MPPED2, 239FB, C11orf8 | metallophosphoesterase domain containing 2 |
C00002514
|
| 4361 | MRE11A, ATLD, HNGS1, MRE11, MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
C00002514
|
| 78988 | MRP63, bMRP63 | mitochondrial ribosomal protein 63 |
C00002514
|
| 51253 | MRPL37, L37mt, MRP-L2, MRP-L37, MRPL2, RPML2 | mitochondrial ribosomal protein L37 |
C00002514
|
| 64949 | MRPS26, C20orf193, GI008, MRP-S13, MRP-S26, MRPS13, NY-BR-87, RPMS13, dJ534B8.3 | mitochondrial ribosomal protein S26 |
C00002514
|
| 23107 | MRPS27, MRP-S27, S27mt | mitochondrial ribosomal protein S27 |
C00002514
|
| 64968 | MRPS6, C21orf101, MRP-S6, RPMS6, S6mt | mitochondrial ribosomal protein S6 |
C00002514
|
| 100009675 |
C00002514
|
||
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00002514
|
| 4437 | MSH3, DUP, MRP1 | mutS homolog 3 |
C00002514
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00002514
|
| 4488 | MSX2, CRS2, FPP, HOX8, MSH, PFM, PFM1 | msh homeobox 2 |
C00002514
|
| 4515 | MTCP1, P13MTCP1, p8MTCP1 | mature T-cell proliferation 1 |
C00002514
|
| 51537 | MTFP1, MTP18 | mitochondrial fission process 1 |
C00002514
|
| 113115 | MTFR2, DUFD1, FAM54A | mitochondrial fission regulator 2 |
C00002514
|
| 92170 | MTG1, GTP, GTPBP7, RP11-108K14.2 | mitochondrial ribosome-associated GTPase 1 |
C00002514
|
| 4522 | MTHFD1, MTHFC, MTHFD | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase (EC:1.5.1.5 6.3.4.3 3.5.4.9) |
C00002514
|
| 9110 | MTMR4, FYVE-DSP2, ZFYVE11 | myotubularin related protein 4 (EC:3.1.3.48) |
C00002514
|
| 57509 | MTUS1, ATBP, ATIP, ICIS, MP44, MTSG1 | microtubule associated tumor suppressor 1 |
C00002514
|
| 9961 | MVP, LRP, VAULT1 | major vault protein |
C00002514
|
| 10608 | MXD4, MAD4, MST149, MSTP149, bHLHc12 | MAX dimerization protein 4 |
C00002514
|
| 4601 | MXI1, MAD2, MXD2, MXI, bHLHc11 | MAX interactor 1, dimerization protein |
C00002514
|
| 439921 | MXRA7, PS1TP1, TMAP1 | matrix-remodelling associated 7 |
C00002514
|
| 4602 | MYB, Cmyb, c-myb, c-myb_CDS, efg | v-myb avian myeloblastosis viral oncogene homolog |
C00002514
|
| 4605 | MYBL2, B-MYB, BMYB | v-myb avian myeloblastosis viral oncogene homolog-like 2 |
C00002514
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00002514
|
| 26292 | MYCBP, AMY-1 | MYC binding protein |
C00002514
|
| 23077 | MYCBP2, PAM | MYC binding protein 2, E3 ubiquitin protein ligase |
C00002514
|
| 79784 | MYH14, DFNA4, DFNA4A, MHC16, MYH17, NMHC_II-C, NMHC-II-C, PNMHH, myosin | myosin, heavy chain 14, non-muscle (EC:3.6.4.1) |
C00002514
|
| 29116 | MYLIP, IDOL, MIR | myosin regulatory light chain interacting protein (EC:6.3.2.-) |
C00002514
|
| 80179 | MYO19, MYOHD1 | myosin XIX |
C00002514
|
| 4430 | MYO1B, myr1 | myosin IB |
C00002514
|
| 4644 | MYO5A, GS1, MYH12, MYO5, MYR12 | myosin VA (heavy chain 12, myoxin) |
C00002514
|
| 4645 | MYO5B | myosin VB |
C00002514
|
| 4646 | MYO6, DFNA22, DFNB37 | myosin VI |
C00002514
|
| 26509 | MYOF, FER1L3 | myoferlin |
C00002514
|
| 440145 | MZT1, C13orf37, MOZART1 | mitotic spindle organizing protein 1 |
C00002514
|
| 9683 | N4BP1 | NEDD4 binding protein 1 |
C00002514
|
| 90634 | N4BP2L1, CG018 | NEDD4 binding protein 2-like 1 |
C00002514
|
| 80218 | NAA50, MAK3, NAT13, NAT13P, NAT5, NAT5P, SAN, hNAT5, hSAN | N(alpha)-acetyltransferase 50, NatE catalytic subunit (EC:2.3.1.-) |
C00002514
|
| 4664 | NAB1 | NGFI-A binding protein 1 (EGR1 binding protein 1) |
C00002514
|
| 133686 | NADK2, C5orf33, MNADK, NADKD1 | NAD kinase 2, mitochondrial (EC:2.7.1.23) |
C00002514
|
| 340719 | NANOS1, NOS1, SPGF12 | nanos homolog 1 (Drosophila) |
C00002514
|
| 140838 | NANP, C20orf147, HDHD4, dJ694B14.3 | N-acetylneuraminic acid phosphatase (EC:3.1.3.29) |
C00002514
|
| 4676 | NAP1L4, NAP1L4b, NAP2, NAP2L, hNAP2 | nucleosome assembly protein 1-like 4 |
C00002514
|
| 100195127 |
C00002514
|
||
| 4678 | NASP, FLB7527, PRO1999 | nuclear autoantigenic sperm protein (histone-binding) |
C00002514
|
| 26960 | NBEA, BCL8B, LYST2 | neurobeachin |
C00002514
|
| 65065 | NBEAL1, A530083I02Rik, ALS2CR16, ALS2CR17 | neurobeachin-like 1 |
C00002514
|
| 55672 | NBPF1, AB13, AB14, AB23, AD2, NBG, NBPF | neuroblastoma breakpoint family, member 1 |
C00002514
|
| 200030 | NBPF11 | neuroblastoma breakpoint family, member 11 |
C00002514
|
| 9918 | NCAPD2, CAP-D2, CNAP1, hCAP-D2 | non-SMC condensin I complex, subunit D2 |
C00002514
|
| 23310 | NCAPD3, CAP-D3, CAPD3, hCAP-D3, hHCP-6, hcp-6 | non-SMC condensin II complex, subunit D3 |
C00002514
|
| 64151 | NCAPG, CAPG, CHCG, NY-MEL-3 | non-SMC condensin I complex, subunit G |
C00002514
|
| 54892 | NCAPG2, CAP-G2, CAPG2, LUZP5, MTB, hCAP-G2 | non-SMC condensin II complex, subunit G2 |
C00002514
|
| 23397 | NCAPH, BRRN1, CAP-H | non-SMC condensin I complex, subunit H |
C00002514
|
| 29781 | NCAPH2, CAPH2 | non-SMC condensin II complex, subunit H2 |
C00002514
|
| 4686 | NCBP1, CBP80, NCBP, Sto1 | nuclear cap binding protein subunit 1, 80kDa |
C00002514
|
| 8648 | NCOA1, F-SRC-1, KAT13A, RIP160, SRC1, bHLHe42, bHLHe74 | nuclear receptor coactivator 1 (EC:2.3.1.48) |
C00002514
|
| 10499 | NCOA2, GRIP1, KAT13C, NCoA-2, SRC2, TIF2, bHLHe75 | nuclear receptor coactivator 2 |
C00002514
|
| 8202 | NCOA3, ACTR, AIB-1, AIB1, CAGH16, CTG26, KAT13B, RAC3, SRC-3, SRC3, TNRC14, TNRC16, TRAM-1, bHLHe42, pCIP | nuclear receptor coactivator 3 (EC:2.3.1.48) |
C00002514
|
| 135112 | NCOA7, ERAP140, ESNA1, Nbla00052, Nbla10993, TLDC4, dJ187J11.3 | nuclear receptor coactivator 7 |
C00002514
|
| 55706 | NDC1, NET3, TMEM48 | NDC1 transmembrane nucleoporin |
C00002514
|
| 54820 | NDE1, HOM-TES-87, LIS4, NDE, NUDE, NUDE1 | nudE neurodevelopment protein 1 |
C00002514
|
| 80762 | NDFIP1, N4WBP5 | Nedd4 family interacting protein 1 |
C00002514
|
| 4728 | NDUFS8, CI-23k, CI23KD, TYKY | NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) (EC:1.6.5.3 1.6.99.3) |
C00002514
|
| 4723 | NDUFV1, CI-51K, CI51KD, UQOR1 | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa (EC:1.6.5.3 1.6.99.3) |
C00002514
|
| 283131 | NEAT1, LINC00084, NCRNA00084, TncRNA, VINC | nuclear paraspeckle assembly transcript 1 (non-protein coding) |
C00002514
|
| 10529 | NEBL, LASP2, LNEBL | nebulette |
C00002514
|
| 23327 | NEDD4L, NEDD4-2, NEDD4.2, RSP5, hNEDD4-2 | neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00002514
|
| 4739 | NEDD9, CAS-L, CAS2, CASL, CASS2, HEF1 | neural precursor cell expressed, developmentally down-regulated 9 |
C00002514
|
| 55247 | NEIL3, FGP2, FPG2, NEI3, hFPG2, hNEI3 | nei endonuclease VIII-like 3 (E. coli) (EC:4.2.99.18) |
C00002514
|
| 4751 | NEK2, HsPK21, NEK2A, NLK1 | NIMA-related kinase 2 (EC:2.7.11.1) |
C00002514
|
| 4753 | NELL2, NRP2 | NEL-like 2 (chicken) |
C00002514
|
| 81831 | NETO2, BTCL2, NEOT2 | neuropilin (NRP) and tolloid (TLL)-like 2 |
C00002514
|
| 4758 | NEU1, NANH, NEU, SIAL1 | sialidase 1 (lysosomal sialidase) (EC:3.2.1.18) |
C00002514
|
| 54492 | NEURL1B, NEURL3, hNeur2, neur2 | neuralized E3 ubiquitin protein ligase 1B (EC:6.3.2.-) |
C00002514
|
| 84901 | NFATC2IP, ESC2, NIP45, RAD60 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein |
C00002514
|
| 4783 | NFIL3, E4BP4, IL3BP1, NF-IL3A, NFIL3A | nuclear factor, interleukin 3 regulated |
C00002514
|
| 4790 | NFKB1, EBP-1, KBF1, NF-kB1, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
C00002514
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00002514
|
| 22981 | NINL, NLP, dJ691N24.1 | ninein-like |
C00002514
|
| 79570 | NKAIN1, FAM77C | Na+/K+ transporting ATPase interacting 1 |
C00002514
|
| 4836 | NMT1, NMT | N-myristoyltransferase 1 (EC:2.3.1.97) |
C00002514
|
| 9397 | NMT2 | N-myristoyltransferase 2 (EC:2.3.1.97) |
C00002514
|
| 10874 | NMU | neuromedin U |
C00002514
|
| 23530 | NNT, GCCD4 | nicotinamide nucleotide transhydrogenase (EC:1.6.1.2) |
C00002514
|
| 64318 | NOC3L, AD24, C10orf117, FAD24 | nucleolar complex associated 3 homolog (S. cerevisiae) |
C00002514
|
| 100380365 |
C00002514
|
||
| 9221 | NOLC1, NOPP130, NOPP140, NS5ATP13, P130 | nucleolar and coiled-body phosphoprotein 1 |
C00002514
|
| 64434 | NOM1, C7orf3, SGD1 | nucleolar protein with MIF4G domain 1 |
C00002514
|
| 4854 | NOTCH3, CADASIL, CASIL, IMF2 | notch 3 |
C00002514
|
| 64067 | NPAS3, MOP6, PASD6, bHLHe12 | neuronal PAS domain protein 3 |
C00002514
|
| 9520 | NPEPPS, AAP-S, MP100, PSA | aminopeptidase puromycin sensitive (EC:3.4.11.14) |
C00002514
|
| 27031 | NPHP3, MKS7, NPH3, RHPD, RHPD1 | nephronophthisis 3 (adolescent) |
C00002514
|
| 10360 | NPM3, PORMIN, TMEM123 | nucleophosmin/nucleoplasmin 3 |
C00002514
|
| 255743 | NPNT, EGFL6L, POEM | nephronectin |
C00002514
|
| 4886 | NPY1R, NPY1-R, NPYR | neuropeptide Y receptor Y1 |
C00002514
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00002514
|
| 4835 | NQO2, DHQV, DIA6, NMOR2, QR2 | NAD(P)H dehydrogenase, quinone 2 (EC:1.10.99.2) |
C00002514
|
| 7181 | NR2C1, TR2 | nuclear receptor subfamily 2, group C, member 1 |
C00002514
|
| 126382 | NR2C2AP, TRA16 | nuclear receptor 2C2-associated protein |
C00002514
|
| 7026 | NR2F2, ARP1, COUPTFB, COUPTFII, NF-E3, NR2F1, SVP40, TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 |
C00002514
|
| 2063 | NR2F6, EAR-2, EAR2, ERBAL2 | nuclear receptor subfamily 2, group F, member 6 |
C00002514
|
| 2908 | NR3C1, GCCR, GCR, GR, GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
C00002514
|
| 4929 | NR4A2, HZF-3, NOT, NURR1, RNR1, TINUR | nuclear receptor subfamily 4, group A, member 2 |
C00002514
|
| 4897 | NRCAM | neuronal cell adhesion molecule |
C00002514
|
| 11270 | NRM, NRM29 | nurim (nuclear envelope membrane protein) |
C00002514
|
| 4907 | NT5E, CALJA, CD73, E5NT, NT, NT5, NTE, eN, eNT | 5'-nucleotidase, ecto (CD73) (EC:3.1.3.5) |
C00002514
|
| 59277 | NTN4, PRO3091 | netrin 4 |
C00002514
|
| 9891 | NUAK1, ARK5 | NUAK family, SNF1-like kinase, 1 (EC:2.7.11.1) |
C00002514
|
| 4521 | NUDT1, MTH1 | nudix (nucleoside diphosphate linked moiety X)-type motif 1 (EC:3.6.1.55 3.6.1.56) |
C00002514
|
| 131870 | NUDT16 | nudix (nucleoside diphosphate linked moiety X)-type motif 16 (EC:3.6.1.62) |
C00002514
|
| 11051 | NUDT21, CFIM25, CPSF5 | nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
C00002514
|
| 57122 | NUP107, NUP84 | nucleoporin 107kDa |
C00002514
|
| 9631 | NUP155, N155 | nucleoporin 155kDa |
C00002514
|
| 23165 | NUP205, C7orf14 | nucleoporin 205kDa |
C00002514
|
| 23225 | NUP210, GP210, POM210 | nucleoporin 210kDa |
C00002514
|
| 129401 | NUP35, MP44, NP44, NUP53 | nucleoporin 35kDa |
C00002514
|
| 79023 | NUP37, p37 | nucleoporin 37kDa |
C00002514
|
| 10762 | NUP50, NPAP60, NPAP60L | nucleoporin 50kDa |
C00002514
|
| 54830 | NUP62CL | nucleoporin 62kDa C-terminal like |
C00002514
|
| 79902 | NUP85, Nup75 | nucleoporin 85kDa |
C00002514
|
| 51203 | NUSAP1, ANKT, BM037, LNP, NUSAP, PRO0310p1, Q0310, SAPL | nucleolar and spindle associated protein 1 |
C00002514
|
| 91775 | NXPE3, FAM55C, MST115 | neurexophilin and PC-esterase domain family, member 3 |
C00002514
|
| 221443 | OARD1, C6orf130, TARG1, dJ34B21.3 | O-acyl-ADP-ribose deacylase 1 |
C00002514
|
| 4947 | OAZ2, AZ2 | ornithine decarboxylase antizyme 2 |
C00002514
|
| 23363 | OBSL1 | obscurin-like 1 (EC:2.7.11.18) |
C00002514
|
| 4957 | ODF2, CT134, ODF2/1, ODF2/2, ODF84 | outer dense fiber of sperm tails 2 |
C00002514
|
| 55239 | OGFOD1, TPA1 | 2-oxoglutarate and iron-dependent oxygenase domain containing 1 (EC:1.14.11.-) |
C00002514
|
| 11339 | OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18beta, hMIS18beta | Opa interacting protein 5 |
C00002514
|
| 10439 | OLFM1, AMY, NOE1, NOELIN1, OlfA | olfactomedin 1 |
C00002514
|
| 10133 | OPTN, ALS12, FIP2, GLC1E, HIP7, HYPL, NRP, TFIIIA-INTP | optineurin |
C00002514
|
| 4998 | ORC1, HSORC1, ORC1L, PARC1 | origin recognition complex, subunit 1 |
C00002514
|
| 716175 |
C00002514
|
||
| 9885 | OSBPL2, ORP-2, ORP2 | oxysterol binding protein-like 2 |
C00002514
|
| 51526 | OSER1, C20orf111, HSPC207, Osr1, Perit1, dJ1183I21.1 | oxidative stress responsive serine-rich 1 |
C00002514
|
| 28962 | OSTM1, GIPN, GL, OPTB5 | osteopetrosis associated transmembrane protein 1 |
C00002514
|
| 220213 | OTUD1, DUBA7, OTDC1 | OTU domain containing 1 (EC:3.4.19.12) |
C00002514
|
| 5019 | OXCT1, OXCT, SCOT | 3-oxoacid CoA transferase 1 (EC:2.8.3.5) |
C00002514
|
| 5021 | OXTR, OT-R | oxytocin receptor |
C00002514
|
| 5034 | P4HB, DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB, PO4DB, PO4HB, PROHB | prolyl 4-hydroxylase, beta polypeptide (EC:5.3.4.1) |
C00002514
|
| 79447 | PAGR1, C16orf53, GAS, PA1 | PAXIP1 associated glutamate-rich protein 1 |
C00002514
|
| 10606 | PAICS, ADE2, ADE2H1, AIRC, PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase (EC:6.3.2.6 4.1.1.21) |
C00002514
|
| 10605 | PAIP1 | poly(A) binding protein interacting protein 1 |
C00002514
|
| 9924 | PAN2, USP52 | PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (EC:3.1.13.4) |
C00002514
|
| 11044 | PAPD7, LAK-1, LAK1, POLK, POLS, TRF4, TRF4-1, TRF41, TUTASE5 | PAP associated domain containing 7 (EC:2.7.7.7) |
C00002514
|
| 10914 | PAPOLA, PAP | poly(A) polymerase alpha (EC:2.7.7.19) |
C00002514
|
| 9060 | PAPSS2, ATPSK2, BCYM4, SK2 | 3'-phosphoadenosine 5'-phosphosulfate synthase 2 (EC:2.7.1.25 2.7.7.4) |
C00002514
|
| 54852 | PAQR5, MPRG | progestin and adipoQ receptor family member V |
C00002514
|
| 56288 | PARD3, ASIP, Baz, PAR3, PAR3alpha, PARD-3, PARD3A, SE2-5L16, SE2-5LT1, SE2-5T2 | par-3 family cell polarity regulator |
C00002514
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00002514
|
| 10038 | PARP2, ADPRT2, ADPRTL2, ADPRTL3, ARTD2, PARP-2, pADPRT-2 | poly (ADP-ribose) polymerase 2 (EC:2.4.2.30) |
C00002514
|
| 55010 | PARPBP, AROM, C12orf48, PARI | PARP1 binding protein |
C00002514
|
| 55742 | PARVA, CH-ILKBP, MXRA2 | parvin, alpha |
C00002514
|
| 5074 | PAWR, PAR4, Par-4 | PRKC, apoptosis, WT1, regulator |
C00002514
|
| 55872 | PBK, CT84, Nori-3, SPK, TOPK | PDZ binding kinase (EC:2.7.12.2) |
C00002514
|
| 5087 | PBX1 | pre-B-cell leukemia homeobox 1 |
C00002514
|
| 301164 |
C00002514
|
||
| 5094 | PCBP2, HNRNPE2, HNRPE2, hnRNP-E2 | poly(rC) binding protein 2 |
C00002514
|
| 57575 | PCDH10, OL-PCDH, PCDH19 | protocadherin 10 |
C00002514
|
| 5098 | PCDHGC3, PC43, PCDH-GAMMA-C3, PCDH2 | protocadherin gamma subfamily C, 3 |
C00002514
|
| 5106 | PCK2, PEPCK, PEPCK-M, PEPCK2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) (EC:4.1.1.32) |
C00002514
|
| 115294 | PCMTD1 | protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
C00002514
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00002514
|
| 5116 | PCNT, KEN, MOPD2, PCN, PCNT2, PCNTB, PCTN2, SCKL4 | pericentrin |
C00002514
|
| 5121 | PCP4, PEP-19 | Purkinje cell protein 4 |
C00002514
|
| 5046 | PCSK6, PACE4, SPC4 | proprotein convertase subtilisin/kexin type 6 |
C00002514
|
| 100380451 |
C00002514
|
||
| 58488 | PCTP, PC-TP, STARD2 | phosphatidylcholine transfer protein |
C00002514
|
| 27250 | PDCD4, H731 | programmed cell death 4 (neoplastic transformation inhibitor) |
C00002514
|
| 10015 | PDCD6IP, AIP1, ALIX, DRIP4, HP95 | programmed cell death 6 interacting protein |
C00002514
|
| 2923 | PDIA3, ER60, ERp57, ERp60, ERp61, GRP57, GRP58, HsT17083, P58, PI-PLC | protein disulfide isomerase family A, member 3 (EC:5.3.4.1) |
C00002514
|
| 10130 | PDIA6, ERP5, P5, TXNDC7 | protein disulfide isomerase family A, member 6 (EC:5.3.4.1) |
C00002514
|
| 9124 | PDLIM1, CLIM1, CLP-36, CLP36, hCLIM1 | PDZ and LIM domain 1 |
C00002514
|
| 27295 | PDLIM3, ALP | PDZ and LIM domain 3 |
C00002514
|
| 54704 | PDP1, PDH, PDP, PDPC, PPM2C | pyruvate dehyrogenase phosphatase catalytic subunit 1 (EC:3.1.3.43) |
C00002514
|
| 23590 | PDSS1, COQ1, COQ10D2, DPS, RP13-16H11.3, SPS, TPRT, TPT, TPT_1, hDPS1 | prenyl (decaprenyl) diphosphate synthase, subunit 1 (EC:2.5.1.91) |
C00002514
|
| 5174 | PDZK1, CAP70, CLAMP, NHERF-3, NHERF3, PDZD1 | PDZ domain containing 1 |
C00002514
|
| 64065 | PERP, KCP1, KRTCAP1, PIGPC1, RP3-496H19.1, THW, dJ496H19.1 | PERP, TP53 apoptosis effector |
C00002514
|
| 5211 | PFKL, PFK-B | phosphofructokinase, liver (EC:2.7.1.11) |
C00002514
|
| 5228 | PGF, D12S1900, PGFL, PLGF, PlGF-2, SHGC-10760 | placental growth factor |
C00002514
|
| 5229 | PGGT1B, BGGI, GGTI | protein geranylgeranyltransferase type I, beta subunit (EC:2.5.1.59) |
C00002514
|
| 5230 | PGK1, MIG10, PGKA | phosphoglycerate kinase 1 (EC:2.7.2.3) |
C00002514
|
| 283209 | PGM2L1, BM32A | phosphoglucomutase 2-like 1 (EC:2.7.1.106) |
C00002514
|
| 5238 | PGM3, AGM1, PAGM, PGM_3 | phosphoglucomutase 3 (EC:5.4.2.3) |
C00002514
|
| 5241 | PGR, NR3C3, PR | progesterone receptor |
C00002514
|
| 10857 | PGRMC1, HPR6.6, MPR | progesterone receptor membrane component 1 |
C00002514
|
| 10424 | PGRMC2, DG6, PMBP | progesterone receptor membrane component 2 |
C00002514
|
| 9749 | PHACTR2, C6orf56 | phosphatase and actin regulator 2 |
C00002514
|
| 55274 | PHF10, BAF45A, XAP135 | PHD finger protein 10 |
C00002514
|
| 26147 | PHF19, MTF2L1, PCL3, TDRD19B | PHD finger protein 19 |
C00002514
|
| 51105 | PHF20L1, TDRD20B | PHD finger protein 20-like 1 |
C00002514
|
| 26227 | PHGDH, 3-PGDH, 3PGDH, PDG, PGAD, PGD, PGDH, SERA | phosphoglycerate dehydrogenase (EC:1.1.1.95) |
C00002514
|
| 7262 | PHLDA2, BRW1C, BWR1C, HLDA2, IPL, TSSC3 | pleckstrin homology-like domain, family A, member 2 |
C00002514
|
| 90102 | PHLDB2, LL5b, LL5beta | pleckstrin homology-like domain, family B, member 2 |
C00002514
|
| 493911 | PHOSPHO2 | phosphatase, orphan 2 (EC:3.1.3.74) |
C00002514
|
| 10745 | PHTF1, PHTF | putative homeodomain transcription factor 1 |
C00002514
|
| 57157 | PHTF2 | putative homeodomain transcription factor 2 |
C00002514
|
| 85007 | PHYKPL, AGXT2L2, PHLU | 5-phosphohydroxy-L-lysine phospho-lyase (EC:4.2.3.134) |
C00002514
|
| 8301 | PICALM, CALM, CLTH, LAP | phosphatidylinositol binding clathrin assembly protein |
C00002514
|
| 80119 | PIF1, C15orf20, PIF | PIF1 5'-to-3' DNA helicase (EC:3.6.4.12) |
C00002514
|
| 55011 | PIH1D1, NOP17 | PIH1 domain containing 1 |
C00002514
|
| 5286 | PIK3C2A, CPK, PI3-K-C2(ALPHA), PI3-K-C2A | phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha (EC:2.7.1.154) |
C00002514
|
| 5289 | PIK3C3, VPS34, hVps34 | phosphatidylinositol 3-kinase, catalytic subunit type 3 (EC:2.7.1.137) |
C00002514
|
| 65018 | PINK1, BRPK, PARK6 | PTEN induced putative kinase 1 (EC:2.7.11.1) |
C00002514
|
| 8394 | PIP5K1A | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha (EC:2.7.1.68) |
C00002514
|
| 5570 | PKIB, PRKACN2 | protein kinase (cAMP-dependent, catalytic) inhibitor beta |
C00002514
|
| 396456 |
C00002514
|
||
| 9088 | PKMYT1, MYT1 | protein kinase, membrane associated tyrosine/threonine 1 (EC:2.7.11.1) |
C00002514
|
| 55344 | PLCXD1 | phosphatidylinositol-specific phospholipase C, X domain containing 1 |
C00002514
|
| 54477 | PLEKHA5, PEPP-2, PEPP2 | pleckstrin homology domain containing, family A member 5 |
C00002514
|
| 55041 | PLEKHB2, EVT2 | pleckstrin homology domain containing, family B (evectins) member 2 |
C00002514
|
| 423589 |
C00002514
|
||
| 79666 | PLEKHF2, EAPF, PHAFIN2, ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
C00002514
|
| 5347 | PLK1, PLK, STPK13 | polo-like kinase 1 (EC:2.7.11.21) |
C00002514
|
| 10733 | PLK4, SAK, STK18 | polo-like kinase 4 (EC:2.7.11.21) |
C00002514
|
| 51090 | PLLP, PMLP, TM4SF11 | plasmolipin |
C00002514
|
| 5352 | PLOD2, LH2, TLH | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (EC:1.14.11.4) |
C00002514
|
| 23654 | PLXNB2, MM1, Nbla00445, PLEXB2, dJ402G11.3 | plexin B2 |
C00002514
|
| 23129 | PLXND1, PLEXD1 | plexin D1 |
C00002514
|
| 5366 | PMAIP1, APR, NOXA | phorbol-12-myristate-13-acetate-induced protein 1 |
C00002514
|
| 56937 | PMEPA1, STAG1, TMEPAI | prostate transmembrane protein, androgen induced 1 |
C00002514
|
| 5376 | PMP22, CMT1A, CMT1E, DSS, GAS-3, HMSNIA, HNPP, Sp110 | peripheral myelin protein 22 |
C00002514
|
| 9512 | PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52 | peptidase (mitochondrial processing) beta (EC:3.4.24.64) |
C00002514
|
| 5378 | PMS1, HNPCC3, PMSL1, hPMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
C00002514
|
| 4860 | PNP, NP, PRO1837, PUNP | purine nucleoside phosphorylase (EC:2.4.2.1) |
C00002514
|
| 50640 | PNPLA8, IPLA2(GAMMA), IPLA2-2, IPLA2G, iPLA2gamma | patatin-like phospholipase domain containing 8 (EC:3.1.1.5) |
C00002514
|
| 10957 | PNRC1, B4-2, PNAS-145, PROL2, PRR2 | proline-rich nuclear receptor coactivator 1 |
C00002514
|
| 25886 | POC1A, PIX2, SOFT, WDR51A | POC1 centriolar protein A |
C00002514
|
| 5422 | POLA1, POLA, p180 | polymerase (DNA directed), alpha 1, catalytic subunit (EC:2.7.7.7) |
C00002514
|
| 23649 | POLA2 | polymerase (DNA directed), alpha 2, accessory subunit |
C00002514
|
| 5424 | POLD1, CDC2, CRCS10, MDPL, POLD | polymerase (DNA directed), delta 1, catalytic subunit (EC:2.7.7.7) |
C00002514
|
| 5425 | POLD2 | polymerase (DNA directed), delta 2, accessory subunit (EC:2.7.7.7) |
C00002514
|
| 10714 | POLD3, P66, P68 | polymerase (DNA-directed), delta 3, accessory subunit |
C00002514
|
| 26073 | POLDIP2, PDIP38, POLD4 | polymerase (DNA-directed), delta interacting protein 2 |
C00002514
|
| 5426 | POLE, CRCS12, FILS, POLE1 | polymerase (DNA directed), epsilon, catalytic subunit (EC:2.7.7.7) |
C00002514
|
| 5427 | POLE2, DPE2 | polymerase (DNA directed), epsilon 2, accessory subunit (EC:2.7.7.7) |
C00002514
|
| 54107 | POLE3, CHARAC17, CHRAC17, YBL1, p17 | polymerase (DNA directed), epsilon 3, accessory subunit (EC:2.7.7.7) |
C00002514
|
| 11201 | POLI, RAD30B, RAD3OB | polymerase (DNA directed) iota (EC:2.7.7.7) |
C00002514
|
| 10721 | POLQ, POLH, PRO0327 | polymerase (DNA directed), theta (EC:2.7.7.7) |
C00002514
|
| 84172 | POLR1B, RPA135, RPA2, Rpo1-2 | polymerase (RNA) I polypeptide B, 128kDa (EC:2.7.7.6) |
C00002514
|
| 5433 | POLR2D, HSRBP4, HSRPB4, RBP4, RPB16 | polymerase (RNA) II (DNA directed) polypeptide D (EC:2.7.7.6) |
C00002514
|
| 5434 | POLR2E, RPABC1, RPB5, XAP4, hRPB25, hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa (EC:2.7.7.6) |
C00002514
|
| 5436 | POLR2G, RPB19, RPB7, hRPB19, hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G (EC:2.7.7.6) |
C00002514
|
| 5437 | POLR2H, RPABC3, RPB17, RPB8 | polymerase (RNA) II (DNA directed) polypeptide H (EC:2.7.7.6) |
C00002514
|
| 8612 | PPAP2C, LPP2, PAP-2c, PAP2-g | phosphatidic acid phosphatase type 2C (EC:3.1.3.4) |
C00002514
|
| 403313 | PPAPDC2, PDP1, PSDP, bA6J24.6 | phosphatidic acid phosphatase type 2 domain containing 2 |
C00002514
|
| 5471 | PPAT, ATASE, GPAT, PRAT | phosphoribosyl pyrophosphate amidotransferase (EC:2.4.2.14) |
C00002514
|
| 60490 | PPCDC | phosphopantothenoylcysteine decarboxylase (EC:4.1.1.36) |
C00002514
|
| 5479 | PPIB, CYP-S1, CYPB, OI9, SCYLP | peptidylprolyl isomerase B (cyclophilin B) (EC:5.2.1.8) |
C00002514
|
| 10105 | PPIF, CYP3, CyP-M, Cyp-D, CypD | peptidylprolyl isomerase F (EC:5.2.1.8) |
C00002514
|
| 5493 | PPL | periplakin |
C00002514
|
| 5494 | PPM1A, PP2C-ALPHA, PP2CA, PP2Calpha | protein phosphatase, Mg2+/Mn2+ dependent, 1A (EC:3.1.3.16) |
C00002514
|
| 22843 | PPM1E, CaMKP-N, POPX1, PP2CH, caMKN | protein phosphatase, Mg2+/Mn2+ dependent, 1E (EC:3.1.3.16) |
C00002514
|
| 5514 | PPP1R10, CAT53, FB19, PNUTS, PP1R10, R111, p99 | protein phosphatase 1, regulatory subunit 10 |
C00002514
|
| 4659 | PPP1R12A, M130, MBS, MYPT1 | protein phosphatase 1, regulatory subunit 12A |
C00002514
|
| 10848 | PPP1R13L, IASPP, NKIP1, RAI, RAI4 | protein phosphatase 1, regulatory subunit 13 like |
C00002514
|
| 23645 | PPP1R15A, GADD34 | protein phosphatase 1, regulatory subunit 15A |
C00002514
|
| 5504 | PPP1R2, IPP-2, IPP2 | protein phosphatase 1, regulatory (inhibitor) subunit 2 (EC:3.1.3.16) |
C00002514
|
| 5520 | PPP2R2A, B55A, B55ALPHA, PR52A, PR55A | protein phosphatase 2, regulatory subunit B, alpha (EC:3.1.3.16) |
C00002514
|
| 5523 | PPP2R3A, PPP2R3, PR130, PR72 | protein phosphatase 2, regulatory subunit B'', alpha (EC:3.1.3.16) |
C00002514
|
| 5529 | PPP2R5E | protein phosphatase 2, regulatory subunit B', epsilon isoform |
C00002514
|
| 5533 | PPP3CC, CALNA3, CNA3, PP2Bgamma | protein phosphatase 3, catalytic subunit, gamma isozyme (EC:3.1.3.16) |
C00002514
|
| 5536 | PPP5C, PP5, PPP5, PPT | protein phosphatase 5, catalytic subunit (EC:3.1.3.16) |
C00002514
|
| 10549 | PRDX4, AOE37-2, PRX-4 | peroxiredoxin 4 (EC:1.11.1.15) |
C00002514
|
| 57580 | PREX1, P-REX1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
C00002514
|
| 5557 | PRIM1, p49 | primase, DNA, polypeptide 1 (49kDa) (EC:2.7.7.7) |
C00002514
|
| 5558 | PRIM2, PRIM2A, p58 | primase, DNA, polypeptide 2 (58kDa) (EC:2.7.7.-) |
C00002514
|
| 5567 | PRKACB, PKACB | protein kinase, cAMP-dependent, catalytic, beta (EC:2.7.11.11) |
C00002514
|
| 5583 | PRKCH, PKC-L, PKCL, PRKCL, nPKC-eta | protein kinase C, eta (EC:2.7.11.13) |
C00002514
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00002514
|
| 10419 | PRMT5, HRMT1L5, IBP72, JBP1, SKB1, SKB1Hs | protein arginine methyltransferase 5 (EC:2.1.1.125) |
C00002514
|
| 150696 | PROM2, PROML2 | prominin 2 |
C00002514
|
| 9128 | PRPF4, HPRP4, HPRP4P, PRP4, Prp4p, SNRNP60 | pre-mRNA processing factor 4 |
C00002514
|
| 5634 | PRPS2, PRSII | phosphoribosyl pyrophosphate synthetase 2 (EC:2.7.6.1) |
C00002514
|
| 78994 | PRR14 | proline rich 14 |
C00002514
|
| 222171 | PRR15 | proline rich 15 |
C00002514
|
| 7916 | PRRC2A, BAT2, D6S51, D6S51E, G2 | proline-rich coiled-coil 2A |
C00002514
|
| 23215 | PRRC2C, BAT2-iso, BAT2D1, BAT2L2, XTP2 | proline-rich coiled-coil 2C |
C00002514
|
| 285368 | PRRT3 | proline-rich transmembrane protein 3 |
C00002514
|
| 11098 | PRSS23, SIG13, SPUVE, ZSIG13 | protease, serine, 23 |
C00002514
|
| 5652 | PRSS8, CAP1, PROSTASIN | protease, serine, 8 |
C00002514
|
| 23362 | PSD3, EFA6R, HCA67 | pleckstrin and Sec7 domain containing 3 |
C00002514
|
| 11168 | PSIP1, DFS70, LEDGF, PAIP, PSIP2, p52, p75 | PC4 and SFRS1 interacting protein 1 |
C00002514
|
| 5702 | PSMC3, TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
C00002514
|
| 29893 | PSMC3IP, GT198, HOP2, HUMGT198A, ODG3, TBPIP | PSMC3 interacting protein |
C00002514
|
| 5721 | PSME2, PA28B, PA28beta, REGbeta | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
C00002514
|
| 84262 | PSMG3, C7orf48, PAC3 | proteasome (prosome, macropain) assembly chaperone 3 |
C00002514
|
| 84722 | PSRC1, DDA3, FP3214 | proline/serine-rich coiled-coil 1 |
C00002514
|
| 5725 | PTBP1, HNRNP-I, HNRNPI, HNRPI, PTB, PTB-1, PTB-T, PTB2, PTB3, PTB4, pPTB | polypyrimidine tract binding protein 1 |
C00002514
|
| 55037 | PTCD3, MRP-S39 | pentatricopeptide repeat domain 3 |
C00002514
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00002514
|
| 9536 | PTGES, MGST-IV, MGST1-L1, MGST1L1, MPGES, PGES, PIG12, PP102, PP1294, TP53I12, mPGES-1 | prostaglandin E synthase (EC:5.3.99.3) |
C00002514
|
| 7803 | PTP4A1, HH72, PRL-1, PRL1, PTP(CAAX1), PTPCAAX1 | protein tyrosine phosphatase type IVA, member 1 (EC:3.1.3.48) |
C00002514
|
| 5770 | PTPN1, PTP1B | protein tyrosine phosphatase, non-receptor type 1 (EC:3.1.3.48) |
C00002514
|
| 5782 | PTPN12, PTP-PEST, PTPG1 | protein tyrosine phosphatase, non-receptor type 12 (EC:3.1.3.48) |
C00002514
|
| 11099 | PTPN21, PTPD1, PTPRL10 | protein tyrosine phosphatase, non-receptor type 21 (EC:3.1.3.48) |
C00002514
|
| 5792 | PTPRF, LAR | protein tyrosine phosphatase, receptor type, F (EC:3.1.3.48) |
C00002514
|
| 5796 | PTPRK, R-PTP-kappa | protein tyrosine phosphatase, receptor type, K (EC:3.1.3.48) |
C00002514
|
| 9232 | PTTG1, EAP1, HPTTG, PTTG, TUTR1 | pituitary tumor-transforming 1 |
C00002514
|
| 83480 | PUS3, 2610020J05Rik | pseudouridylate synthase 3 (EC:5.4.99.45) |
C00002514
|
| 5819 | PVRL2, CD112, HVEB, PRR2, PVRR2 | poliovirus receptor-related 2 (herpesvirus entry mediator B) |
C00002514
|
| 79912 | PYROXD1 | pyridine nucleotide-disulphide oxidoreductase domain 1 (EC:1.8.1.-) |
C00002514
|
| 79832 | QSER1 | glutamine and serine rich 1 |
C00002514
|
| 22864 | R3HDM2, PR01365 | R3H domain containing 2 |
C00002514
|
| 80223 | RAB11FIP1, NOEL1A, RCP, rab11-FIP1 | RAB11 family interacting protein 1 (class I) |
C00002514
|
| 51552 | RAB14, FBP, RAB-14 | RAB14, member RAS oncogene family |
C00002514
|
| 35577 |
C00002514
|
||
| 53917 | RAB24 | RAB24, member RAS oncogene family |
C00002514
|
| 11031 | RAB31, Rab22B | RAB31, member RAS oncogene family |
C00002514
|
| 10981 | RAB32 | RAB32, member RAS oncogene family |
C00002514
|
| 83452 | RAB33B, SMC2 | RAB33B, member RAS oncogene family |
C00002514
|
| 10966 | RAB40B, RAR, SEC4L | RAB40B, member RAS oncogene family |
C00002514
|
| 5870 | RAB6A, RAB6 | RAB6A, member RAS oncogene family |
C00002514
|
| 8934 | RAB7L1, RAB7L | RAB7, member RAS oncogene family-like 1 |
C00002514
|
| 4218 | RAB8A, MEL, RAB8 | RAB8A, member RAS oncogene family |
C00002514
|
| 29127 | RACGAP1, CYK4, HsCYK-4, ID-GAP, MgcRacGAP | Rac GTPase activating protein 1 |
C00002514
|
| 5810 | RAD1, HRAD1, REC1 | RAD1 homolog (S. pombe) (EC:3.1.11.2) |
C00002514
|
| 56852 | RAD18, RNF73 | RAD18 homolog (S. cerevisiae) |
C00002514
|
| 5885 | RAD21, CDLS4, HR21, HRAD21, MCD1, NXP1, SCC1, hHR21 | RAD21 homolog (S. pombe) |
C00002514
|
| 5888 | RAD51, BRCC5, HRAD51, HsRad51, HsT16930, MRMV2, RAD51A, RECA | RAD51 recombinase |
C00002514
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00002514
|
| 5889 | RAD51C, BROVCA3, FANCO, R51H3, RAD51L2 | RAD51 paralog C |
C00002514
|
| 25788 | RAD54B, RDH54 | RAD54 homolog B (S. cerevisiae) |
C00002514
|
| 8438 | RAD54L, HR54, RAD54A, hHR54, hRAD54 | RAD54-like (S. cerevisiae) |
C00002514
|
| 8480 | RAE1, MIG14, MRNP41, Mnrp41, dJ481F12.3, dJ800J21.1 | ribonucleic acid export 1 |
C00002514
|
| 10743 | RAI1, SMCR, SMS | retinoic acid induced 1 |
C00002514
|
| 253959 | RALGAPA1, GARNL1, GRIPE, RalGAPalpha1, TULIP1, p240 | Ral GTPase activating protein, alpha subunit 1 (catalytic) |
C00002514
|
| 5902 | RANBP1, HTF9A | RAN binding protein 1 |
C00002514
|
| 5905 | RANGAP1, Fug1, RANGAP, SD | Ran GTPase activating protein 1 |
C00002514
|
| 9693 | RAPGEF2, CNrasGEF, NRAPGEP, PDZ-GEF1, PDZGEF1, RA-GEF, RA-GEF-1, Rap-GEP, nRap_GEP | Rap guanine nucleotide exchange factor (GEF) 2 |
C00002514
|
| 51195 | RAPGEFL1, Link-GEFII | Rap guanine nucleotide exchange factor (GEF)-like 1 |
C00002514
|
| 5921 | RASA1, CM-AVM, CMAVM, GAP, PKWS, RASA, RASGAP, p120GAP, p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 |
C00002514
|
| 51655 | RASD1, AGS1, DEXRAS1, MGC:26290 | RAS, dexamethasone-induced 1 |
C00002514
|
| 158158 | RASEF, RAB45 | RAS and EF-hand domain containing |
C00002514
|
| 10125 | RASGRP1, CALDAG-GEFI, CALDAG-GEFII, RASGRP, V, hRasGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
C00002514
|
| 65997 | RASL11B | RAS-like, family 11, member B |
C00002514
|
| 283349 | RASSF3, RASSF5 | Ras association (RalGDS/AF-6) domain family member 3 |
C00002514
|
| 125950 | RAVER1 | ribonucleoprotein, PTB-binding 1 |
C00002514
|
| 9821 | RB1CC1, CC1, FIP200 | RB1-inducible coiled-coil 1 |
C00002514
|
| 5928 | RBBP4, NURF55, RBAP48 | retinoblastoma binding protein 4 |
C00002514
|
| 5932 | RBBP8, COM1, CTIP, JWDS, RIM, SAE2, SCKL2 | retinoblastoma binding protein 8 |
C00002514
|
| 10741 | RBBP9, BOG, RBBP10 | retinoblastoma binding protein 9 |
C00002514
|
| 23543 | RBFOX2, FOX2, Fox-2, HNRBP2, HRNBP2, RBM9, RTA, dJ106I20.3, fxh | RNA binding protein, fox-1 homolog (C. elegans) 2 |
C00002514
|
| 5934 | RBL2, P130, Rb2 | retinoblastoma-like 2 (p130) |
C00002514
|
| 8241 | RBM10, DXS8237E, GPATC9, GPATCH9, S1-1, TARPS, ZRANB5 | RNA binding motif protein 10 |
C00002514
|
| 64783 | RBM15, OTT, OTT1, SPEN | RNA binding motif protein 15 |
C00002514
|
| 221662 | RBM24, RNPC6, dJ259A10.1 | RNA binding motif protein 24 |
C00002514
|
| 55285 | RBM41 | RNA binding motif protein 41 |
C00002514
|
| 5937 | RBMS1, C2orf12, HCC-4, MSSP, MSSP-1, MSSP-2, MSSP-3, SCR2, YC1 | RNA binding motif, single stranded interacting protein 1 |
C00002514
|
| 27316 | RBMX, HNRNPG, HNRPG, RBMXP1, RBMXRT, RNMX, hnRNP-G | RNA binding motif protein, X-linked |
C00002514
|
| 55213 | RCBTB1, CLLD7, CLLL7, GLP, RP11-185C18.1 | regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
C00002514
|
| 91433 | RCCD1 | RCC1 domain containing 1 |
C00002514
|
| 55758 | RCOR3, RP11-318L16.1 | REST corepressor 3 |
C00002514
|
| 112724 | RDH13, SDR7C3 | retinol dehydrogenase 13 (all-trans/9-cis) (EC:1.1.1.-) |
C00002514
|
| 201299 | RDM1, RAD52B | RAD52 motif 1 |
C00002514
|
| 9401 | RECQL4, RECQ4 | RecQ protein-like 4 (EC:3.6.4.12) |
C00002514
|
| 221035 | REEP3, C10orf74 | receptor accessory protein 3 |
C00002514
|
| 80346 | REEP4, C8orf20 | receptor accessory protein 4 |
C00002514
|
| 7905 | REEP5, C5orf18, D5S346, DP1, TB2, YOP1 | receptor accessory protein 5 |
C00002514
|
| 85004 | RERG | RAS-like, estrogen-regulated, growth inhibitor |
C00002514
|
| 5979 | RET, CDHF12, CDHR16, HSCR1, MEN2A, MEN2B, MTC1, PTC, RET-ELE1, RET51 | ret proto-oncogene (EC:2.7.10.1) |
C00002514
|
| 5982 | RFC2, RFC40 | replication factor C (activator 1) 2, 40kDa |
C00002514
|
| 5983 | RFC3, RFC38 | replication factor C (activator 1) 3, 38kDa |
C00002514
|
| 5984 | RFC4, A1, RFC37 | replication factor C (activator 1) 4, 37kDa |
C00002514
|
| 5985 | RFC5, RFC36 | replication factor C (activator 1) 5, 36.5kDa |
C00002514
|
| 23180 | RFTN1, PIB10, PIG9, RAFTLIN | raftlin, lipid raft linker 1 |
C00002514
|
| 5997 | RGS2, G0S8 | regulator of G-protein signaling 2, 24kDa |
C00002514
|
| 64285 | RHBDF1, C16orf8, Dist1, EGFR-RS, gene-89, gene-90, hDist1 | rhomboid 5 homolog 1 (Drosophila) |
C00002514
|
| 83695 | RHNO1, C12orf32, RHINO | RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
C00002514
|
| 9886 | RHOBTB1 | Rho-related BTB domain containing 1 |
C00002514
|
| 23433 | RHOQ, ARHQ, RASL7A, TC10, TC10A | ras homolog family member Q |
C00002514
|
| 55288 | RHOT1, ARHT1, MIRO-1, MIRO1 | ras homolog family member T1 |
C00002514
|
| 26150 | RIBC2, C22orf11 | RIB43A domain with coiled-coils 2 |
C00002514
|
| 253260 | RICTOR, AVO3, PIA, hAVO3 | RPTOR independent companion of MTOR, complex 2 |
C00002514
|
| 151393 | RMDN2, FAM82A, FAM82A1, PRO34163, PYST9371, RMD-2, RMD2, RMD4 | regulator of microtubule dynamics 2 |
C00002514
|
| 80010 | RMI1, BLAP75, C9orf76, FAAP75, RP11-346I8.1 | RecQ mediated genome instability 1 |
C00002514
|
| 116028 | RMI2, BLAP18, C16orf75 | RecQ mediated genome instability 2 |
C00002514
|
| 10535 | RNASEH2A, AGS4, JUNB, RNASEHI, RNHIA, RNHL | ribonuclease H2, subunit A (EC:3.1.26.4) |
C00002514
|
| 390 | RND3, ARHE, Rho8, RhoE, memB | Rho family GTPase 3 |
C00002514
|
| 255488 | RNF144B, IBRDC2, PIR2, bA528A10.3, p53RFP | ring finger protein 144B |
C00002514
|
| 149603 | RNF187, RACO-1, RACO1 | ring finger protein 187 |
C00002514
|
| 551328 |
C00002514
|
||
| 79102 | RNF26 | ring finger protein 26 |
C00002514
|
| 9475 | ROCK2, ROCK-II | Rho-associated, coiled-coil containing protein kinase 2 (EC:2.7.11.1) |
C00002514
|
| 6117 | RPA1, HSSB, MST075, REPA1, RF-A, RP-A, RPA70 | replication protein A1, 70kDa |
C00002514
|
| 6119 | RPA3, REPA3 | replication protein A3, 14kDa |
C00002514
|
| 6160 | RPL31, L31 | ribosomal protein L31 |
C00002514
|
| 6168 | RPL37A, L37A | ribosomal protein L37a |
C00002514
|
| 116832 | RPL39L, RPL39L1 | ribosomal protein L39-like |
C00002514
|
| 6184 | RPN1, OST1, RBPH1 | ribophorin I (EC:2.4.99.18) |
C00002514
|
| 56475 | RPRM, REPRIMO | reprimo, TP53 dependent G2 arrest mediator candidate |
C00002514
|
| 64121 | RRAGC, GTR2, RAGC, TIB929 | Ras-related GTP binding C |
C00002514
|
| 6240 | RRM1, R1, RIR1, RR1 | ribonucleotide reductase M1 (EC:1.17.4.1) |
C00002514
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00002514
|
| 51018 | RRP15, KIAA0507 | ribosomal RNA processing 15 homolog (S. cerevisiae) |
C00002514
|
| 51773 | RSF1, HBXAP, RSF-1, XAP8, p325 | remodeling and spacing factor 1 |
C00002514
|
| 51319 | RSRC1, SFRS21, SRrp53 | arginine/serine-rich coiled-coil 1 |
C00002514
|
| 6251 | RSU1, RSP-1 | Ras suppressor protein 1 |
C00002514
|
| 6253 | RTN2, NSP2, NSPL1, NSPLI, SPG12 | reticulon 2 |
C00002514
|
| 10900 | RUNDC3A, RAP2IP, RPIP-8, RPIP8 | RUN domain containing 3A |
C00002514
|
| 154661 | RUNDC3B, RPIB9, RPIP9 | RUN domain containing 3B |
C00002514
|
| 10856 | RUVBL2, ECP51, INO80J, REPTIN, RVB2, TIH2, TIP48, TIP49B | RuvB-like AAA ATPase 2 (EC:3.6.4.12) |
C00002514
|
| 6259 | RYK, D3S3195, JTK5, JTK5A, RYK1 | receptor-like tyrosine kinase (EC:2.7.10.1) |
C00002514
|
| 6281 | S100A10, 42C, ANX2L, ANX2LG, CAL1L, CLP11, Ca[1], GP11, P11, p10 | S100 calcium binding protein A10 |
C00002514
|
| 6277 | S100A6, 2A9, 5B10, CABP, CACY, PRA | S100 calcium binding protein A6 |
C00002514
|
| 1903 | S1PR3, EDG-3, EDG3, LPB3, S1P3 | sphingosine-1-phosphate receptor 3 |
C00002514
|
| 113174 | SAAL1, SPACIA1 | serum amyloid A-like 1 |
C00002514
|
| 29901 | SAC3D1, HSU79266, SHD1 | SAC3 domain containing 1 |
C00002514
|
| 26278 | SACS, ARSACS, DNAJC29 | spastic ataxia of Charlevoix-Saguenay (sacsin) |
C00002514
|
| 10055 | SAE1, AOS1, HSPC140, SUA1, UBLE1A | SUMO1 activating enzyme subunit 1 |
C00002514
|
| 6294 | SAFB, HAP, HET, SAF-B1, SAFB1 | scaffold attachment factor B |
C00002514
|
| 57167 | SALL4, DRRS, HSAL4, ZNF797, dJ1112F19.1 | spalt-like transcription factor 4 |
C00002514
|
| 23034 | SAMD4A, SAMD4, SMAUG, SMAUG1, SMG, SMGA | sterile alpha motif domain containing 4A |
C00002514
|
| 23328 | SASH1, SH3D6A, dJ323M4, dJ323M4.1 | SAM and SH3 domain containing 1 |
C00002514
|
| 6303 | SAT1, DC21, KFSD, KFSDX, SAT, SSAT, SSAT-1 | spermidine/spermine N1-acetyltransferase 1 (EC:2.3.1.57) |
C00002514
|
| 23314 | SATB2 | SATB homeobox 2 |
C00002514
|
| 949 | SCARB1, CD36L1, CLA-1, CLA1, HDLQTL6, SR-BI, SRB1 | scavenger receptor class B, member 1 |
C00002514
|
| 950 | SCARB2, AMRF, CD36L2, EPM4, HLGP85, LGP85, LIMP-2, LIMPII, SR-BII | scavenger receptor class B, member 2 |
C00002514
|
| 51097 | SCCPDH, NET11, RP11-439E19.2 | saccharopine dehydrogenase (putative) (EC:1.5.1.9) |
C00002514
|
| 6319 | SCD, FADS5, MSTP008, SCD1, SCDOS | stearoyl-CoA desaturase (delta-9-desaturase) (EC:1.14.19.1) |
C00002514
|
| 6383 | SDC2, CD362, HSPG, HSPG1, SYND2 | syndecan 2 |
C00002514
|
| 10483 | SEC23B, CDA-II, CDAII, CDAN2, HEMPAS | Sec23 homolog B (S. cerevisiae) |
C00002514
|
| 9871 | SEC24D | SEC24 family member D |
C00002514
|
| 29927 | SEC61A1, HSEC61, SEC61, SEC61A | Sec61 alpha 1 subunit (S. cerevisiae) |
C00002514
|
| 8991 | SELENBP1, LPSB, SBP56, SP56, hSBP | selenium binding protein 1 |
C00002514
|
| 362713 |
C00002514
|
||
| 51714 | SELT | selenoprotein T |
C00002514
|
| 10512 | SEMA3C, SEMAE, SemE | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
C00002514
|
| 29843 | SENP1, SuPr-2 | SUMO1/sentrin specific peptidase 1 (EC:3.4.22.68) |
C00002514
|
| 22929 | SEPHS1, SELD, SPS, SPS1 | selenophosphate synthetase 1 (EC:2.7.9.3) |
C00002514
|
| 6414 | SEPP1, SELP, SeP | selenoprotein P, plasma, 1 |
C00002514
|
| 23176 | SEPT8, SEP2 | septin 8 |
C00002514
|
| 10801 | SEPT9, AF17q25, MSF, MSF1, NAPB, PNUTL4, SINT1, SeptD1 | septin 9 |
C00002514
|
| 94009 | SERHL, BK126B4.1, BK126B4.2, HS126B42, dJ222E13.1 | serine hydrolase-like |
C00002514
|
| 57515 | SERINC1, TDE1L, TDE2, TMS-2, TMS2 | serine incorporator 1 |
C00002514
|
| 256987 | SERINC5, C5orf12, TPO1 | serine incorporator 5 |
C00002514
|
| 1992 | SERPINB1, EI, ELANH2, LEI, M/NEI, MNEI, PI-2, PI2 | serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
C00002514
|
| 83667 | SESN2, HI95, SES2, SEST2 | sestrin 2 |
C00002514
|
| 7536 | SF1, BBP, D11S636, MBBP, ZCCHC25, ZFM1, ZNF162 | splicing factor 1 |
C00002514
|
| 23450 | SF3B3, RSE1, SAP130, SF3b130, STAF130 | splicing factor 3b, subunit 3, 130kDa |
C00002514
|
| 2810 | SFN, YWHAS | stratifin |
C00002514
|
| 119392 | SFR1, C10orf78, MEI5, MEIR5, bA373N18.1 | SWI5-dependent recombination repair 1 |
C00002514
|
| 100380358 |
C00002514
|
||
| 100049528 |
C00002514
|
||
| 100305297 |
C00002514
|
||
| 94081 | SFXN1 | sideroflexin 1 |
C00002514
|
| 118980 | SFXN2 | sideroflexin 2 |
C00002514
|
| 6445 | SGCG, A4, DAGA4, DMDA, DMDA1, LGMD2C, MAM, SCARMD2, SCG3, TYPE | sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
C00002514
|
| 23678 | SGK3, CISK, SGK2, SGKL | serum/glucocorticoid regulated kinase family, member 3 (EC:2.7.11.1) |
C00002514
|
| 151648 | SGOL1, NY-BR-85, SGO, Sgo1 | shugoshin-like 1 (S. pombe) |
C00002514
|
| 151246 | SGOL2, SGO2, TRIPIN | shugoshin-like 2 (S. pombe) |
C00002514
|
| 6451 | SH3BGRL, SH3BGR | SH3 domain binding glutamic acid-rich protein like |
C00002514
|
| 23677 | SH3BP4, BOG25, TTP | SH3-domain binding protein 4 |
C00002514
|
| 152503 | SH3D19, EBP, EVE1, Kryn, SH3P19 | SH3 domain containing 19 |
C00002514
|
| 30011 | SH3KBP1, CD2BP3, CIN85, GIG10, HSB-1, HSB1, MIG18 | SH3-domain kinase binding protein 1 |
C00002514
|
| 57630 | SH3RF1, POSH, RNF142, SH3MD2 | SH3 domain containing ring finger 1 |
C00002514
|
| 26751 | SH3YL1, RAY | SH3 and SYLF domain containing 1 |
C00002514
|
| 79801 | SHCBP1, PAL | SHC SH2-domain binding protein 1 |
C00002514
|
| 387914 | SHISA2, C13orf13, PRO28631, TMEM46, WGAR9166, bA398O19.2, hShisa | shisa family member 2 |
C00002514
|
| 6470 | SHMT1, CSHMT, SHMT | serine hydroxymethyltransferase 1 (soluble) (EC:2.1.2.1) |
C00002514
|
| 6472 | SHMT2, GLYA, SHMT | serine hydroxymethyltransferase 2 (mitochondrial) (EC:2.1.2.1) |
C00002514
|
| 57619 | SHROOM3, APXL3, SHRM, ShrmL | shroom family member 3 |
C00002514
|
| 23411 | SIRT1, SIR2L1 | sirtuin 1 |
C00002514
|
| 220134 | SKA1, C18orf24 | spindle and kinetochore associated complex subunit 1 |
C00002514
|
| 348235 | SKA2, FAM33A | spindle and kinetochore associated complex subunit 2 |
C00002514
|
| 221150 | SKA3, C13orf3, RAMA1 | spindle and kinetochore associated complex subunit 3 |
C00002514
|
| 8935 | SKAP2, PRAP, RA70, SAPS, SCAP2, SKAP-HOM, SKAP55R | src kinase associated phosphoprotein 2 |
C00002514
|
| 6498 | SKIL, SNO, SnoA, SnoI, SnoN | SKI-like oncogene |
C00002514
|
| 6502 | SKP2, FBL1, FBXL1, FLB1, p45 | S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
C00002514
|
| 7884 | SLBP, HBP | stem-loop binding protein |
C00002514
|
| 6558 | SLC12A2, BSC, BSC2, NKCC1 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
C00002514
|
| 6560 | SLC12A4, KCC1 | solute carrier family 12 (potassium/chloride transporter), member 4 |
C00002514
|
| 84561 | SLC12A8, CCC9 | solute carrier family 12, member 8 |
C00002514
|
| 26503 | SLC17A5, AST, ISSD, NSD, SD, SIALIN, SIASD, SLD | solute carrier family 17 (acidic sugar transporter), member 5 |
C00002514
|
| 6509 | SLC1A4, ASCT1, SATT | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
C00002514
|
| 63027 | SLC22A23, C6orf85 | solute carrier family 22, member 23 |
C00002514
|
| 8402 | SLC25A11, OGC, SLC20A4 | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
C00002514
|
| 10166 | SLC25A15, D13S327, HHH, ORC1, ORNT1 | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
C00002514
|
| 123096 | SLC25A29, C14orf69, CACL, ORNT3 | solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
C00002514
|
| 51312 | SLC25A37, MFRN, MFRN1, MSC, MSCP, PRO1278, PRO1584, PRO2217 | solute carrier family 25 (mitochondrial iron transporter), member 37 |
C00002514
|
| 51629 | SLC25A39, CGI69 | solute carrier family 25, member 39 |
C00002514
|
| 284129 | SLC26A11 | solute carrier family 26 (anion exchanger), member 11 |
C00002514
|
| 1836 | SLC26A2, D5S1708, DTD, DTDST, EDM4, MST153, MSTP157 | solute carrier family 26 (anion exchanger), member 2 |
C00002514
|
| 11001 | SLC27A2, ACSVL1, FACVL1, FATP2, HsT17226, VLACS, VLCS, hFACVL1 | solute carrier family 27 (fatty acid transporter), member 2 (EC:6.2.1.3) |
C00002514
|
| 11000 | SLC27A3, ACSVL3, FATP3, VLCS-3 | solute carrier family 27 (fatty acid transporter), member 3 (EC:6.2.1.-) |
C00002514
|
| 2030 | SLC29A1, ENT1 | solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
C00002514
|
| 56731 | SLC2A4RG, GEF, HDBP-1, HDBP1, Si-1-2, Si-1-2-19 | SLC2A4 regulator |
C00002514
|
| 7779 | SLC30A1, ZNT1, ZRC1 | solute carrier family 30 (zinc transporter), member 1 |
C00002514
|
| 1317 | SLC31A1, COPT1, CTR1 | solute carrier family 31 (copper transporter), member 1 |
C00002514
|
| 23443 | SLC35A3 | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
C00002514
|
| 11046 | SLC35D2, HFRC1, SQV7L, UGTrel8, hfrc | solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
C00002514
|
| 79939 | SLC35E1 | solute carrier family 35, member E1 |
C00002514
|
| 2542 | SLC37A4, G6PT1, G6PT2, G6PT3, GSD1b, GSD1c, GSD1d, TRG-19, TRG19 | solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
C00002514
|
| 55630 | SLC39A4, AEZ, AWMS2, ZIP4 | solute carrier family 39 (zinc transporter), member 4 |
C00002514
|
| 64116 | SLC39A8, BIGM103, LZT-Hs6, ZIP8 | solute carrier family 39 (zinc transporter), member 8 |
C00002514
|
| 6526 | SLC5A3, SMIT, SMIT1, SMIT2 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
C00002514
|
| 11254 | SLC6A14, BMIQ11 | solute carrier family 6 (amino acid transporter), member 14 |
C00002514
|
| 6541 | SLC7A1, ATRC1, CAT-1, ERR, HCAT1, REC1L | solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
C00002514
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00002514
|
| 8140 | SLC7A5, 4F2LC, CD98, D16S469E, E16, LAT1, MPE16, hLAT1 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
C00002514
|
| 9368 | SLC9A3R1, EBP50, NHERF, NHERF-1, NHERF1, NPHLOP2 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
C00002514
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002514
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00002514
|
| 8243 | SMC1A, CDLS2, DXS423E, SB1.8, SMC1, SMC1L1, SMC1alpha, SMCB | structural maintenance of chromosomes 1A |
C00002514
|
| 10592 | SMC2, CAP-E, CAPE, SMC-2, SMC2L1 | structural maintenance of chromosomes 2 |
C00002514
|
| 10051 | SMC4, CAP-C, CAPC, SMC-4, SMC4L1, hCAP-C | structural maintenance of chromosomes 4 |
C00002514
|
| 23137 | SMC5, SMC5L1 | structural maintenance of chromosomes 5 |
C00002514
|
| 201895 | SMIM14, C4orf34 | small integral membrane protein 14 |
C00002514
|
| 56950 | SMYD2, HSKM-B, KMT3C, ZMYND14 | SET and MYND domain containing 2 (EC:2.1.1.43) |
C00002514
|
| 8773 | SNAP23, HsT17016, SNAP-23, SNAP23A, SNAP23B | synaptosomal-associated protein, 23kDa |
C00002514
|
| 6619 | SNAPC3, PTFbeta, SNAP50 | small nuclear RNA activating complex, polypeptide 3, 50kDa |
C00002514
|
| 84973 | SNHG7, NCRNA00061 | small nucleolar RNA host gene 7 (non-protein coding) |
C00002514
|
| 8303 | SNN | stannin |
C00002514
|
| 677809 | SNORA24, ACA24 | small nucleolar RNA, H/ACA box 24 |
C00002514
|
| 26797 | SNORD52, RNU52, U52 | small nucleolar RNA, C/D box 52 |
C00002514
|
| 79622 | SNRNP25, C16orf33 | small nuclear ribonucleoprotein 25kDa (U11/U12) |
C00002514
|
| 6626 | SNRPA, Mud1, U1-A, U1A | small nuclear ribonucleoprotein polypeptide A |
C00002514
|
| 6627 | SNRPA1, Lea1 | small nuclear ribonucleoprotein polypeptide A' |
C00002514
|
| 6628 | SNRPB, COD, SNRPB1, Sm-B/B', SmB/B', SmB/SmB', snRNP-B | small nuclear ribonucleoprotein polypeptides B and B1 |
C00002514
|
| 6632 | SNRPD1, HsT2456, SMD1, SNRPD, Sm-D1 | small nuclear ribonucleoprotein D1 polypeptide 16kDa |
C00002514
|
| 29887 | SNX10, OPTB8 | sorting nexin 10 |
C00002514
|
| 23161 | SNX13, RGS-PX1 | sorting nexin 13 |
C00002514
|
| 28966 | SNX24, PRO1284 | sorting nexin 24 |
C00002514
|
| 83891 | SNX25, SBBI31 | sorting nexin 25 |
C00002514
|
| 92017 | SNX29, A-388D4.1, RUNDC2A | sorting nexin 29 |
C00002514
|
| 58533 | SNX6, MSTP010, TFAF2 | sorting nexin 6 |
C00002514
|
| 51429 | SNX9, SDP1, SH3PX1, SH3PXD3A, WISP | sorting nexin 9 |
C00002514
|
| 8835 | SOCS2, CIS2, Cish2, SOCS-2, SSI-2, SSI2, STATI2 | suppressor of cytokine signaling 2 |
C00002514
|
| 9655 | SOCS5, CIS6, CISH6, Cish5, SOCS-5 | suppressor of cytokine signaling 5 |
C00002514
|
| 6658 | SOX3, GHDX, MRGH, PHP, PHPX, SOXB | SRY (sex determining region Y)-box 3 |
C00002514
|
| 6659 | SOX4, EVI16 | SRY (sex determining region Y)-box 4 |
C00002514
|
| 6662 | SOX9, CMD1, CMPD1, SRA1 | SRY (sex determining region Y)-box 9 |
C00002514
|
| 10615 | SPAG5, DEEPEST, MAP126, hMAP126 | sperm associated antigen 5 |
C00002514
|
| 64847 | SPATA20, SSP411, Tisp78 | spermatogenesis associated 20 |
C00002514
|
| 124044 | SPATA2L, C16orf76, tamo | spermatogenesis associated 2-like |
C00002514
|
| 55812 | SPATA7, HSD-3.1, HSD3, LCA3 | spermatogenesis associated 7 |
C00002514
|
| 147841 | SPC24, SPBC24 | SPC24, NDC80 kinetochore complex component |
C00002514
|
| 57405 | SPC25, SPBC25, hSpc25 | SPC25, NDC80 kinetochore complex component |
C00002514
|
| 25803 | SPDEF, PDEF, RP11-375E1__A.3, bA375E1.3 | SAM pointed domain containing ETS transcription factor |
C00002514
|
| 54908 | SPDL1, CCDC99 | spindle apparatus coiled-coil protein 1 |
C00002514
|
| 23111 | SPG20, SPARTIN, TAHCCP1 | spastic paraplegia 20 (Troyer syndrome) |
C00002514
|
| 139886 | SPIN4 | spindlin family, member 4 |
C00002514
|
| 84501 | SPIRE2, Spir-2 | spire-type actin nucleation factor 2 |
C00002514
|
| 200734 | SPRED2, Spred-2 | sprouty-related, EVH1 domain containing 2 |
C00002514
|
| 10558 | SPTLC1, HSAN1, HSN1, LBC1, LCB1, SPT1, SPTI | serine palmitoyltransferase, long chain base subunit 1 (EC:2.3.1.50) |
C00002514
|
| 58472 | SQRDL, PRO1975 | sulfide quinone reductase-like (yeast) |
C00002514
|
| 8878 | SQSTM1, A170, OSIL, PDB3, ZIP3, p60, p62, p62B | sequestosome 1 |
C00002514
|
| 79644 | SRD5A3, CDG1P, CDG1Q, KRIZI, SRD5A2L, SRD5A2L1 | steroid 5 alpha-reductase 3 (EC:1.3.1.22 1.3.1.94) |
C00002514
|
| 6717 | SRI, CP-22, CP22, SCN, V19 | sorcin |
C00002514
|
| 6723 | SRM, PAPT, SPDSY, SPS1, SRML1 | spermidine synthase (EC:2.5.1.16) |
C00002514
|
| 58477 | SRPRB, APMCF1 | signal recognition particle receptor, B subunit |
C00002514
|
| 63826 | SRR, ILV1, ISO1 | serine racemase (EC:4.3.1.17 5.1.1.18 4.3.1.18) |
C00002514
|
| 23524 | SRRM2, 300-KD, CWF21, Cwc21, SRL300, SRm300 | serine/arginine repetitive matrix 2 |
C00002514
|
| 10772 | SRSF10, FUSIP1, FUSIP2, NSSR, SFRS13, SFRS13A, SRp38, SRrp40, TASR, TASR1, TASR2 | serine/arginine-rich splicing factor 10 |
C00002514
|
| 23635 | SSBP2, HSPC116, SOSS-B2 | single-stranded DNA binding protein 2 |
C00002514
|
| 6749 | SSRP1, FACT, FACT80, T160 | structure specific recognition protein 1 |
C00002514
|
| 117178 | SSX2IP, ADIP | synovial sarcoma, X breakpoint 2 interacting protein |
C00002514
|
| 6482 | ST3GAL1, Gal-NAc6S, SIAT4A, SIATFL, ST3GalA, ST3GalA.1, ST3GalIA, ST3GalIA,1, ST3O | ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (EC:2.4.99.4) |
C00002514
|
| 7903 | ST8SIA4, PST, PST1, SIAT8D, ST8SIA-IV | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (EC:2.4.99.-) |
C00002514
|
| 338596 | ST8SIA6, SIA8F, SIAT8F, ST8SIA-VI | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 (EC:2.4.99.-) |
C00002514
|
| 10274 | STAG1, SA1, SCC3A | stromal antigen 1 |
C00002514
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00002514
|
| 6781 | STC1, STC | stanniocalcin 1 |
C00002514
|
| 8614 | STC2, STC-2, STCRP | stanniocalcin 2 |
C00002514
|
| 6491 | STIL, MCPH7, SIL | SCL/TAL1 interrupting locus |
C00002514
|
| 10963 | STIP1, HOP, IEF-SSP-3521, P60, STI1, STI1L | stress-induced-phosphoprotein 1 |
C00002514
|
| 23012 | STK38L, NDR2 | serine/threonine kinase 38 like (EC:2.7.11.1) |
C00002514
|
| 6789 | STK4, KRS2, MST1, TIIAC, YSK3 | serine/threonine kinase 4 (EC:2.7.11.1) |
C00002514
|
| 412271 |
C00002514
|
||
| 3925 | STMN1, C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | stathmin 1 |
C00002514
|
| 85439 | STON2, STN2, STNB, STNB2 | stonin 2 |
C00002514
|
| 92335 | STRADA, LYK5, NY-BR-96, PMSE, STRAD, Stlk | STE20-related kinase adaptor alpha |
C00002514
|
| 29966 | STRN3, SG2NA | striatin, calmodulin binding protein 3 |
C00002514
|
| 55014 | STX17 | syntaxin 17 |
C00002514
|
| 6809 | STX3, STX3A | syntaxin 3 |
C00002514
|
| 8417 | STX7 | syntaxin 7 |
C00002514
|
| 23213 | SULF1, HSULF-1, SULF-1 | sulfatase 1 |
C00002514
|
| 55959 | SULF2, HSULF-2 | sulfatase 2 |
C00002514
|
| 6820 | SULT2B1, HSST2 | sulfotransferase family, cytosolic, 2B, member 1 (EC:2.8.2.2) |
C00002514
|
| 11198 | SUPT16H, CDC68, FACTP140, SPT16/CDC68 | suppressor of Ty 16 homolog (S. cerevisiae) |
C00002514
|
| 6839 | SUV39H1, H3-K9-HMTase_1, KMT1A, MG44, SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00002514
|
| 79723 | SUV39H2, KMT1B | suppressor of variegation 3-9 homolog 2 (Drosophila) (EC:2.1.1.43) |
C00002514
|
| 51111 | SUV420H1, CGI85, KMT5B | suppressor of variegation 4-20 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00002514
|
| 23075 | SWAP70, SWAP-70 | SWAP switching B-cell complex 70kDa subunit |
C00002514
|
| 55638 | SYBU, GOLSYN, OCSYN, SNPHL | syntabulin (syntaxin-interacting) |
C00002514
|
| 256126 | SYCE2, CESC1 | synaptonemal complex central element protein 2 |
C00002514
|
| 84144 | SYDE2, RP11-33E12.1 | synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
C00002514
|
| 10492 | SYNCRIP, GRY-RBP, GRYRBP, HNRNPQ, HNRPQ1, NSAP1, PP68, hnRNP-Q | synaptotagmin binding, cytoplasmic RNA interacting protein |
C00002514
|
| 23224 | SYNE2, EDMD5, NUA, NUANCE, Nesp2, Nesprin-2, SYNE-2, TROPH | spectrin repeat containing, nuclear envelope 2 |
C00002514
|
| 9143 | SYNGR3 | synaptogyrin 3 |
C00002514
|
| 55333 | SYNJ2BP, ARIP2, OMP25 | synaptojanin 2 binding protein |
C00002514
|
| 51760 | SYT17 | synaptotagmin XVII |
C00002514
|
| 54843 | SYTL2, CHR11SYT, EXO4, SGA72M, SLP2, SLP2A | synaptotagmin-like 2 |
C00002514
|
| 94121 | SYTL4, SLP4 | synaptotagmin-like 4 |
C00002514
|
| 94122 | SYTL5, slp5 | synaptotagmin-like 5 |
C00002514
|
| 23118 | TAB2, CHTD2, MAP3K7IP2 | TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
C00002514
|
| 10460 | TACC3, ERIC-1, ERIC1 | transforming, acidic coiled-coil containing protein 3 |
C00002514
|
| 4070 | TACSTD2, EGP-1, EGP1, GA733-1, GA7331, GP50, M1S1, TROP2 | tumor-associated calcium signal transducer 2 |
C00002514
|
| 6883 | TAF12, TAF2J, TAFII20 | TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
C00002514
|
| 6877 | TAF5, TAF2D, TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
C00002514
|
| 26115 | TANC2, ROLSA, rols | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
C00002514
|
| 57551 | TAOK1, KFC-B, MAP3K16, MARKK, PSK-2, PSK2, TAO1, hKFC-B, hTAOK1 | TAO kinase 1 (EC:2.7.11.1) |
C00002514
|
| 51347 | TAOK3, DPK, JIK, MAP3K18 | TAO kinase 3 (EC:2.7.11.1) |
C00002514
|
| 80222 | TARS2, TARSL1, thrRS | threonyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.3) |
C00002514
|
| 9797 | TATDN2 | TatD DNase domain containing 2 |
C00002514
|
| 57465 | TBC1D24, EIEE16, FIME, TLDC6 | TBC1 domain family, member 24 |
C00002514
|
| 6904 | TBCD, SSD-1, tfcD | tubulin folding cofactor D |
C00002514
|
| 6907 | TBL1X, EBI, SMAP55, TBL1 | transducin (beta)-like 1X-linked |
C00002514
|
| 123036 | TC2N, C14orf47, C2CD1, MTAC2D1, Tac2-N | tandem C2 domains, nuclear |
C00002514
|
| 6941 | TCF19, SC1, TCF-19 | transcription factor 19 |
C00002514
|
| 83439 | TCF7L1, TCF-3, TCF3 | transcription factor 7-like 1 (T-cell specific, HMG-box) |
C00002514
|
| 55775 | TDP1 | tyrosyl-DNA phosphodiesterase 1 (EC:3.1.4.-) |
C00002514
|
| 26136 | TES, TESS, TESS-2 | testis derived transcript (3 LIM domains) |
C00002514
|
| 93081 | TEX30, C13orf27 | testis expressed 30 |
C00002514
|
| 7022 | TFAP2C, AP2-GAMMA, ERF1, TFAP2G, hAP-2g | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
C00002514
|
| 7027 | TFDP1, DP1, DRTF1, Dp-1 | transcription factor Dp-1 |
C00002514
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00002514
|
| 7033 | TFF3, ITF, P1B, TFI | trefoil factor 3 (intestinal) |
C00002514
|
| 7035 | TFPI, EPI, LACI, TFI, TFPI1 | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
C00002514
|
| 7037 | TFRC, CD71, T9, TFR, TFR1, TR, TRFR, p90 | transferrin receptor |
C00002514
|
| 7042 | TGFB2, LDS4, TGF-beta2 | transforming growth factor, beta 2 |
C00002514
|
| 7046 | TGFBR1, AAT5, ACVRLK4, ALK-5, ALK5, LDS1A, LDS2A, MSSE, SKR4, TGFR-1 | transforming growth factor, beta receptor 1 (EC:2.7.11.30) |
C00002514
|
| 7050 | TGIF1, HPE4, TGIF | TGFB-induced factor homeobox 1 |
C00002514
|
| 60436 | TGIF2 | TGFB-induced factor homeobox 2 |
C00002514
|
| 7057 | THBS1, THBS, THBS-1, TSP, TSP-1, TSP1 | thrombospondin 1 |
C00002514
|
| 51337 | THEM6, C8orf55, DSCD75 | thioesterase superfamily member 6 |
C00002514
|
| 8563 | THOC5, C22orf19, Fmip, PK1.3, fSAP79 | THO complex 5 |
C00002514
|
| 7064 | THOP1, EP24.15, MEPD_HUMAN, MP78, TOP | thimet oligopeptidase 1 (EC:3.4.24.15) |
C00002514
|
| 79875 | THSD4, ADAMTSL-6, ADAMTSL6, FVSY9334, PRO34005 | thrombospondin, type I, domain containing 4 |
C00002514
|
| 90381 | TICRR, C15orf42, SLD3, Treslin | TOPBP1-interacting checkpoint and replication regulator |
C00002514
|
| 8914 | TIMELESS, TIM, TIM1, hTIM | timeless circadian clock |
C00002514
|
| 29090 | TIMM21, C18orf55, TIM21 | translocase of inner mitochondrial membrane 21 homolog (yeast) |
C00002514
|
| 7077 | TIMP2, CSC-21K, DDC8 | TIMP metallopeptidase inhibitor 2 |
C00002514
|
| 7078 | TIMP3, HSMRK222, K222, K222TA2, SFD | TIMP metallopeptidase inhibitor 3 |
C00002514
|
| 54962 | TIPIN | TIMELESS interacting protein |
C00002514
|
| 7083 | TK1, TK2 | thymidine kinase 1, soluble (EC:2.7.1.21) |
C00002514
|
| 116238 | TLCD1 | TLC domain containing 1 |
C00002514
|
| 7088 | TLE1, ESG, ESG1, GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
C00002514
|
| 11011 | TLK2, HsHPK, PKU-ALPHA | tousled-like kinase 2 (EC:2.7.11.1) |
C00002514
|
| 4071 | TM4SF1, H-L6, L6, M3S1, TAAL6 | transmembrane 4 L six family member 1 |
C00002514
|
| 53346 | TM6SF1 | transmembrane 6 superfamily member 1 |
C00002514
|
| 147798 | TMC4 | transmembrane channel-like 4 |
C00002514
|
| 23023 | TMCC1 | transmembrane and coiled-coil domain family 1 |
C00002514
|
| 283578 | TMED8, FAM15B | transmembrane emp24 protein transport domain containing 8 |
C00002514
|
| 54664 | TMEM106B | transmembrane protein 106B |
C00002514
|
| 79022 | TMEM106C | transmembrane protein 106C |
C00002514
|
| 84314 | TMEM107, GRVS638, PRO1268 | transmembrane protein 107 |
C00002514
|
| 129303 | TMEM150A, TM6P1, TMEM150 | transmembrane protein 150A |
C00002514
|
| 441027 | TMEM150C | transmembrane protein 150C |
C00002514
|
| 84187 | TMEM164, bB360B22.3 | transmembrane protein 164 |
C00002514
|
| 64418 | TMEM168 | transmembrane protein 168 |
C00002514
|
| 57583 | TMEM181, GPR178, KIAA1423 | transmembrane protein 181 |
C00002514
|
| 55266 | TMEM19 | transmembrane protein 19 |
C00002514
|
| 23306 | TMEM194A, TMEM194 | transmembrane protein 194A |
C00002514
|
| 29058 | TMEM230, C20orf30, HSPC274, dJ1116H23.2.1 | transmembrane protein 230 |
C00002514
|
| 57393 | TMEM27, NX-17, NX17 | transmembrane protein 27 |
C00002514
|
| 55754 | TMEM30A, C6orf67, CDC50A | transmembrane protein 30A |
C00002514
|
| 55151 | TMEM38B, C9orf87, D4Ertd89e, OI14, TRIC-B, TRICB, bA219P18.1 | transmembrane protein 38B |
C00002514
|
| 120224 | TMEM45B | transmembrane protein 45B |
C00002514
|
| 10329 | TMEM5, HP10481, MDDGA10 | transmembrane protein 5 |
C00002514
|
| 169200 | TMEM64 | transmembrane protein 64 |
C00002514
|
| 157378 | TMEM65 | transmembrane protein 65 |
C00002514
|
| 27346 | TMEM97, MAC30 | transmembrane protein 97 |
C00002514
|
| 7112 | TMPO, CMD1T, LAP2, LEMD4, PRO0868, TP | thymopoietin |
C00002514
|
| 64699 | TMPRSS3, DFNB10, DFNB8, ECHOS1, TADG12 | transmembrane protease, serine 3 (EC:3.4.21.-) |
C00002514
|
| 735524 |
C00002514
|
||
| 160335 | TMTC2, IBDBP1 | transmembrane and tetratricopeptide repeat containing 2 |
C00002514
|
| 81542 | TMX1, PDIA11, TMX, TXNDC, TXNDC1 | thioredoxin-related transmembrane protein 1 |
C00002514
|
| 56255 | TMX4, DJ971N18.2, PDIA14, TXNDC13 | thioredoxin-related transmembrane protein 4 |
C00002514
|
| 7126 | TNFAIP1, B12, B61, BTBD34, EDP1, hBACURD2 | tumor necrosis factor, alpha-induced protein 1 (endothelial) |
C00002514
|
| 51330 | TNFRSF12A, CD266, FN14, TWEAKR | tumor necrosis factor receptor superfamily, member 12A |
C00002514
|
| 27242 | TNFRSF21, BM-018, CD358, DR6 | tumor necrosis factor receptor superfamily, member 21 |
C00002514
|
| 10766 | TOB2, TOB4, TOBL, TROB2 | transducer of ERBB2, 2 |
C00002514
|
| 114034 | TOE1 | target of EGR1, member 1 (nuclear) |
C00002514
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00002514
|
| 11073 | TOPBP1, TOP2BP1 | topoisomerase (DNA) II binding protein 1 |
C00002514
|
| 94241 | TP53INP1, SIP, TP53DINP1, TP53INP1A, TP53INP1B, Teap, p53DINP1 | tumor protein p53 inducible nuclear protein 1 |
C00002514
|
| 58476 | TP53INP2, C20orf110, DOR, PINH, dJ1181N3.1 | tumor protein p53 inducible nuclear protein 2 |
C00002514
|
| 7163 | TPD52, D52, N8L, PC-1, PrLZ, hD52 | tumor protein D52 |
C00002514
|
| 7164 | TPD52L1, D53, hD53 | tumor protein D52-like 1 |
C00002514
|
| 7167 | TPI1, TIM, TPI, TPID | triosephosphate isomerase 1 (EC:5.3.1.1) |
C00002514
|
| 7168 | TPM1, C15orf13, CMD1Y, CMH3, HTM-alpha, LVNC9, TMSA | tropomyosin 1 (alpha) |
C00002514
|
| 127262 | TPRG1L, FAM79A, RP11-46F15.3, h-mover, mover | tumor protein p63 regulated 1-like |
C00002514
|
| 7179 | TPTE, CT44, PTEN2 | transmembrane phosphatase with tensin homology (EC:3.1.3.48) |
C00002514
|
| 22974 | TPX2, C20orf1, C20orf2, DIL-2, DIL2, FLS353, GD:C20orf1, HCA519, HCTP4, REPP86, p100 | TPX2, microtubule-associated |
C00002514
|
| 6434 | TRA2B, Htra2-beta, SFRS10, SRFS10, TRA2-BETA, TRAN2B | transformer 2 beta homolog (Drosophila) |
C00002514
|
| 9618 | TRAF4, CART1, MLN62, RNF83 | TNF receptor-associated factor 4 |
C00002514
|
| 6399 | TRAPPC2, MIP2A, SEDL, SEDT, TRAPPC2P1, TRS20, ZNF547L, hYP38334 | trafficking protein particle complex 2 |
C00002514
|
| 22878 | TRAPPC8, GSG1, HsT2706, KIAA1012, TRS85 | trafficking protein particle complex 8 |
C00002514
|
| 55809 | TRERF1, BCAR2, HSA277276, RAPA, RP1-139D8.5, TREP132, TReP-132, dJ139D8.5 | transcriptional regulating factor 1 |
C00002514
|
| 10221 | TRIB1, C8FW, GIG-2, GIG2, SKIP1, TRB-1, TRB1 | tribbles pseudokinase 1 |
C00002514
|
| 286827 | TRIM59, MRF1, RNF104, TRIM57, TSBF1 | tripartite motif containing 59 |
C00002514
|
| 11078 | TRIOBP, DFNB28, TARA, dJ37E16.4 | TRIO and F-actin binding protein |
C00002514
|
| 9319 | TRIP13, 16E1BP | thyroid hormone receptor interactor 13 |
C00002514
|
| 93587 | TRMT10A, RG9MTD2, TRM10 | tRNA methyltransferase 10 homolog A (S. cerevisiae) |
C00002514
|
| 57570 | TRMT5, KIAA1393, TRM5 | tRNA methyltransferase 5 (EC:2.1.1.228) |
C00002514
|
| 51605 | TRMT6, GCD10, Gcd10p | tRNA methyltransferase 6 homolog (S. cerevisiae) |
C00002514
|
| 10024 | TROAP, TASTIN | trophinin associated protein |
C00002514
|
| 7249 | TSC2, LAM, TSC4 | tuberous sclerosis 2 |
C00002514
|
| 1831 | TSC22D3, DIP, DSIPI, GILZ, TSC-22R, hDIP | TSC22 domain family, member 3 |
C00002514
|
| 116461 | TSEN15, C1orf19, sen15 | TSEN15 tRNA splicing endonuclease subunit |
C00002514
|
| 7247 | TSN, BCLF-1, C3PO, RCHF1, REHF-1, TBRBP, TRSLN | translin |
C00002514
|
| 10103 | TSPAN1, NET1, TM4C, TM4SF | tetraspanin 1 |
C00002514
|
| 6302 | TSPAN31, SAS | tetraspanin 31 |
C00002514
|
| 7259 | TSPYL1, TSPYL | TSPY-like 1 |
C00002514
|
| 158219 | TTC39B, C9orf52 | tetratricopeptide repeat domain 39B |
C00002514
|
| 23508 | TTC9, TTC9A | tetratricopeptide repeat domain 9 |
C00002514
|
| 8458 | TTF2, HuF2 | transcription termination factor, RNA polymerase II |
C00002514
|
| 7272 | TTK, CT96, ESK, MPH1, MPS1, MPS1L1, PYT | TTK protein kinase (EC:2.7.12.1) |
C00002514
|
| 406303 |
C00002514
|
||
| 203068 | TUBB, M40, OK/SW-cl.56, TUBB1, TUBB5 | tubulin, beta class I |
C00002514
|
| 7280 | TUBB2A, TUBB, TUBB2, dJ40E16.7 | tubulin, beta 2A class IIa |
C00002514
|
| 100691897 |
C00002514
|
||
| 51174 | TUBD1, TUBD | tubulin, delta 1 |
C00002514
|
| 7283 | TUBG1, CDCBM4, GCP-1, TUBG, TUBGCP1 | tubulin, gamma 1 |
C00002514
|
| 27229 | TUBGCP4, 76P, GCP-4, GCP4, Grip76 | tubulin, gamma complex associated protein 4 |
C00002514
|
| 7286 | TUFT1 | tuftelin 1 |
C00002514
|
| 55000 | TUG1, LINC00080, NCRNA00080, TI-227H | taurine up-regulated 1 (non-protein coding) |
C00002514
|
| 56995 | TULP4, TUSP | tubby like protein 4 |
C00002514
|
| 7291 | TWIST1, ACS3, BPES2, BPES3, CRS1, SCS, TWIST, bHLHa38 | twist family bHLH transcription factor 1 |
C00002514
|
| 7298 | TYMS, HST422, TMS, TS | thymidylate synthetase (EC:2.1.1.45) |
C00002514
|
| 283991 | UBALD2, FAM100B | UBA-like domain containing 2 |
C00002514
|
| 7320 | UBE2B, E2-17kDa, HHR6B, HR6B, RAD6B, UBC2 | ubiquitin-conjugating enzyme E2B (EC:6.3.2.19) |
C00002514
|
| 7321 | UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A | ubiquitin-conjugating enzyme E2D 1 (EC:6.3.2.19) |
C00002514
|
| 7322 | UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B | ubiquitin-conjugating enzyme E2D 2 (EC:6.3.2.19) |
C00002514
|
| 7328 | UBE2H, E2-20K, GID3, UBC8, UBCH, UBCH2 | ubiquitin-conjugating enzyme E2H (EC:6.3.2.19) |
C00002514
|
| 51465 | UBE2J1, HSPC153, HSPC205, HSU93243, NCUBE-1, NCUBE1, UBC6, Ubc6p | ubiquitin-conjugating enzyme E2, J1 (EC:6.3.2.19) |
C00002514
|
| 7334 | UBE2N, UBC13, UbcH-ben, UbcH13 | ubiquitin-conjugating enzyme E2N (EC:6.3.2.19) |
C00002514
|
| 134111 | UBE2QL1 | ubiquitin-conjugating enzyme E2Q family-like 1 (EC:6.3.2.19) |
C00002514
|
| 29089 | UBE2T, PIG50 | ubiquitin-conjugating enzyme E2T (putative) (EC:6.3.2.19) |
C00002514
|
| 55284 | UBE2W, UBC-16, UBC16 | ubiquitin-conjugating enzyme E2W (putative) (EC:6.3.2.19) |
C00002514
|
| 55148 | UBR7, C14orf130 | ubiquitin protein ligase E3 component n-recognin 7 (putative) |
C00002514
|
| 51377 | UCHL5, CGI-70, INO80R, UCH-L5, UCH37 | ubiquitin carboxyl-terminal hydrolase L5 (EC:3.4.19.12) |
C00002514
|
| 7371 | UCK2, TSA903, UK, UMPK | uridine-cytidine kinase 2 (EC:2.7.1.48) |
C00002514
|
| 7358 | UGDH, GDH, UDP-GlcDH, UDPGDH, UGD | UDP-glucose 6-dehydrogenase (EC:1.1.1.22) |
C00002514
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00002514
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00002514
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00002514
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00002514
|
| 54600 | UGT1A9, HLUGP4, LUGP4, UDPGT, UDPGT_1-9, UGT-1I, UGT1-09, UGT1-9, UGT1.9, UGT1AI, UGT1I | UDP glucuronosyltransferase 1 family, polypeptide A9 (EC:2.4.1.17) |
C00002514
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00002514
|
| 127933 | UHMK1, KIS, KIST, P-CIP2 | U2AF homology motif (UHM) kinase 1 (EC:2.7.11.1) |
C00002514
|
| 29128 | UHRF1, ICBP90, Np95, RNF106, hNP95, hUHRF1 | ubiquitin-like with PHD and ring finger domains 1 (EC:6.3.2.19) |
C00002514
|
| 7372 | UMPS, OPRT | uridine monophosphate synthetase (EC:2.4.2.10 4.1.1.23) |
C00002514
|
| 7374 | UNG, DGU, HIGM4, HIGM5, UDG, UNG1, UNG15, UNG2 | uracil-DNA glycosylase (EC:3.2.2.27) |
C00002514
|
| 65109 | UPF3B, HUPF3B, MRXS14, RENT3B, UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) |
C00002514
|
| 84300 | UQCC2, C6orf125, Cbp6, M19, MNF1, bA6B20.2 | ubiquinol-cytochrome c reductase complex assembly factor 2 (EC:2.7.4.21) |
C00002514
|
| 7398 | USP1, UBP | ubiquitin specific peptidase 1 (EC:3.4.19.12) |
C00002514
|
| 9960 | USP3, SIH003, UBP | ubiquitin specific peptidase 3 (EC:3.4.19.12) |
C00002514
|
| 7375 | USP4, UNP, Unph | ubiquitin specific peptidase 4 (proto-oncogene) (EC:3.4.19.12) |
C00002514
|
| 55230 | USP40 | ubiquitin specific peptidase 40 (EC:3.4.19.12) |
C00002514
|
| 8078 | USP5, ISOT | ubiquitin specific peptidase 5 (isopeptidase T) (EC:3.4.19.12) |
C00002514
|
| 159195 | USP54, C10orf29, bA137L10.3, bA137L10.4 | ubiquitin specific peptidase 54 |
C00002514
|
| 10090 | UST, 2OST | uronyl-2-sulfotransferase |
C00002514
|
| 7407 | VARS, G7A, VARS1, VARS2 | valyl-tRNA synthetase (EC:6.1.1.9) |
C00002514
|
| 57176 | VARS2, G7a, VARS2L, VARSL | valyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.9) |
C00002514
|
| 10493 | VAT1, VATI | vesicle amine transport 1 |
C00002514
|
| 7416 | VDAC1, PORIN, VDAC-1 | voltage-dependent anion channel 1 |
C00002514
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00002514
|
| 9686 | VGLL4, VGL-4 | vestigial like 4 (Drosophila) |
C00002514
|
| 79001 | VKORC1, EDTP308, IMAGE3455200, MST134, MST576, VKCFD2, VKOR | vitamin K epoxide reductase complex, subunit 1 (EC:1.1.4.1) |
C00002514
|
| 54832 | VPS13C | vacuolar protein sorting 13 homolog C (S. cerevisiae) |
C00002514
|
| 40542 |
C00002514
|
||
| 27072 | VPS41, HVPS41, HVSP41, hVps41p | vacuolar protein sorting 41 homolog (S. cerevisiae) |
C00002514
|
| 11311 | VPS45, H1, H1VPS45, SCN5, VPS45A, VPS45B, VPS54A, VSP45, VSP45A | vacuolar protein sorting 45 homolog (S. cerevisiae) |
C00002514
|
| 7443 | VRK1, PCH1, PCH1A | vaccinia related kinase 1 (EC:2.7.11.1) |
C00002514
|
| 374666 | WASH3P, FAM39DP | WAS protein family homolog 3 pseudogene |
C00002514
|
| 23559 | WBP1, WBP-1 | WW domain binding protein 1 |
C00002514
|
| 11169 | WDHD1, AND-1, CHTF4, CTF4 | WD repeat and HMG-box DNA binding protein 1 |
C00002514
|
| 89891 | WDR34, DIC5, FAP133, bA216B9.3 | WD repeat domain 34 |
C00002514
|
| 10785 | WDR4, TRM82, TRMT82 | WD repeat domain 4 |
C00002514
|
| 84058 | WDR54 | WD repeat domain 54 |
C00002514
|
| 79968 | WDR76, CDW14 | WD repeat domain 76 |
C00002514
|
| 33865 |
C00002514
|
||
| 7468 | WHSC1, MMSET, NSD2, REIIBP, TRX5, WHS | Wolf-Hirschhorn syndrome candidate 1 (EC:2.1.1.43) |
C00002514
|
| 55062 | WIPI1, ATG18, ATG18A, WIPI49 | WD repeat domain, phosphoinositide interacting 1 |
C00002514
|
| 26100 | WIPI2, ATG18B, Atg21, WIPI-2 | WD repeat domain, phosphoinositide interacting 2 |
C00002514
|
| 8839 | WISP2, CCN5, CT58, CTGF-L | WNT1 inducible signaling pathway protein 2 |
C00002514
|
| 58525 | WIZ, ZNF803 | widely interspaced zinc finger motifs |
C00002514
|
| 55884 | WSB2, SBA2 | WD repeat and SOCS box containing 2 |
C00002514
|
| 80014 | WWC2, BOMB | WW and C2 domain containing 2 |
C00002514
|
| 55841 | WWC3, BM042 | WWC family member 3 |
C00002514
|
| 11059 | WWP1, AIP5, Tiul1, hSDRP1 | WW domain containing E3 ubiquitin protein ligase 1 |
C00002514
|
| 23039 | XPO7, EXP7, RANBP16 | exportin 7 |
C00002514
|
| 2547 | XRCC6, CTC75, CTCBF, G22P1, KU70, ML8, TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 |
C00002514
|
| 91419 | XRCC6BP1, KUB3 | XRCC6 binding protein 1 (EC:3.4.24.-) |
C00002514
|
| 4904 | YBX1, BP-8, CSDA2, CSDB, DBPB, MDR-NF1, NSEP-1, NSEP1, YB-1, YB1 | Y box binding protein 1 |
C00002514
|
| 388403 | YPEL2 | yippee-like 2 (Drosophila) |
C00002514
|
| 51646 | YPEL5 | yippee-like 5 (Drosophila) |
C00002514
|
| 7529 | YWHAB, GW128, HS1, KCIP-1, YWHAA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
C00002514
|
| 7531 | YWHAE, 14-3-3E, KCIP-1, MDCR, MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
C00002514
|
| 22890 | ZBTB1, ZNF909 | zinc finger and BTB domain containing 1 |
C00002514
|
| 65986 | ZBTB10, RINZF | zinc finger and BTB domain containing 10 |
C00002514
|
| 57684 | ZBTB26, ZNF481, bioref | zinc finger and BTB domain containing 26 |
C00002514
|
| 253461 | ZBTB38, CIBZ, ZNF921 | zinc finger and BTB domain containing 38 |
C00002514
|
| 57659 | ZBTB4, KAISO-L1, ZNF903 | zinc finger and BTB domain containing 4 |
C00002514
|
| 140685 | ZBTB46, BTBD4, BZEL, RINZF, ZNF340, dJ583P15.7, dJ583P15.8 | zinc finger and BTB domain containing 46 |
C00002514
|
| 339487 | ZBTB8OS, ARCH, ARCH2 | zinc finger and BTB domain containing 8 opposite strand |
C00002514
|
| 51101 | ZC2HC1A, C8orf70, FAM164A | zinc finger, C2HC-type containing 1A |
C00002514
|
| 80149 | ZC3H12A, MCPIP, MCPIP1, RP3-423B22.1, dJ423B22.1 | zinc finger CCCH-type containing 12A |
C00002514
|
| 54819 | ZCCHC10 | zinc finger, CCHC domain containing 10 |
C00002514
|
| 23390 | ZDHHC17, HIP14, HIP3, HYPH | zinc finger, DHHC-type containing 17 (EC:2.3.1.225) |
C00002514
|
| 64429 | ZDHHC6, ZNF376 | zinc finger, DHHC-type containing 6 (EC:2.3.1.225) |
C00002514
|
| 55625 | ZDHHC7, DHHC7, SERZ-B, SERZ1, ZNF370 | zinc finger, DHHC-type containing 7 (EC:2.3.1.225) |
C00002514
|
| 90637 | ZFAND2A, AIRAP | zinc finger, AN1-type domain 2A |
C00002514
|
| 161882 | ZFPM1, FOG, FOG1, ZC2HC11A, ZNF408, ZNF89A | zinc finger protein, FOG family member 1 |
C00002514
|
| 53349 | ZFYVE1, DFCP1, SR3, TAFF1, ZNFN2A1 | zinc finger, FYVE domain containing 1 |
C00002514
|
| 124220 | ZG16B, EECP, HRPE773, JCLN2, PAUF, PRO1567 | zymogen granule protein 16B |
C00002514
|
| 7545 | ZIC1, ZIC, ZNF201 | Zic family member 1 |
C00002514
|
| 7586 | ZKSCAN1, 9130423L19Rik, KOX18, PHZ-37, ZNF139, ZNF36, ZSCAN33 | zinc finger with KRAB and SCAN domains 1 |
C00002514
|
| 57178 | ZMIZ1, MIZ, RAI17, TRAFIP10, ZIMP10, hZIMP10 | zinc finger, MIZ-type containing 1 |
C00002514
|
| 9205 | ZMYM5, HSPC050, MYM, ZNF198L1, ZNF237 | zinc finger, MYM-type 5 |
C00002514
|
| 51351 | ZNF117, H-plk, HPF9 | zinc finger protein 117 |
C00002514
|
| 7559 | ZNF12, GIOT-3, HZF11, KOX3, ZNF325 | zinc finger protein 12 |
C00002514
|
| 7678 | ZNF124, HZF-16, HZF16, ZK7 | zinc finger protein 124 |
C00002514
|
| 90338 | ZNF160, F11, HKr18, HZF5, KR18 | zinc finger protein 160 |
C00002514
|
| 7769 | ZNF226 | zinc finger protein 226 |
C00002514
|
| 339324 | ZNF260, OZRF1, PEX1, ZFP260 | zinc finger protein 260 |
C00002514
|
| 11179 | ZNF277, NRIF4, ZNF277P | zinc finger protein 277 |
C00002514
|
| 23036 | ZNF292, Nbla00365, ZFP292, ZN-16, Zn-15, bA393I2.3 | zinc finger protein 292 |
C00002514
|
| 59348 | ZNF350, ZBRK1, ZFQR | zinc finger protein 350 |
C00002514
|
| 6940 | ZNF354A, EZNF, HKL1, KID-1, KID1, TCF17 | zinc finger protein 354A |
C00002514
|
| 195828 | ZNF367, AFF29, CDC14B, ZFF29 | zinc finger protein 367 |
C00002514
|
| 151126 | ZNF385B, ZNF533 | zinc finger protein 385B |
C00002514
|
| 55893 | ZNF395, HDBP-2, HDBP2, HDRF-2, PBF, PRF-1, PRF1, Si-1-8-14 | zinc finger protein 395 |
C00002514
|
| 51710 | ZNF44, GIOT-2, KOX7, ZNF, ZNF504, ZNF55, ZNF58 | zinc finger protein 44 |
C00002514
|
| 168544 | ZNF467, EZI, Zfp467 | zinc finger protein 467 |
C00002514
|
| 84838 | ZNF496, NIZP1, ZFP496, ZKSCAN17, ZSCAN49 | zinc finger protein 496 |
C00002514
|
| 93134 | ZNF561 | zinc finger protein 561 |
C00002514
|
| 84914 | ZNF587, ZF6 | zinc finger protein 587 |
C00002514
|
| 9831 | ZNF623 | zinc finger protein 623 |
C00002514
|
| 199692 | ZNF627 | zinc finger protein 627 |
C00002514
|
| 121274 | ZNF641 | zinc finger protein 641 |
C00002514
|
| 55279 | ZNF654 | zinc finger protein 654 |
C00002514
|
| 339500 | ZNF678 | zinc finger protein 678 |
C00002514
|
| 80139 | ZNF703, ZEPPO1, ZNF503L, ZPO1 | zinc finger protein 703 |
C00002514
|
| 255403 | ZNF718 | zinc finger protein 718 |
C00002514
|
| 79755 | ZNF750, ZFP750 | zinc finger protein 750 |
C00002514
|
| 284309 | ZNF776 | zinc finger protein 776 |
C00002514
|
| 146540 | ZNF785 | zinc finger protein 785 |
C00002514
|
| 7637 | ZNF84, HPF2 | zinc finger protein 84 |
C00002514
|
| 7644 | ZNF91, HPF7, HTF10 | zinc finger protein 91 |
C00002514
|
| 30834 | ZNRD1, HTEX-6, Rpa12, TEX6, ZR14, hZR14, tctex-6 | zinc ribbon domain containing 1 (EC:2.7.7.6) |
C00002514
|
| 23053 | ZSWIM8, KIAA0913 | zinc finger, SWIM-type containing 8 |
C00002514
|
| 55055 | ZWILCH, KNTC1AP, hZwilch | zwilch kinetochore protein |
C00002514
|
| 11130 | ZWINT, HZwint-1, KNTC2AP, ZWINT1 | ZW10 interacting kinetochore protein |
C00002514
|
| 158586 | ZXDB, ZNF905, dJ83L6.1 | zinc finger, X-linked, duplicated B |
C00002514
|
| 7791 | ZYX, ESP-2, HED-2 | zyxin |
C00002514
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (68)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300018 | 46,xy sex reversal 2; srxy2 |
P51843
|
| #300200 | Adrenal hypoplasia, congenital; ahc |
P51843
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #614490 | Blood group, junior system; jr |
Q9UNQ0
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #613630 | Cocoon syndrome |
O15111
|
| #114500 | Colorectal cancer; crc |
Q14191
|
| #127750 | Dementia, lewy body; dlb |
P37840
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #612132 | Ectodermal dysplasia, anhidrotic, with t-cell immunodeficiency, autosomal dominant |
P25963
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #133239 | Esophageal cancer |
P04637
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #612219 | Ewing sarcoma; es |
P11308
|
| #301500 | Fabry disease |
P06280
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237450 | Hyperbilirubinemia, rotor type; hblrr |
Q9NPD5
Q9Y6L6 |
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #146300 | Hypophosphatasia, adult |
P05186
|
| #241510 | Hypophosphatasia, childhood |
P05186
|
| #241500 | Hypophosphatasia, infantile |
P05186
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #211980 | Lung cancer |
P04637
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #158580 | Neuronopathy, distal hereditary motor, type viia; hmn7a |
Q9GZV3
|
| #257200 | Niemann-pick disease, type a |
P17405
|
| #607616 | Niemann-pick disease, type b |
P17405
|
| #613705 | Orofacial cleft 10; ofc10 |
P63165
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #168601 | Parkinson disease 1, autosomal dominant; park1 |
P37840
|
| #605543 | Parkinson disease 4, autosomal dominant; park4 |
P37840
|
| #168600 | Parkinson disease, late-onset; pd |
P37840
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
P04637
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #138900 | Uric acid concentration, serum, quantitative trait locus 1; uaqtl1 |
Q9UNQ0
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #277700 | Werner syndrome; wrn |
Q14191
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (92)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00882 | Cocoon syndrome |
O15111
(related)
|
| H00081 | Hashimoto's thyroiditis |
P01215
(marker)
|
| H00082 | Graves' disease |
P01215
(marker)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P01215
(marker)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04637 (related) Q92731 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
P04637
(related)
Q01196 (related) |
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
|
| H00006 | Hairy-cell leukemia |
P04637
(related)
|
| H00008 | Burkitt lymphoma |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00010 | Multiple myeloma |
P04637
(related)
|
| H00013 | Small cell lung cancer |
P04637
(related)
|
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00016 | Oral cancer |
P04637
(related)
P04637 (marker) |
| H00017 | Esophageal cancer |
P04637
(related)
P04637 (marker) |
| H00018 | Gastric cancer |
P04637
(related)
|
| H00019 | Pancreatic cancer |
P04637
(related)
P04637 (marker) |
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) |
| H00022 | Bladder cancer |
P04637
(related)
|
| H00025 | Penile cancer |
P04637
(related)
P04637 (marker) P08253 (related) P14780 (related) |
| H00027 | Ovarian cancer |
P04637
(related)
|
| H00028 | Choriocarcinoma |
P04637
(related)
P08253 (related) |
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00031 | Breast cancer |
P04637
(related)
|
| H00032 | Thyroid cancer |
P04637
(related)
|
| H00036 | Osteosarcoma |
P04637
(related)
P08684 (marker) |
| H00038 | Malignant melanoma |
P04637
(related)
|
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00041 | Kaposi's sarcoma |
P04637
(related)
|
| H00042 | Glioma |
P04637
(related)
P04637 (marker) |
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00046 | Cholangiocarcinoma |
P04637
(related)
|
| H00047 | Gallbladder cancer |
P04637
(related)
|
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00055 | Laryngeal cancer |
P04637
(related)
P04637 (marker) |
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00021 | Renal cell carcinoma |
P04637
(marker)
|
| H00213 | Hypophosphatasia |
P05186
(related)
|
| H00125 | Fabry disease |
P06280
(related)
|
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00024 | Prostate cancer |
P10275
(related)
P11308 (related) |
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00023 | Testicular cancer |
P10696
(marker)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00035 | Ewing's sarcoma |
P11308
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00137 | Niemann-Pick disease (NPD) typeA and B |
P17405
(related)
|
| H00424 | Defects in the degradation of sphingomyelin |
P17405
(related)
|
| H00007 | Hodgkin lymphoma |
P25963
(related)
|
| H00095 | Ectodermal dysplasia associated immunodeficiency (EDA-ID) |
P25963
(related)
|
| H00057 | Parkinson's disease (PD) |
P37840
(related)
|
| H00066 | Lewy body dementia (LBD) |
P37840
(related)
|
| H00552 | Glycerol kinase deficiency (GKD) |
P51843
(related)
|
| H00607 | 46,XY disorders of sex development (Disorders of gonadal development) |
P51843
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
Q14191 (related) |
| H00516 | Isolated orofacial clefts |
P63165
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
Q9NUW8 (related) |
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|