Xylocarpus granatum
Species
KNApSAcK Entry
| Organism name | Xylocarpus granatum |
|---|---|
| Genus | Xylocarpus |
| Family | Meliaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Xylocarpus granatum |
|---|---|
| Linked NCBI taxonomy ID | 241841 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Meliaceae |
|---|---|
| ID | 43707 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (67)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
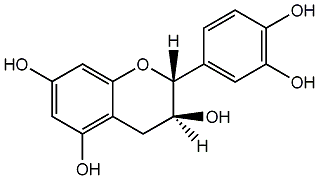
C00000947
|
D-Catechol
/ D-Catechin / Teafuran 30A / (+)-Catechin / Dexcyanidanol / (+)-3',4',5,7-Tetrahydroxy-2,3-trans-flavan-3-ol |
CHEMBL80941
CHEMBL311498 CHEMBL129482 CHEMBL200715 CHEMBL206452 CHEMBL583912 |
D002392
|
85 / 58 / 54 | 91 / 13 | No. 52 | No. 14 |
|
|
C00048205
|
Tigloylseneganolide A
|
No. 102 |

|
|||||
|
C00040073
|
Proceranolide
|
CHEMBL1081393
|
No. 102 |

|
||||
|
C00047911
|
Granatumin D
/ (-)-Granatumin D |
No. 102 |

|
|||||
|
C00047909
|
Granatumin B
/ (+)-Granatumin B |
No. 102 |

|
|||||
|
C00047908
|
Granatumin A
/ (+)-Granatumin A |
No. 102 |

|
|||||
|
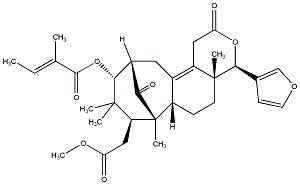
C00037373
|
Khayasin T
|
No. 102 |
|
|||||
|
C00047910
|
Granatumin C
/ (+)-Granatumin C |
No. 102 |

|
|||||
|
C00024641
|
Dihydrochelerythrine
/ 12,13-Dihydrochelerythrine |
CHEMBL400359
|
No. 195 | No. 4 |

|
|||
|
C00040717
|
Xylocarpin I
/ (+)-Xylocarpin I |
No. 196 |

|
|||||
|
C00047901
|
Gedunin
|
CHEMBL465226
CHEMBL1335340 CHEMBL2004035 |
C106014
|
16 / 33 / 55 | 83 / 1 | No. 204 | No. 51 |

|
|
C00034343
|
Xyloccensin K
|
CHEMBL1082138
CHEMBL1966058 |
No. 255 |

|
||||
|
C00032531
|
Xyloccensin Q
|
No. 378 |

|
|||||
|
C00032530
|
Xyloccensin P
|
No. 378 |

|
|||||
|
C00047913
|
Granatumin F
/ (+)-Granatumin F |
No. 378 |

|
|||||
|
C00047914
|
Granatumin G
/ (+)-Granatumin G |
No. 378 |

|
|||||
|
C00035430
|
Xyloccensin R
/ (+)-Xyloccensin R |
No. 378 |

|
|||||
|
C00048236
|
Xyloccensin O
|
No. 378 |

|
|||||
|
C00035432
|
Xyloccensin T
/ (+)-Xyloccensin T |
No. 378 |

|
|||||
|
C00035431
|
Xyloccensin S
/ (+)-Xyloccensin S |
No. 378 |

|
|||||
|
C00047790
|
Chisocheton F
|
CHEMBL1079490
CHEMBL1774404 |
No. 453 |

|
||||
|
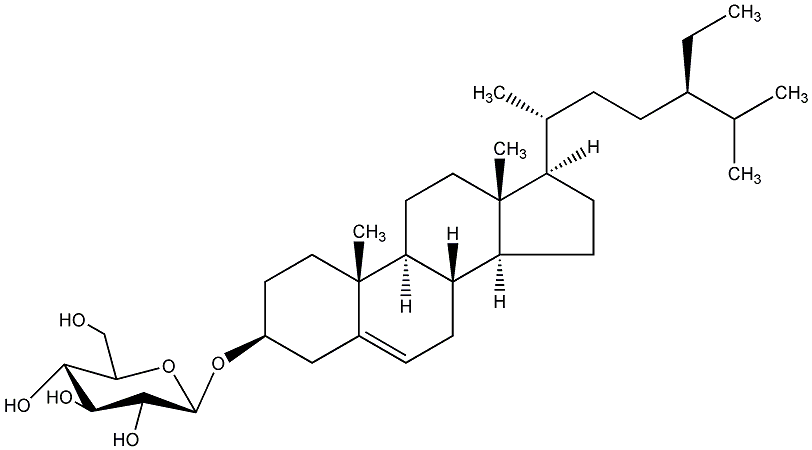
C00019308
|
Doursterol
/ Daucosterin / 3-O-beta-D-Glucopyranosyl sitosterol / beta-Sitosterol 3-O-beta-D-glucopyranoside |
CHEMBL197711
CHEMBL506678 CHEMBL2304043 |
C011015
|
5 / 4 / 2 | 0 / 3 | No. 520 |
|
|
|
C00024103
|
Swietemahonolide
|
No. 535 |

|
|||||
|
C00039289
|
Granaxylocarpin C
/ (-)-Granaxylocarpin C |
CHEMBL388137
|
No. 535 |

|
||||
|
C00002189
|
N-Methylflindersine
|
CHEMBL400130
|
No. 799 | No. 7 |

|
|||
|
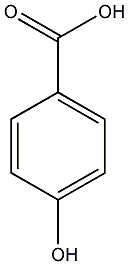
C00000856
|
4-Hydroxybenzoic acid
/ p-Hydroxybenzoic acid |
CHEMBL441343
|
C038193
|
21 / 7 / 16 | 2 / 1 | No. 817 | No. 81 |
|
|
C00040710
|
Xylocarpin B
|
No. 833 |

|
|||||
|
C00040709
|
Xylocarpin A
|
CHEMBL388296
|
No. 833 |

|
||||
|
C00040711
|
Xylocarpin C
|
No. 833 |

|
|||||
|
C00040712
|
Xylocarpin D
|
CHEMBL225041
|
No. 833 |

|
||||
|
C00040713
|
Xylocarpin E
|
No. 833 |

|
|||||
|
C00041971
|
Xyloccensin Z2
|
No. 833 |

|
|||||
|
C00041970
|
Xyloccensin Z1
|
No. 833 |

|
|||||
|
C00035433
|
Xyloccensin U
|
No. 833 |

|
|||||
|
C00039290
|
Granaxylocarpin D
|
CHEMBL225041
|
No. 833 |

|
||||
|
C00040715
|
Xylocarpin G
|
No. 873 |

|
|||||
|
C00032532
|
Xylogranatin A
|
No. 873 |

|
|||||
|
C00040714
|
Xylocarpin F
|
CHEMBL1555153
|
8 / 17 / 44 | No. 873 |

|
|||
|
C00048918
|
Xylogranatin L
/ (-)-Xylogranatin L |
No. 975 |

|
|||||
|
C00048917
|
Xylogranatin K
/ (-)-Xylogranatin K |
No. 975 |

|
|||||
|
C00048916
|
Xylogranatin J
/ (-)-Xylogranatin J |
No. 975 |

|
|||||
|
C00048919
|
Xylogranatin M
/ (-)-Xylogranatin M |
No. 975 |

|
|||||
|
C00048922
|
Xylogranatin P
/ (-)-Xylogranatin P |
No. 975 |

|
|||||
|
C00048921
|
Xylogranatin O
/ (-)-Xylogranatin O |
No. 975 |

|
|||||
|
C00048915
|
Xylogranatin I
/ (-)-Xylogranatin I |
No. 975 |

|
|||||
|
C00048923
|
Xylogranatin Q
/ (-)-Xylogranatin Q |
No. 975 |

|
|||||
|
C00048920
|
Xylogranatin N
/ (-)-Xylogranatin N |
No. 975 |

|
|||||
|
C00048072
|
Protoxylocarpin H
|
CHEMBL1075702
|
No. 1247 |

|
||||
|
C00048070
|
Protoxylocarpin F
|
CHEMBL1075700
CHEMBL1075701 |
No. 1247 |

|
||||
|
C00048071
|
Protoxylocarpin G
|
CHEMBL1075700
CHEMBL1075701 |
No. 1247 |

|
||||
|
C00003368
|
Spicatin
|
No. 1532 | No. 38 |

|
||||
|
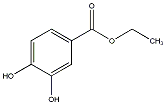
C00032950
|
Ethyl protocatechuate
/ Ethyl 3,4-dihydroxybenzoate |
CHEMBL486216
|
C018063
|
7 / 3 / 5 | No. 1722 |
|
||
|
C00039288
|
Granaxylocarpin B
/ (-)-Granaxylocarpin B |
CHEMBL390119
|
No. 1754 |

|
||||
|
C00032535
|
Xylogranatin D
|
No. 1754 |

|
|||||
|
C00039287
|
Granaxylocarpin A
/ (+)-Granaxylocarpin A |
CHEMBL389681
|
No. 1754 |

|
||||
|
C00032533
|
Xylogranatin B
|
No. 1754 |

|
|||||
|
C00040716
|
Xylocarpin H
/ (+)-Xylocarpin H |
CHEMBL390119
|
No. 1754 |

|
||||
|
C00032534
|
Xylogranatin C
|
No. 1754 |

|
|||||
|
C00041367
|
Aurantiamide
|
CHEMBL475827
|
No. 2605 |

|
||||
|
C00003170
|
Phaseic acid
|
No. 2610 | No. 38 |

|
||||
|
C00048913
|
Xylogranatin G
/ (+)-Xylogranatin G |
No. 4179 |

|
|||||
|
C00048912
|
Xylogranatin F
/ (+)-Xylogranatin F |
No. 4179 |

|
|||||
|
C00048914
|
Xylogranatin H
/ (+)-Xylogranatin H |
No. 4179 |

|
|||||
|
C00047912
|
Granatumin E
/ (-)-Granatumin E |
No. 5312 |

|
|||||
|
C00047876
|
Febrifugin A
|
No. 5312 |

|
|||||
|
C00048924
|
Xylogranatin R
/ (+)-Xylogranatin R |
No. 6267 |

|
|||||
|
C00048237
|
Xylogranatinin
|
No. 8323 |

|
Human Protein / Gene in interactions
108 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P00918 | Carbonic anhydrase 2 | Lyase | C00000856 C00000947 C00032950 | 1 / 2 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00000947 C00040714 C00047901 | 2 / 3 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00000856 C00000947 C00032950 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00000856 C00000947 C00032950 | 1 / 2 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00000947 C00040714 C00047901 | 4 / 1 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00000856 C00000947 C00032950 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00000947 C00040714 C00047901 | 4 / 3 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00000947 C00040714 C00047901 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000947 C00040714 C00047901 | 0 / 1 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00000856 C00000947 C00032950 | 0 / 1 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00000856 C00000947 C00032950 | 0 / 0 |
| P00734 | Prothrombin | S1A | C00000947 C00019308 | 4 / 2 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00000856 C00000947 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00000856 C00000947 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00000947 C00019308 | 0 / 0 |
| Q6W5P4 | Neuropeptide S receptor | Neuropeptide receptor | C00000947 C00047901 | 1 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00000856 C00000947 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00000947 C00047901 | 2 / 0 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00040714 C00047901 | 7 / 37 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00000856 C00000947 | 1 / 1 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00000856 C00000947 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00000947 C00040714 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00000947 C00019308 | 0 / 0 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | C00000947 | 5 / 1 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00000947 | 1 / 1 |
| P02768 | Serum albumin | Secreted protein | C00000947 | 0 / 0 |
| P49327 | Fatty acid synthase | Transferase | C00000947 | 0 / 0 |
| Q9UQ49 | Sialidase-3 | Enzyme | C00000856 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00000947 | 0 / 1 |
| Q9GZT9 | Egl nine homolog 1 | Enzyme | C00032950 | 1 / 1 |
| P54132 | Bloom syndrome protein | Enzyme | C00000947 | 1 / 2 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00000856 | 0 / 0 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00000856 | 1 / 8 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00000947 | 0 / 0 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00000947 | 3 / 2 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00000947 | 0 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00000947 | 0 / 0 |
| P08047 | Transcription factor Sp1 | Unclassified protein | C00019308 | 0 / 0 |
| Q9Y3R4 | Sialidase-2 | Enzyme | C00000947 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00047901 | 11 / 10 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00000947 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00040714 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00000947 | 0 / 0 |
| P07998 | Ribonuclease pancreatic | Enzyme | C00000947 | 0 / 0 |
| P28907 | ADP-ribosyl cyclase 1 | Enzyme | C00000947 | 0 / 1 |
| O75496 | Geminin | Unclassified protein | C00047901 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00000856 | 0 / 0 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00000947 | 0 / 0 |
| Q8N1Q1 | Carbonic anhydrase 13 | Lyase | C00000947 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00019308 | 0 / 0 |
| P07711 | Cathepsin L1 | C1A | C00000947 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00047901 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00000947 | 1 / 1 |
| P09923 | Intestinal-type alkaline phosphatase | Enzyme | C00000947 | 0 / 0 |
| P05186 | Alkaline phosphatase, tissue-nonspecific isozyme | Enzyme | C00000947 | 3 / 1 |
| P52209 | 6-phosphogluconate dehydrogenase, decarboxylating | Enzyme | C00000947 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00000947 | 1 / 1 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00000856 | 1 / 1 |
| P04745 | Alpha-amylase 1 | Enzyme | C00000947 | 0 / 0 |
| P08581 | Hepatocyte growth factor receptor | TK tyrosine-protein kinase MET subfamily | C00000947 | 2 / 3 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00000947 | 0 / 0 |
| P10696 | Alkaline phosphatase, placental-like | Enzyme | C00000947 | 0 / 1 |
| P56817 | Beta-secretase 1 | A1A | C00000947 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00000947 | 0 / 0 |
| Q13093 | Platelet-activating factor acetylhydrolase | Enzyme | C00000947 | 3 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00000947 | 3 / 3 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00000947 | 2 / 2 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00000947 | 5 / 2 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00047901 | 0 / 0 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00000856 | 0 / 0 |
| P00374 | Dihydrofolate reductase | Oxidoreductase | C00000947 | 1 / 1 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00000947 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00000947 | 0 / 0 |
| P08238 | Heat shock protein HSP 90-beta | Other cytosolic protein | C00047901 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00000947 | 0 / 0 |
| P51580 | Thiopurine S-methyltransferase | Enzyme | C00000856 | 1 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00000947 | 1 / 1 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00000947 | 0 / 3 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00000947 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00047901 | 1 / 1 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00000947 | 0 / 0 |
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00000856 | 1 / 1 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00000856 | 0 / 0 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00000856 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00047901 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00000947 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000947 | 1 / 0 |
| P10415 | Apoptosis regulator Bcl-2 | Other cytosolic protein | C00000947 | 0 / 7 |
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | C00000947 | 3 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00000947 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00000947 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00000947 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00000947 | 1 / 1 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00047901 | 1 / 0 |
| O00255 | Menin | Unclassified protein | C00000947 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00000947 | 1 / 2 |
| Q05513 | Protein kinase C zeta type | Iota | C00000947 | 0 / 0 |
| Q04759 | Protein kinase C theta type | Delta | C00000947 | 0 / 1 |
| Q02156 | Protein kinase C epsilon type | Eta | C00000947 | 0 / 0 |
| O94806 | Serine/threonine-protein kinase D3 | Pkd | C00000947 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00000947 | 0 / 0 |
| Q05655 | Protein kinase C delta type | Delta | C00000947 | 0 / 0 |
| P05129 | Protein kinase C gamma type | Alpha | C00000947 | 1 / 1 |
| P05771 | Protein kinase C beta type | Alpha | C00000947 | 0 / 0 |
| P24723 | Protein kinase C eta type | Eta | C00000947 | 1 / 0 |
| P41743 | Protein kinase C iota type | Iota | C00000947 | 0 / 0 |
| Q15139 | Serine/threonine-protein kinase D1 | Pkd | C00000947 | 0 / 0 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00000947 | 0 / 0 |
169 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00000856
C00000947
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00000947
C00047901
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00000947
C00047901
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00000947
C00047901
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00000947
C00047901
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00000947
C00047901
|
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative |
C00000947
C00047901
|
| 3683 | ITGAL, CD11A, LFA-1, LFA1A | integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
C00000947
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00047901
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00047901
|
| 84159 | ARID5B, DESRT, MRF-2, MRF2 | AT rich interactive domain 5B (MRF1-like) |
C00047901
|
| 8209 | C21orf33, ES1, GT335, HES1, KNPH, KNPI | chromosome 21 open reading frame 33 |
C00047901
|
| 27101 | CACYBP, GIG5, RP1-102G20.6, S100A6BP, SIP | calcyclin binding protein |
C00047901
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00047901
|
| 1646 | AKR1C2, AKR1C-pseudo, BABP, DD, DD2, DDH2, HAKRD, HBAB, MCDR2, SRXY8 | aldo-keto reductase family 1, member C2 (EC:1.3.1.20 1.1.1.357) |
C00047901
|
| 1645 | AKR1C1, 2-ALPHA-HSD, 20-ALPHA-HSD, C9, DD1, DD1/DD2, DDH, DDH1, H-37, HAKRC, HBAB, MBAB | aldo-keto reductase family 1, member C1 (EC:1.3.1.20 1.1.1.149 1.1.1.112) |
C00047901
|
| 874 | CBR3, SDR21C2, hCBR3 | carbonyl reductase 3 (EC:1.1.1.184) |
C00047901
|
| 8317 | CDC7, CDC7L1, HsCDC7, Hsk1, huCDC7 | cell division cycle 7 (EC:2.7.11.1) |
C00047901
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00047901
|
| 26973 | CHORDC1, CHP1 | cysteine and histidine-rich domain (CHORD) containing 1 |
C00047901
|
| 10370 | CITED2, ASD8, MRG-1, MRG1, P35SRJ, VSD2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
C00047901
|
| 10598 | AHSA1, AHA1, C14orf3, p38 | AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
C00047901
|
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00047901
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00047901
|
| 3301 | DNAJA1, DJ-2, DjA1, HDJ2, HSDJ, HSJ2, HSPF4, NEDD7, hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 |
C00047901
|
| 11080 | DNAJB4, DNAJW, DjB4, HLJ1 | DnaJ (Hsp40) homolog, subfamily B, member 4 |
C00047901
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00047901
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00047901
|
| 11153 | FICD, HIP13, HYPE, UNQ3041 | FIC domain containing (EC:2.7.7.n1) |
C00047901
|
| 2322 | FLT3, CD135, FLK-2, FLK2, STK1 | fms-related tyrosine kinase 3 (EC:2.7.10.1) |
C00047901
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00047901
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00047901
|
| 54676 | GTPBP2 | GTP binding protein 2 |
C00047901
|
| 29923 | HILPDA, C7orf68, HIG-2, HIG2 | hypoxia inducible lipid droplet-associated |
C00047901
|
| 3157 | HMGCS1, HMGCS | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) (EC:2.3.3.10) |
C00047901
|
| 3952 | LEP, LEPD, OB, OBS | leptin |
C00000856
|
| 3320 | HSP90AA1, EL52, HSP86, HSP89A, HSP90A, HSP90N, HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 |
C00047901
|
| 3303 | HSPA1A, HSP70-1, HSP70-1A, HSP70I, HSP72, HSPA1 | heat shock 70kDa protein 1A |
C00047901
|
| 3304 | HSPA1B, HSP70-1B, HSP70-2 | heat shock 70kDa protein 1B |
C00047901
|
| 3309 | HSPA5, BIP, GRP78, MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
C00047901
|
| 10808 | HSPH1, HSP105, HSP105A, HSP105B, NY-CO-25 | heat shock 105kDa/110kDa protein 1 |
C00047901
|
| 3557 | IL1RN, DIRA, ICIL-1RA, IL-1RN, IL-1ra, IL-1ra3, IL1F3, IL1RA, IRAP, MVCD4 | interleukin 1 receptor antagonist |
C00047901
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00047901
|
| 182 | JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1 | jagged 1 |
C00047901
|
| 3781 | KCNN2, KCa2.2, SK2, SKCA2, SKCa_2, hSK2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
C00047901
|
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00047901
|
| 23175 | LPIN1, PAP1 | lipin 1 (EC:3.1.3.4) |
C00047901
|
| 26046 | LTN1, C21orf10, C21orf98, RNF160, ZNF294 | listerin E3 ubiquitin protein ligase 1 |
C00047901
|
| 9935 | MAFB, KRML, MCTO | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
C00047901
|
| 4191 | MDH2, M-MDH, MDH, MGC:3559, MOR1 | malate dehydrogenase 2, NAD (mitochondrial) (EC:1.1.1.37) |
C00047901
|
| 4199 | ME1, HUMNDME, MES | malic enzyme 1, NADP(+)-dependent, cytosolic (EC:1.1.1.40) |
C00047901
|
| 4335 | MNT, MAD6, MXD6, ROX, bHLHd3 | MAX network transcriptional repressor |
C00047901
|
| 29074 | MRPL18, L18mt, MRP-L18 | mitochondrial ribosomal protein L18 |
C00047901
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00047901
|
| 6307 | MSMO1, DESP4, ERG25, SC4MOL | methylsterol monooxygenase 1 (EC:1.14.13.72) |
C00047901
|
| 23077 | MYCBP2, PAM | MYC binding protein 2, E3 ubiquitin protein ligase |
C00047901
|
| 55892 | MYNN, OSZF, ZBTB31, ZNF902 | myoneurin |
C00047901
|
| 4683 | NBN, AT-V1, AT-V2, ATV, NBS, NBS1, P95 | nibrin |
C00047901
|
| 4824 | NKX3-1, BAPX2, NKX3, NKX3.1, NKX3A | NK3 homeobox 1 |
C00047901
|
| 9315 | NREP, C5orf13, D4S114, P311, PRO1873, PTZ17, SEZ17 | neuronal regeneration related protein |
C00047901
|
| 4986 | OPRK1, K-OR-1, KOR, KOR-1, OPRK | opioid receptor, kappa 1 |
C00047901
|
| 51526 | OSER1, C20orf111, HSPC207, Osr1, Perit1, dJ1183I21.1 | oxidative stress responsive serine-rich 1 |
C00047901
|
| 5152 | PDE9A, HSPDE9A2 | phosphodiesterase 9A (EC:3.1.4.35) |
C00047901
|
| 5293 | PIK3CD, P110DELTA, PI3K, p110D | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta (EC:2.7.1.153) |
C00047901
|
| 5295 | PIK3R1, AGM7, GRB1, p85, p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
C00047901
|
| 10105 | PPIF, CYP3, CyP-M, Cyp-D, CypD | peptidylprolyl isomerase F (EC:5.2.1.8) |
C00047901
|
| 5813 | PURA, PUR-ALPHA, PUR1, PURALPHA | purine-rich element binding protein A |
C00047901
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00047901
|
| 27230 | SERP1, RAMP4 | stress-associated endoplasmic reticulum protein 1 |
C00047901
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00047901
|
| 6520 | SLC3A2, 4F2, 4F2HC, 4T2HC, CD98, CD98HC, MDU1, NACAE | solute carrier family 3 (amino acid transporter heavy chain), member 2 |
C00047901
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00047901
|
| 79811 | SLTM, Met | SAFB-like, transcription modulator |
C00047901
|
| 6659 | SOX4, EVI16 | SRY (sex determining region Y)-box 4 |
C00047901
|
| 100380807 |
C00047901
|
||
| 10963 | STIP1, HOP, IEF-SSP-3521, P60, STI1, STI1L | stress-induced-phosphoprotein 1 |
C00047901
|
| 51347 | TAOK3, DPK, JIK, MAP3K18 | TAO kinase 3 (EC:2.7.11.1) |
C00047901
|
| 7113 | TMPRSS2, PP9284, PRSS10 | transmembrane protease, serine 2 (EC:3.4.21.-) |
C00047901
|
| 10628 | TXNIP, EST01027, HHCPA78, THIF, VDUP1 | thioredoxin interacting protein |
C00047901
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00047901
|
| 7320 | UBE2B, E2-17kDa, HHR6B, HR6B, RAD6B, UBC2 | ubiquitin-conjugating enzyme E2B (EC:6.3.2.19) |
C00047901
|
| 7358 | UGDH, GDH, UDP-GlcDH, UDPGDH, UGD | UDP-glucose 6-dehydrogenase (EC:1.1.1.22) |
C00047901
|
| 79084 | WDR77, MEP-50, MEP50, Nbla10071, RP11-552M11.3, p44, p44/Mep50 | WD repeat domain 77 |
C00047901
|
| 23099 | ZBTB43, ZBTB22B, ZNF-X, ZNF297B | zinc finger and BTB domain containing 43 |
C00047901
|
| 80149 | ZC3H12A, MCPIP, MCPIP1, RP3-423B22.1, dJ423B22.1 | zinc finger CCCH-type containing 12A |
C00047901
|
| 9429 | ABCG2, ABC15, ABCP, BCRP, BCRP1, BMDP, CD338, CDw338, EST157481, GOUT1, MRX, MXR, MXR1, UAQTL1 | ATP-binding cassette, sub-family G (WHITE), member 2 |
C00000947
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00000947
|
| 302 | ANXA2, ANX2, ANX2L4, CAL1H, LIP2, LPC2, LPC2D, P36, PAP-IV | annexin A2 |
C00000947
|
| 393 | ARHGAP4, C1, RGC1, RhoGAP4, SrGAP4, p115 | Rho GTPase activating protein 4 |
C00000947
|
| 468 | ATF4, CREB-2, CREB2, TAXREB67, TXREB | activating transcription factor 4 |
C00000947
|
| 472 | ATM, AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | ataxia telangiectasia mutated (EC:2.7.11.1) |
C00000947
|
| 2683 | B4GALT1, B4GAL-T1, CDG2D, GGTB2, GT1, GTB, beta4Gal-T1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (EC:2.4.1.22 2.4.1.90 2.4.1.38) |
C00000947
|
| 56751 | BARHL1 | BarH-like homeobox 1 |
C00000947
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00000947
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00000947
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00000947
|
| 388962 | BOLA3, MMDS2 | bolA family member 3 |
C00000947
|
| 79184 | BRCC3, BRCC36, C6.1A, CXorf53 | BRCA1/BRCA2-containing complex, subunit 3 (EC:3.1.2.15) |
C00000947
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00000947
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00000947
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00000947
|
| 930 | CD19, B4, CVID3 | CD19 molecule |
C00000947
|
| 975 | CD81, CVID6, S5.7, TAPA1, TSPAN28 | CD81 molecule |
C00000947
|
| 928 | CD9, BTCC-1, DRAP-27, MIC3, MRP-1, TSPAN-29, TSPAN29 | CD9 molecule |
C00000947
|
| 1054 | CEBPG, GPE1BP, IG/EBP-1 | CCAAT/enhancer binding protein (C/EBP), gamma |
C00000947
|
| 10978 | CLP1, HEAB, hClp1 | cleavage and polyadenylation factor I subunit 1 (EC:2.7.1.78) |
C00000947
|
| 10063 | COX17 | COX17 cytochrome c oxidase copper chaperone |
C00000947
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00000947
|
| 54205 | CYCS, CYC, HCS, THC4 | cytochrome c, somatic |
C00000947
|
| 7913 | DEK, D6S231E | DEK oncogene |
C00000947
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00000947
|
| 2130 | EWSR1, EWS, bK984G1.4 | EWS RNA-binding protein 1 |
C00000947
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00000947
|
| 2323 | FLT3LG, FL, FLT3L | fms-related tyrosine kinase 3 ligand |
C00000947
|
| 2495 | FTH1, FHC, FTH, FTHL6, PIG15, PLIF | ferritin, heavy polypeptide 1 (EC:1.16.3.1) |
C00000947
|
| 2896 | GRN, CLN11, GEP, GP88, PCDGF, PEPI, PGRN | granulin |
C00000947
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00000947
|
| 3054 | HCFC1, CFF, HCF-1, HCF1, HFC1, MRX3, VCAF | host cell factor C1 (VP16-accessory protein) |
C00000947
|
| 3232 | HOXD3, HOX1D, HOX4, HOX4A, Hox-4.1 | homeobox D3 |
C00000947
|
| 3329 | HSPD1, CPN60, GROEL, HLD4, HSP-60, HSP60, HSP65, HuCHA60, SPG13 | heat shock 60kDa protein 1 (chaperonin) |
C00000947
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00000947
|
| 3586 | IL10, CSIF, GVHDS, IL-10, IL10A, TGIF | interleukin 10 |
C00000947
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00000947
|
| 3559 | IL2RA, CD25, IDDM10, IL2R, TCGFR | interleukin 2 receptor, alpha |
C00000947
|
| 9235 | IL32, IL-32alpha, IL-32beta, IL-32delta, IL-32gamma, NK4, TAIF, TAIFa, TAIFb, TAIFc, TAIFd | interleukin 32 |
C00000947
|
| 3662 | IRF4, LSIRF, MUM1, NF-EM5 | interferon regulatory factor 4 |
C00000947
|
| 467 | ATF3 | activating transcription factor 3 |
C00047901
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00000947
|
| 4067 | LYN, JTK8, p53Lyn, p56Lyn | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (EC:2.7.10.2) |
C00000947
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00000947
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00000947
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00000947
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00000947
|
| 4282 | MIF, GIF, GLIF, MMIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) (EC:5.3.3.12 5.3.2.1) |
C00000947
|
| 64975 | MRPL41, BMRP, MRP-L27, MRPL27, RPML27 | mitochondrial ribosomal protein L41 |
C00000947
|
| 4493 | MT1E, MT1, MTD | metallothionein 1E |
C00000947
|
| 653361 | NCF1, NCF1A, NOXO2, SH3PXD1A, p47phox | neutrophil cytosolic factor 1 |
C00000947
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00000947
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00000947
|
| 4851 | NOTCH1, TAN1, hN1 | notch 1 |
C00000947
|
| 4869 | NPM1, B23, NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
C00000947
|
| 5175 | PECAM1, CD31, CD31/EndoCAM, GPIIA', PECA1, PECAM-1, endoCAM | platelet/endothelial cell adhesion molecule 1 |
C00000947
|
| 5187 | PER1, PER, RIGUI, hPER | period circadian clock 1 |
C00000947
|
| 5327 | PLAT, T-PA, TPA | plasminogen activator, tissue (EC:3.4.21.68) |
C00000947
|
| 5328 | PLAU, ATF, BDPLT5, QPD, UPA, URK, u-PA | plasminogen activator, urokinase (EC:3.4.21.73) |
C00000947
|
| 7916 | PRRC2A, BAT2, D6S51, D6S51E, G2 | proline-rich coiled-coil 2A |
C00000947
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00000947
|
| 5747 | PTK2, FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK | protein tyrosine kinase 2 (EC:2.7.10.2) |
C00000947
|
| 5879 | RAC1, Rac-1, TC-25, p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
C00000947
|
| 5915 | RARB, HAP, NR1B2, RRB2 | retinoic acid receptor, beta |
C00000947
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00000947
|
| 6128 | RPL6, L6, SHUJUN-2, TAXREB107, TXREB1 | ribosomal protein L6 |
C00000947
|
| 6285 | S100B, NEF, S100, S100-B, S100beta | S100 calcium binding protein B |
C00000947
|
| 6286 | S100P, MIG9 | S100 calcium binding protein P |
C00000947
|
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00000947
|
| 7536 | SF1, BBP, D11S636, MBBP, ZCCHC25, ZFM1, ZNF162 | splicing factor 1 |
C00000947
|
| 6574 | SLC20A1, GLVR1, Glvr-1, PIT1, PiT-1 | solute carrier family 20 (phosphate transporter), member 1 |
C00000947
|
| 6699 | SPRR1B, CORNIFIN, GADD33, SPRR1 | small proline-rich protein 1B |
C00000947
|
| 6772 | STAT1, CANDF7, ISGF-3, STAT91 | signal transducer and activator of transcription 1, 91kDa |
C00000947
|
| 6777 | STAT5B, STAT5 | signal transducer and activator of transcription 5B |
C00000947
|
| 6778 | STAT6, D12S1644, IL-4-STAT, STAT6B, STAT6C | signal transducer and activator of transcription 6, interleukin-4 induced |
C00000947
|
| 6932 | TCF7, TCF-1 | transcription factor 7 (T-cell specific, HMG-box) |
C00000947
|
| 7083 | TK1, TK2 | thymidine kinase 1, soluble (EC:2.7.1.21) |
C00000947
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00000947
|
| 943 | TNFRSF8, CD30, D1S166E, Ki-1 | tumor necrosis factor receptor superfamily, member 8 |
C00000947
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00000947
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00000947
|
| 2829 | XCR1, CCXCR1, GPR5 | chemokine (C motif) receptor 1 |
C00000947
|
| 51042 | ZNF593, ZT86 | zinc finger protein 593 |
C00000947
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (82)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #600807 | Asthma, susceptibility to |
Q13093
|
| #608584 | Asthma-related traits, susceptibility to, 2 |
Q6W5P4
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #114500 | Colorectal cancer; crc |
P84022
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #609820 | Erythrocytosis, familial, 3; ecyt3 |
Q9GZT9
|
| #133239 | Esophageal cancer |
P04637
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #114550 | Hepatocellular carcinoma |
P08581
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #146300 | Hypophosphatasia, adult |
P05186
|
| #241510 | Hypophosphatasia, childhood |
P05186
|
| #241500 | Hypophosphatasia, infantile |
P05186
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #147050 | Ige responsiveness, atopic; iger |
Q13093
|
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #211980 | Lung cancer |
P00533
P04637 |
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613839 | Megaloblastic anemia due to dihydrofolate reductase deficiency |
P00374
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #614278 | Platelet-activating factor acetylhydrolase deficiency; pafad |
Q13093
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #605074 | Renal cell carcinoma, papillary, 1; rccp1 |
P08581
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #605361 | Spinocerebellar ataxia 14; sca14 |
P05129
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
P04637
|
| #601367 | Stroke, ischemic |
P00734
P24723 |
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #610460 | Thiopurine s-methyltransferase deficiency |
P51580
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (93)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
P04637 (marker) P08581 (related) Q16790 (marker) |
| H01197 | Dihydrofolate reductase (DHFR) deficiency |
P00374
(related)
|
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P04637 (related) P04637 (marker) P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
P04637 (related) P08581 (related) P10415 (related) |
| H00022 | Bladder cancer |
P00533
(related)
P04637 (related) |
| H00028 | Choriocarcinoma |
P00533
(related)
P04637 (related) P10415 (related) |
| H00030 | Cervical cancer |
P00533
(related)
P10415 (related) |
| H00042 | Glioma |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) |
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
Q01453 (related) |
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00004 | Chronic myeloid leukemia (CML) |
P04637
(related)
|
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
P10415 (related) P28907 (marker) |
| H00006 | Hairy-cell leukemia |
P04637
(related)
|
| H00008 | Burkitt lymphoma |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00010 | Multiple myeloma |
P04637
(related)
|
| H00013 | Small cell lung cancer |
P04637
(related)
P10415 (related) |
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00019 | Pancreatic cancer |
P04637
(related)
P04637 (marker) |
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) |
| H00025 | Penile cancer |
P04637
(related)
P04637 (marker) P35354 (related) |
| H00026 | Endometrial Cancer |
P04637
(related)
|
| H00027 | Ovarian cancer |
P04637
(related)
|
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00031 | Breast cancer |
P04637
(related)
|
| H00032 | Thyroid cancer |
P04637
(related)
|
| H00036 | Osteosarcoma |
P04637
(related)
P08684 (marker) |
| H00038 | Malignant melanoma |
P04637
(related)
|
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00041 | Kaposi's sarcoma |
P04637
(related)
P10415 (related) |
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00046 | Cholangiocarcinoma |
P04637
(related)
P08581 (related) P35354 (related) |
| H00047 | Gallbladder cancer |
P04637
(related)
|
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
P05129
(related)
Q99700 (related) Q9NUW8 (related) |
| H00213 | Hypophosphatasia |
P05186
(related)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00054 | Nasopharyngeal cancer |
P10415
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00023 | Testicular cancer |
P10696
(marker)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00964 | Thiopurine S-methyltransferase deficiency (TPMT deficiency) |
P51580
(related)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q03164
(related)
Q03164 (marker) |
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00408 | Type I diabetes mellitus |
Q04759
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00236 | Congenital polycythemia |
Q9GZT9
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
18 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D002493 | Central Nervous System Diseases |
C00019308
|
| D003072 | Cognition Disorders |
C00019308
|
| D013118 | Spinal Cord Diseases |
C00019308
|
| D012220 | Rhinitis |
C00000856
|
| D007938 | Leukemia |
C00047901
|
| D050197 | Atherosclerosis |
C00000947
|
| D001943 | Breast Neoplasms |
C00000947
|
| D002471 | Cell Transformation, Neoplastic |
C00000947
|
| D003556 | Cystitis |
C00000947
|
| D056486 | Drug-Induced Liver Injury |
C00000947
|
| D034381 | Hearing Loss |
C00000947
|
| D006331 | Heart Diseases |
C00000947
|
| D020138 | HYPERHOMOCYSTEINEMIA |
C00000947
|
| D008107 | Liver Diseases |
C00000947
|
| D017114 | Liver Failure, Acute |
C00000947
|
| D008113 | Liver Neoplasms |
C00000947
|
| D008325 | Mammary Neoplasms, Experimental |
C00000947
|
| D009362 | Neoplasm Metastasis |
C00000947
|