Morus australis
Species
KNApSAcK Entry
| Organism name | Morus australis |
|---|---|
| Genus | Morus |
| Family | Moraceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Morus australis |
|---|---|
| Linked NCBI taxonomy ID | 66392 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Moraceae |
|---|---|
| ID | 3487 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (15)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00005374
|
Quercetin
|
CHEMBL82242
CHEMBL479232 CHEMBL1437696 |
C012526
|
14 / 2 / 2 | 2 / 1 | No. 2 | No. 15 |
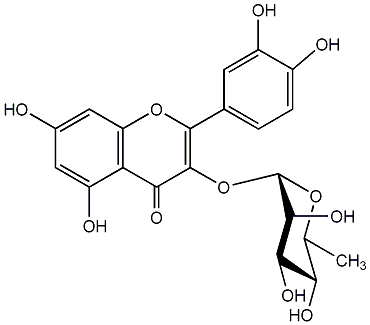
|
|
C00004028
|
Mulberrin
/ Kuwanon C / Norartocarpin / 2-(2,4-Dihydroxyphenyl)-5,7-dihydroxy-3,8-bis(3-methyl-2-butenyl)-4H-1-benzopyran-4-one |
CHEMBL518543
|
3 / 0 / 3 | No. 14 | No. 15 |

|
||
|
C00013442
|
Australone A
/ 2H,(-)-8-(2,4-Dihydroxyphenyl)-5-hydroxy-2-methyl-2-(4-methyl-3-pentenyl)-6H-benzo[1,2-b:5,4-b']dipyran-6-one |
No. 18 | No. 15 |

|
||||
|
C00004029
|
Cyclomorusin
/ Cyclomulberrochromene / 6,11-Dihydroxy-3,3-dimethyl-8-(2-methyl-1-propenyl)-3H,7H,8H-bis[1]benzopyrano[4,3-b:6',5'-e]pyran-7-one |
CHEMBL1770313
|
1 / 0 / 0 | No. 18 | No. 15 |

|
||
|
C00001070
|
Morusin
/ Mulberrochromene / 2-(2,4-Dihydroxyphenyl)-5-hydroxy-8,8-dimethyl-3-(3-methyl-2-butenyl)-4H,8H-benzo[1,2-b:3,4-b']dipyran-4-one |
CHEMBL464006
|
C057451
|
16 / 17 / 10 | No. 18 | No. 15 |
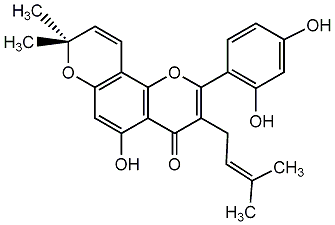
|
|
|
C00004059
|
Morusinol
/ Oxydihydromorusin / 2-(2,4-Dihydroxyphenyl)-5-hydroxy-3-(3-hydroxy-3-methylbutyl)-8,8-dimethyl-4H,8H-benzo[1,2-b:3,4-b']dipyran-4-one |
CHEMBL1719948
|
10 / 5 / 2 | No. 18 | No. 15 |

|
||
|
C00003738
|
beta-Amyrin
/ beta-Amirin / beta-Amyrine / beta-Amyrenol |
C036380
|
0 / 4 | No. 23 | No. 51 |
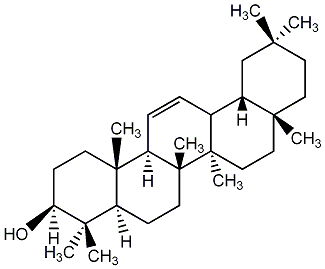
|
||
|
C00003741
|
Betulinic acid
|
CHEMBL269277
CHEMBL71690 CHEMBL519059 CHEMBL1318530 CHEMBL2005635 |
C002070
|
34 / 17 / 14 | 10 / 2 | No. 23 | No. 51 |
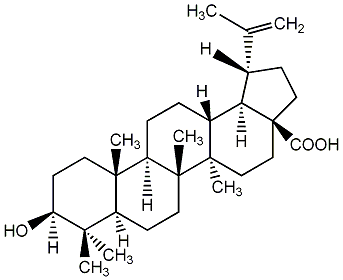
|
|
C00014246
|
Sanggenol N
/ (2S)-5,7,2'-Trihydroxy-5'-prenyl-6'',6''-dimethylpyrano[2'',3'':4',3']flavanone |
No. 39 |

|
|||||
|
C00001063
|
Albanin F
/ Kuwanone G |
CHEMBL444942
|
C095194
|
0 / 2 | No. 81 | No. 20 |
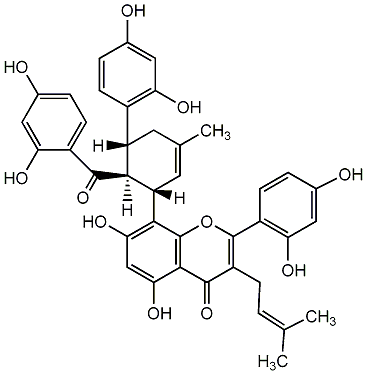
|
|
|
C00001064
|
Kuwanone H
|
CHEMBL506234
|
C095195
|
No. 81 | No. 20 |

|
||
|
C00029633
|
Ursolic acid
/ Acetylursolic acid |
CHEMBL55086
CHEMBL410525 |
4 / 2 / 2 | No. 177 |
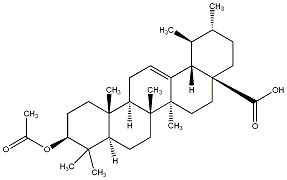
|
|||
|
C00036166
|
Mulberrofuran D
|
CHEMBL517247
|
1 / 0 / 0 | No. 319 | No. 15 |

|
||
|
C00019453
|
Sanggenofuran A
/ 6,5'-Dihydroxy-3'-methoxy-7-[(2E)-3,7-dimetyl-2,6-octadienyl]-2'-prenyl-2-arylbezofuran |
No. 319 | No. 15 |

|
||||
|
C00014245
|
Sanggenol O
/ (2S)-5,7-Dihydroxy-bis(6'',6''-dimethylpyrano[2'',3'':2',3'][2'',3'':4',5']flavanone |
No. 881 |

|
Human Protein / Gene in interactions
60 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P56817 | Beta-secretase 1 | A1A | C00001070 C00004028 C00004029 C00004059 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00001070 C00003741 C00004059 | 1 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00001070 C00003741 C00004059 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00001070 C00004059 C00005374 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00001070 C00004059 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00005374 C00029633 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00001070 C00004059 | 1 / 1 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00001070 C00004028 | 0 / 3 |
| P06746 | DNA polymerase beta | Enzyme | C00003741 C00029633 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00003741 C00005374 | 0 / 0 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00001070 C00004059 | 1 / 1 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00003741 C00005374 | 1 / 1 |
| Q8IUX4 | DNA dC->dU-editing enzyme APOBEC-3F | Enzyme | C00003741 C00004059 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00003741 C00005374 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00003741 C00004059 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00001070 C00004059 | 2 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00001070 C00005374 | 0 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00001070 C00004028 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00003741 | 1 / 1 |
| P11387 | DNA topoisomerase 1 | Isomerase | C00003741 | 0 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00003741 | 0 / 0 |
| P15559 | NAD(P)H dehydrogenase [quinone] 1 | Enzyme | C00003741 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00005374 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00003741 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00003741 | 0 / 1 |
| P08151 | Zinc finger protein GLI1 | Unclassified protein | C00003741 | 0 / 0 |
| Q02156 | Protein kinase C epsilon type | Eta | C00003741 | 0 / 0 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00005374 | 1 / 1 |
| P37840 | Alpha-synuclein | Unclassified protein | C00001070 | 4 / 2 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00003741 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00003741 | 11 / 10 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00001070 | 7 / 3 |
| Q04206 | Transcription factor p65 | Transcription Factor | C00003741 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00003741 | 0 / 0 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00005374 | 0 / 0 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | C00003741 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00029633 | 2 / 2 |
| P18054 | Arachidonate 12-lipoxygenase, 12S-type | Enzyme | C00001070 | 2 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00003741 | 0 / 0 |
| Q13133 | Oxysterols receptor LXR-alpha | NR1H3 | C00003741 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00003741 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00003741 | 0 / 0 |
| P55055 | Oxysterols receptor LXR-beta | NR1H3 | C00003741 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00003741 | 0 / 0 |
| P05771 | Protein kinase C beta type | Alpha | C00003741 | 0 / 0 |
| P07237 | Protein disulfide-isomerase | Enzyme | C00005374 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00003741 | 0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00005374 | 0 / 0 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00001070 | 0 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00003741 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00001070 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00003741 | 2 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00005374 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00029633 | 0 / 0 |
| Q9P1W9 | Serine/threonine-protein kinase pim-2 | Pim | C00005374 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00036166 | 0 / 0 |
| Q9UBT2 | SUMO-activating enzyme subunit 2 | Enzyme | C00003741 | 0 / 0 |
| Q9UBE0 | SUMO-activating enzyme subunit 1 | Unclassified protein | C00003741 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00005374 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00003741 | 1 / 0 |
12 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00003741
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00003741
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00003741
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00003741
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00003741
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00003741
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00003741
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00003741
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00003741
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00003741
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00005374
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00005374
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (36)
| OMIM | preferred title | UniProt |
|---|---|---|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #114500 | Colorectal cancer; crc |
P18054
P84022 |
| #127750 | Dementia, lewy body; dlb |
P37840
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #133239 | Esophageal cancer |
P18054
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #168601 | Parkinson disease 1, autosomal dominant; park1 |
P37840
|
| #605543 | Parkinson disease 4, autosomal dominant; park4 |
P37840
|
| #168600 | Parkinson disease, late-onset; pd |
P37840
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (27)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00017 | Esophageal cancer |
P35354
(related)
|
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00057 | Parkinson's disease (PD) |
P37840
(related)
|
| H00066 | Lewy body dementia (LBD) |
P37840
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
Q13148
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
8 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D010146 | Pain |
C00005374
C00003738 |
| D006930 | Hyperalgesia |
C00003738
|
| D008103 | Liver Cirrhosis |
C00003741
|
| D005157 | Facial Pain |
C00003738
|
| D008545 | Melanoma |
C00003741
|
| D007249 | Inflammation |
C00003738
|
| D003731 | Dental Caries |
C00001063
|
| D010510 | Periodontal Diseases |
C00001063
|