Fraxinus americana
Species
KNApSAcK Entry
| Organism name | Fraxinus americana |
|---|---|
| Genus | Fraxinus |
| Family | Oleaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Fraxinus americana |
|---|---|
| Linked NCBI taxonomy ID | 38872 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Oleaceae |
|---|---|
| ID | 4144 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (14)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
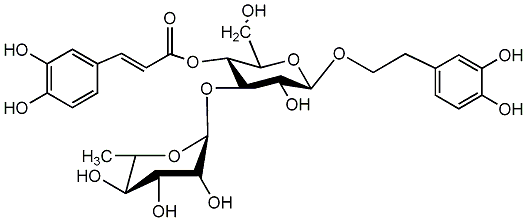
C00002783
|
Kusaginin
/ Acteoside / Verbascoside |
CHEMBL231853
CHEMBL444478 CHEMBL1364101 |
C058956
|
12 / 1 / 1 | 34 / 3 | No. 33 |
|
|
|
C00036871
|
Campneoside I
|
No. 33 |

|
|||||
|
C00037161
|
Fraxamoside
/ (-)-Fraxamoside |
No. 243 |

|
|||||
|
C00037421
|
Ligstroside
/ (-)-Ligstroside |
CHEMBL1086877
|
1 / 1 / 0 | No. 243 |

|
|||
|
C00037025
|
Demethylligstroside
/ (-)-Demethylligstroside |
No. 243 |

|
|||||
|
C00003093
|
Oleuropein
|
CHEMBL1911053
|
C002769
|
8 / 5 | No. 243 |

|
||
|
C00036342
|
(2''R)-2''-Hydroxyoleuropein
|
No. 243 |

|
|||||
|
C00036343
|
(2''S)-2''-Hydroxyoleuropein
|
No. 243 |

|
|||||
|
C00037160
|
Frameroside
|
No. 389 |

|
|||||
|
C00033258
|
Nucenide
/ Nuezhenide |
CHEMBL1076818
|
C083178
|
2 / 1 / 0 | No. 545 |

|
||
|
C00033266
|
Oleoside dimethyl ester
/ (-)-Oleoside dimethyl ester |
CHEMBL1087778
|
2 / 1 / 0 | No. 924 |

|
|||
|
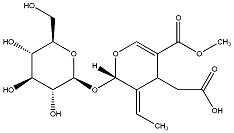
C00037580
|
Oleoside 11-methyl ester
/ (-)-Oleoside 11-methyl ester |
CHEMBL1086878
|
1 / 1 / 0 | No. 924 |
|
|||
|
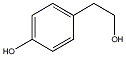
C00029515
|
p-Tyrosol
/ 4-(2-Hydroxyethyl)phenol / p-Hydroxyphenethyl alcohol / 2-(4-Hydroxyphenyl)ethanol |
CHEMBL53566
|
C011867
|
6 / 3 / 5 | No. 936 | No. 6 |
|
|
|
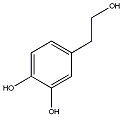
C00032635
|
3-Hydroxytyrosol
/ 3,4-Dihydroxyphenethyl alcohol |
CHEMBL1950045
|
C005975
|
26 / 4 | No. 1114 | No. 6 |
|
Human Protein / Gene in interactions
19 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P14672 | Solute carrier family 2, facilitated glucose transporter member 4 | Unclassified protein | C00033258 C00033266 C00037421 C00037580 | 1 / 0 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | C00033258 C00033266 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002783 C00029515 | 0 / 0 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00002783 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00002783 | 0 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00029515 | 1 / 2 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002783 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00002783 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00029515 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00029515 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002783 | 0 / 0 |
| P08581 | Hepatocyte growth factor receptor | TK tyrosine-protein kinase MET subfamily | C00029515 | 2 / 3 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002783 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00002783 | 0 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00029515 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002783 | 1 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002783 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002783 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00002783 | 0 / 0 |
53 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00002783
C00003093
C00032635
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002783
C00003093
C00032635
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002783
C00003093
C00032635
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002783
C00003093
C00032635
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002783
C00003093
C00032635
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00003093
C00032635
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00003093
C00032635
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002783
C00032635
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002783
C00032635
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002783
C00032635
|
| 7019 | TFAM, MTTF1, MTTFA, TCF6, TCF6L1, TCF6L2, TCF6L3 | transcription factor A, mitochondrial |
C00032635
|
| 993 | CDC25A, CDC25A2 | cell division cycle 25A (EC:3.1.3.48) |
C00032635
|
| 37618 |
C00032635
|
||
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00032635
|
| 10922 | FASTK, FAST | Fas-activated serine/threonine kinase (EC:2.7.11.8) |
C00032635
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00032635
|
| 2936 | GSR | glutathione reductase (EC:1.8.1.7) |
C00032635
|
| 3567 | IL5, EDF, IL-5, TRF | interleukin 5 (colony-stimulating factor, eosinophil) |
C00032635
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00032635
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00032635
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00032635
|
| 1588 | CYP19A1, ARO, ARO1, CPV1, CYAR, CYP19, CYPXIX, P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00003093
|
| 4779 | NFE2L1, LCR-F1, NRF1, TCF11 | nuclear factor, erythroid 2-like 1 |
C00032635
|
| 5862 | RAB2A, LHX, RAB2 | RAB2A, member RAS oncogene family |
C00032635
|
| 6119 | RPA3, REPA3 | replication protein A3, 14kDa |
C00032635
|
| 6773 | STAT2, ISGF-3, P113, STAT113 | signal transducer and activator of transcription 2, 113kDa |
C00032635
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00032635
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00002783
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00002783
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002783
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002783
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00002783
|
| 896 | CCND3 | cyclin D3 |
C00002783
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00002783
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00002783
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00002783
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00002783
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002783
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00002783
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00002783
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002783
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002783
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00002783
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00002783
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002783
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00002783
|
| 27035 | NOX1, GP91-2, MOX1, NOH-1, NOH1 | NADPH oxidase 1 |
C00002783
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00002783
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002783
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00002783
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00002783
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00002783
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00002783
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (5)
| OMIM | preferred title | UniProt |
|---|---|---|
| #125853 | Diabetes mellitus, noninsulin-dependent; niddm |
P14672
|
| #114550 | Hepatocellular carcinoma |
P08581
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #605074 | Renal cell carcinoma, papillary, 1; rccp1 |
P08581
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
KEGG DISEASE (6)
| KEGG | name | UniProt |
|---|---|---|
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00018 | Gastric cancer |
P08581
(related)
|
| H00021 | Renal cell carcinoma |
P08581
(related)
|
| H00046 | Cholangiocarcinoma |
P08581
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
Diseases related to CTD interactions
12 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D064420 | Drug-Related Side Effects and Adverse Reactions |
C00003093
|
| D006331 | Heart Diseases |
C00003093
|
| D006937 | Hypercholesterolemia |
C00003093
|
| D006949 | Hyperlipidemias |
C00003093
|
| D015228 | Hypertriglyceridemia |
C00003093
|
| D003928 | Diabetic Nephropathies |
C00032635
|
| D050171 | Dyslipidemias |
C00032635
|
| D006943 | Hyperglycemia |
C00032635
|
| D008107 | Liver Diseases |
C00032635
|
| D007249 | Inflammation |
C00002783
|
| D007938 | Leukemia |
C00002783
|
| D014947 | Wounds and Injuries |
C00002783
|