Clerodendrum inerme
Species
KNApSAcK Entry
| Organism name | Clerodendrum inerme |
|---|---|
| Genus | Clerodendrum |
| Family | Labiatae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Clerodendrum inerme |
|---|---|
| Linked NCBI taxonomy ID | 49994 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Lamiaceae |
|---|---|
| ID | 4136 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (23)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
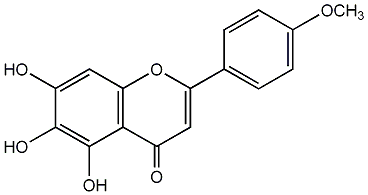
C00003836
|
Scutellarein 4'-methyl ether
|
No. 3 | No. 15 |
|
||||
|
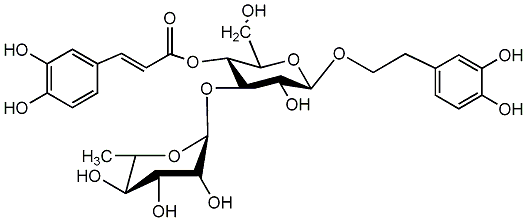
C00002783
|
Kusaginin
/ Acteoside / Verbascoside |
CHEMBL231853
CHEMBL444478 CHEMBL1364101 |
C058956
|
12 / 1 / 1 | 34 / 3 | No. 33 |
|
|
|
C00030657
|
Leucosceptoside A
|
CHEMBL450121
|
1 / 0 / 0 | No. 33 |

|
|||
|
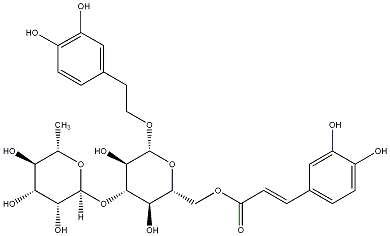
C00030516
|
Isoacteoside
/ Isoverbascoside |
CHEMBL504873
|
C064683
|
6 / 5 / 4 | No. 33 |
|
||
|
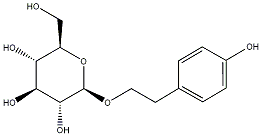
C00031274
|
Salidroside
/ Rhodioloside / (-)-Rhodioloside / Tyrosol alpha-(beta-D-glucopyranoside) |
CHEMBL465208
CHEMBL1899113 |
C009172
|
3 / 0 / 0 | No. 45 | No. 72 |
|
|
|
C00010619
|
Monomelittoside
|
No. 100 | No. 36 |

|
||||
|
C00036634
|
6-Nonacosanone
|
No. 115 |

|
|||||
|
C00036403
|
11-Pentacosanone
|
No. 115 |

|
|||||
|
C00037004
|
Darendoside B
|
CHEMBL462940
|
C080861
|
No. 128 | No. 72 |
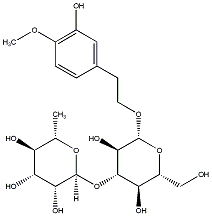
|
||
|
C00030098
|
Verbasoside
/ (-)-Verbasoside / Decaffeoylverbascoside / (-)-Decaffeoylverbascoside |
CHEMBL1779146
|
No. 128 | No. 72 |
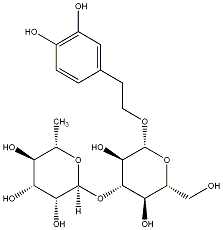
|
|||
|
C00032760
|
(-)-Benzyl beta-primeveroside
/ Benzyl alcohol beta-D-xylopyranosyl (1->6)-beta-D-glucopyranoside |
CHEMBL1371061
|
C069904
|
3 / 1 / 1 | No. 128 | No. 72 |
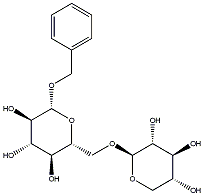
|
|
|
C00031281
|
Sammangaoside B
/ Summangaoside B / (-)-Sammangaoside B |
No. 153 |

|
|||||
|
C00031280
|
Sammangaoside A
/ Staphylionoside H / (-)-Sammangaoside A |
CHEMBL1814430
|
No. 153 |

|
||||
|
C00035203
|
14,15-Dihydro-15beta-methoxy-3-epicaryoptin
|
No. 172 | No. 43 |

|
||||
|
C00035204
|
14,15-Dihydro-15-hydroxy-3-epicaryoptin
|
No. 172 | No. 43 |

|
||||
|
C00036351
|
(7S,8R)-Dehydrodiconiferyl alcohol 4-O-beta-glucopyranoside
|
CHEMBL1086907
|
No. 193 |

|
||||
|
C00036352
|
(7S,8R)-Dehydrodiconiferyl alcohol 9-O-beta-glucopyranoside
|
CHEMBL466931
|
No. 193 |

|
||||
|
C00010621
|
Melittoside
|
No. 222 |

|
|||||
|
C00029346
|
(Z)-3-Hexenyl beta-D-glucopyranoside
/ (-)-(Z)-3-Hexenyl-beta-glucopyranoside / (Z)-3-Hexen-1-ol-beta-D-glucopyranoside |
CHEMBL2152486
|
C058860
|
No. 258 | No. 73 |

|
||
|
C00036919
|
Clerodermic acid
|
CHEMBL228501
|
No. 405 |

|
||||
|
C00037809
|
Seguinoside K
|
No. 411 |

|
|||||
|
C00036467
|
Leonuriside A
/ 2,6-Dimethoxy-p-hydroquinone 1-O-beta-glucopyranoside |
CHEMBL464406
|
2 / 4 / 2 | No. 678 | No. 6 |
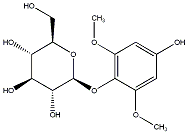
|
||
|
C00037783
|
Sammangaoside C
|
No. 1212 |

|
Human Protein / Gene in interactions
23 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| O75496 | Geminin | Unclassified protein | C00002783 C00031274 C00032760 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002783 C00031274 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00002783 C00030657 | 0 / 0 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00002783 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00036467 | 0 / 0 |
| P39900 | Macrophage metalloelastase | M10A | C00030516 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00030516 | 2 / 2 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00031274 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002783 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002783 | 1 / 1 |
| P03956 | Interstitial collagenase | M10A | C00030516 | 0 / 1 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00032760 | 1 / 1 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00002783 | 0 / 0 |
| P00734 | Prothrombin | S1A | C00036467 | 4 / 2 |
| P45452 | Collagenase 3 | M10A | C00030516 | 1 / 1 |
| P08253 | 72 kDa type IV collagenase | M10A | C00030516 | 1 / 3 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002783 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00002783 | 0 / 0 |
| P08254 | Stromelysin-1 | M10A | C00030516 | 1 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002783 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00032760 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002783 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002783 | 0 / 0 |
34 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00002783
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00002783
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002783
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002783
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00002783
|
| 896 | CCND3 | cyclin D3 |
C00002783
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00002783
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00002783
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00002783
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00002783
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002783
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00002783
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00002783
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002783
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002783
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00002783
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00002783
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00002783
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002783
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002783
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00002783
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002783
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002783
|
| 27035 | NOX1, GP91-2, MOX1, NOH-1, NOH1 | NADPH oxidase 1 |
C00002783
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00002783
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00002783
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002783
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002783
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00002783
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00002783
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00002783
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00002783
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002783
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002783
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (11)
| OMIM | preferred title | UniProt |
|---|---|---|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #602111 | Spondyloepimetaphyseal dysplasia, missouri type |
P45452
|
| #601367 | Stroke, ischemic |
P00734
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
KEGG DISEASE (8)
| KEGG | name | UniProt |
|---|---|---|
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00028 | Choriocarcinoma |
P03956
(related)
P08253 (related) |
| H00025 | Penile cancer |
P08253
(related)
P14780 (related) |
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
P45452 (related) |
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|