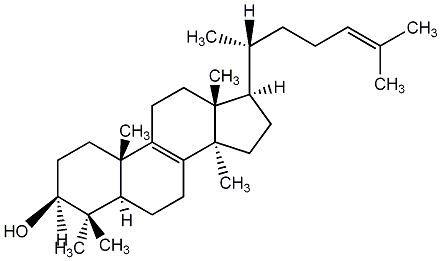
Metabolite
KNApSAcK Entry
| id | C00003657 |
|---|---|
| Name | Lanosterol / Lanosta-8,24-dien-3beta-ol |
| CAS RN | 79-63-0 |
| Standard InChI | InChI=1S/C30H50O/c1-20(2)10-9-11-21(3)22-14-18-30(8)24-12-13-25-27(4,5)26(31)16-17-28(25,6)23(24)15-19-29(22,30)7/h10,21-22,25-26,31H,9,11-19H2,1-8H3/t21-,22-,25+,26+,28-,29-,30+/m1/s1 |
| Standard InChI (Main Layer) | InChI=1S/C30H50O/c1-20(2)10-9-11-21(3)22-14-18-30(8)24-12-13-25-27(4,5)26(31)16-17-28(25,6)23(24)15-19-29(22,30)7/h10,21-22,25-26,31H,9,11-19H2,1-8H3 |
Cluster
| Phytochemical cluster | No. 51 |
|---|---|
| KCF-S cluster | No. 218 |
Link
ChEMBL
| By standard InChI | CHEMBL225111 |
|---|---|
| By standard InChI Main Layer | CHEMBL225111 CHEMBL465181 CHEMBL1397369 |
KEGG
| By LinkDB | C01724 |
|---|
CTD
| By CAS RN | D007810 |
|---|
Species
Summary
Family (Top 10)
| family name | count |
|---|---|
| Euphorbiaceae | 3 |
| Blastocladiaceae | 3 |
| Coriolaceae | 3 |
| Saccharomycetaceae | 2 |
| Physaraceae | 2 |
| Rhizidiomycetaceae | 1 |
| Pucciniaceae | 1 |
| Chytriomycetaceae | 1 |
| Phaffomycetaceae | 1 |
| Catenariaceae | 1 |
List (35)
* NCBI
Human Protein / Gene in interaction
10 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | CHEMBL1397369 |
CHEMBL1614331
(1)
|
0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | CHEMBL1397369 |
CHEMBL1614544
(1)
|
11 / 10 |
| Q16850 | Lanosterol 14-alpha demethylase | Cytochrome P450 51A1 | CHEMBL225111 |
CHEMBL934980
(1)
|
0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | CHEMBL1397369 |
CHEMBL1794311
(1)
|
2 / 3 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | CHEMBL1397369 |
CHEMBL1614458
(1)
|
0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | CHEMBL1397369 |
CHEMBL1794467
(1)
|
0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | CHEMBL1397369 |
CHEMBL1614240
(1)
|
0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | CHEMBL1397369 |
CHEMBL1614108
(1)
CHEMBL1613886
(1)
|
0 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | CHEMBL1397369 |
CHEMBL1614421
(1)
|
4 / 3 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | CHEMBL225111 |
CHEMBL2114915
(1)
|
0 / 0 |
CTD interaction (1)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| D007810 | 1593 |
CYP27A1
CP27 CTX CYP27 |
cytochrome P450, family 27, subfamily A, polypeptide 1 (EC:1.14.13.15) | CYP27A1 protein results in increased metabolism of Lanosterol |
increases metabolic processing
|
protein |
14622972
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (17)
| OMIM | preferred title | UniProt |
|---|---|---|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (17)
| KEGG | disease name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|