Flavobacterium sp. Strain R1534
Species
KNApSAcK Entry
| Organism name | Flavobacterium sp. Strain R1534 |
|---|---|
| Genus | Flavobacterium |
| Family | Flavobacteriaceae |
| Kingdom | Bacteria |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Flavobacterium |
|---|---|
| Linked NCBI taxonomy ID | 237 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Flavobacteriaceae |
|---|---|
| ID | 49546 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Bacteria |
|---|---|
| ID | 2 |
Plant class
| Plant class | |
|---|---|
| ID |
Metabolite list (12)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
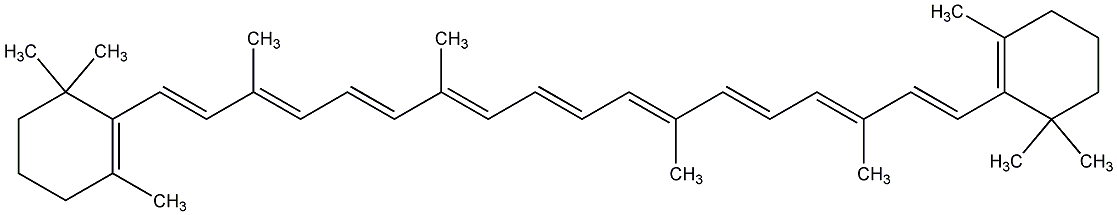
C00000919
|
beta-Carotene
|
CHEMBL1293
CHEMBL1966639 |
D019207
|
7 / 12 / 11 | 104 / 19 | No. 26 | No. 59 |
|
|
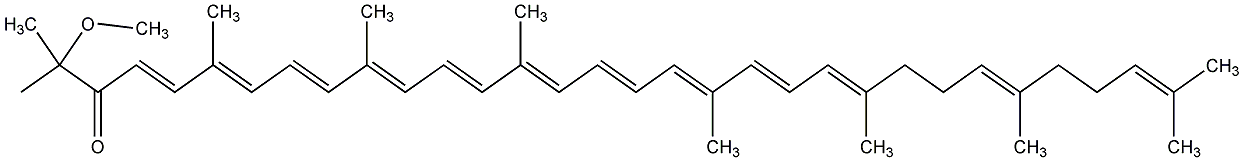
C00000914
|
Spheroidenone
|
C104880
|
No. 97 | No. 59 |
|
|||
|
C00000906
|
7,8,11,12-Tetrahydrolycopene
|
No. 97 | No. 59 |
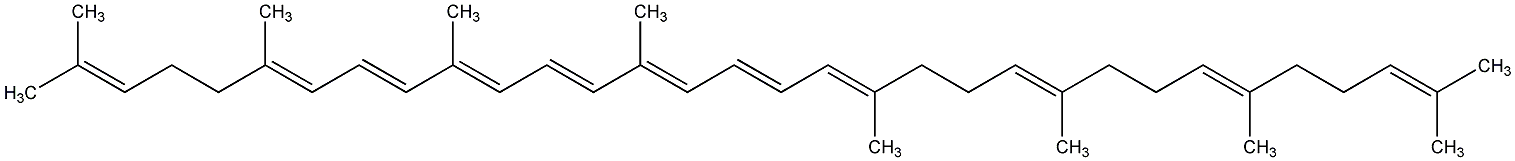
|
||||
|
C00000911
|
Lycopene
/ all-trans-Lycopene |
CHEMBL501174
CHEMBL1984187 |
C015329
|
6 / 13 / 13 | 84 / 13 | No. 97 | No. 59 |
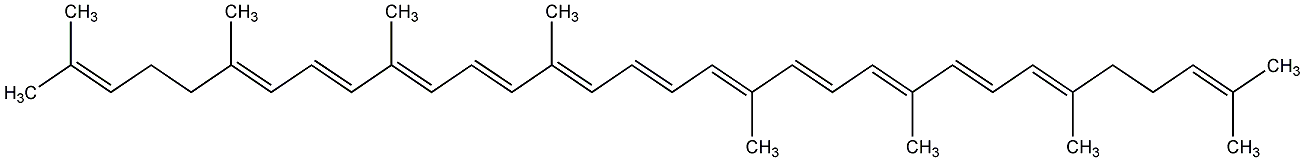
|
|
C00000913
|
Phytofluene
|
No. 97 | No. 59 |
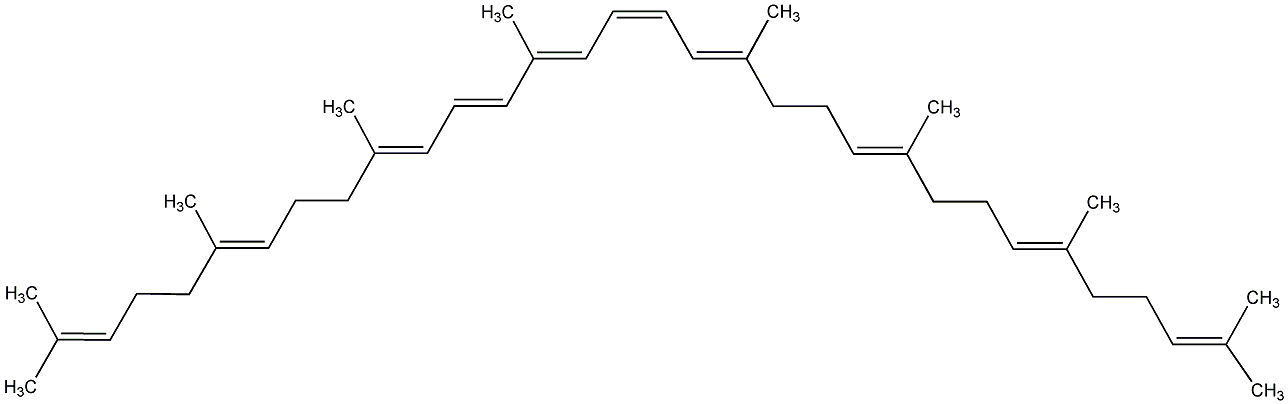
|
||||
|
C00000912
|
Neurosporene
|
C008604
|
No. 97 | No. 59 |
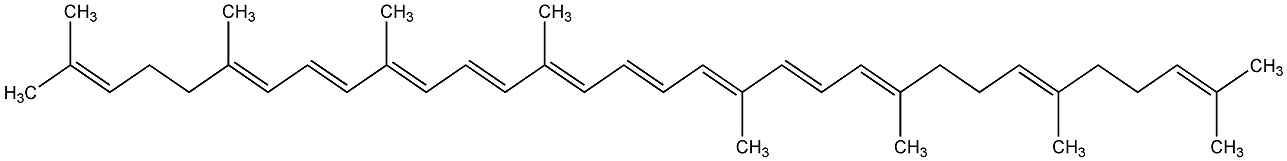
|
|||
|
C00000926
|
gamma-Carotene
|
No. 131 | No. 59 |
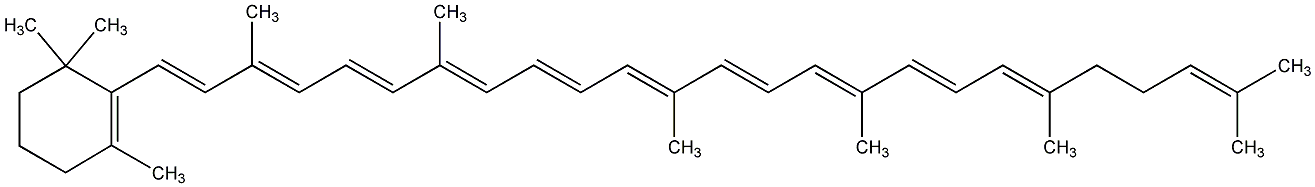
|
||||
|
C00000905
|
Phytoene
/ 15-cis-Phytoene / 15,15'-cis-Phytoene |
CHEMBL1988265
|
No. 2614 | No. 59 |
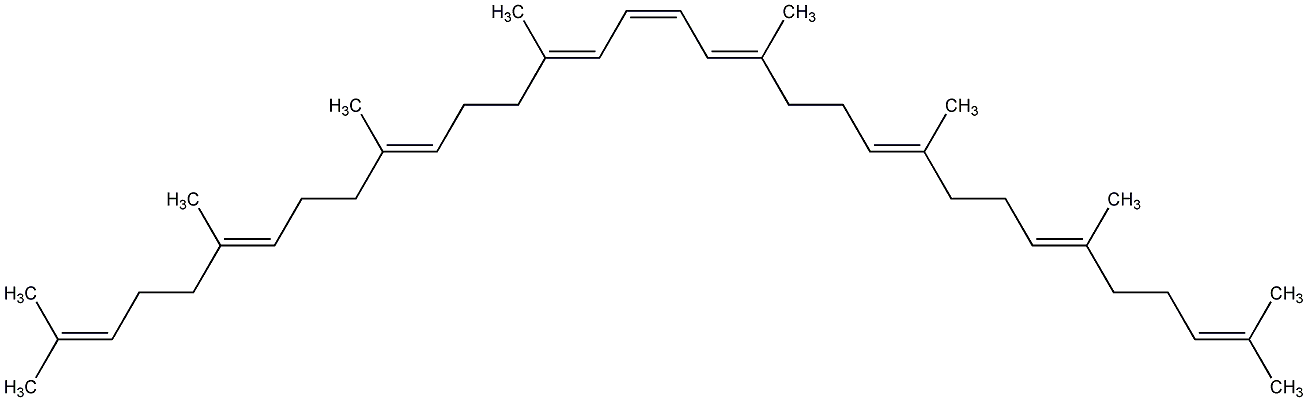
|
|||
|
C00000846
|
Geranyl diphosphate
/ Geranyl pyrophosphate |
CHEMBL41342
|
C015234
|
1 / 0 / 0 | 2 / 0 | No. 3475 | No. 34 |
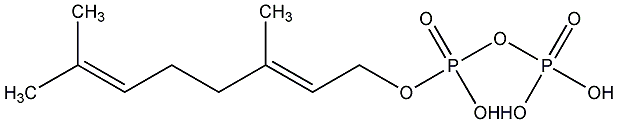
|
|
C00000908
|
GGPP
/ Geranylgeranyl diphosphate / Geranylgeranyl pyrophosphate |
CHEMBL1160059
CHEMBL1160061 CHEMBL1229266 |
C002963
|
3 / 0 / 0 | 34 / 0 | No. 4178 | No. 38 |
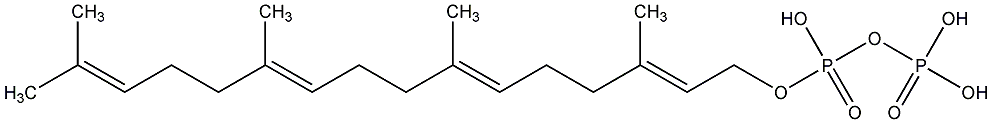
|
|
C00000907
|
FPP
/ Farnesyl pyrophosphate |
CHEMBL69330
CHEMBL1160056 CHEMBL1160060 |
C004808
|
5 / 0 / 0 | 11 / 1 | No. 4178 | No. 38 |
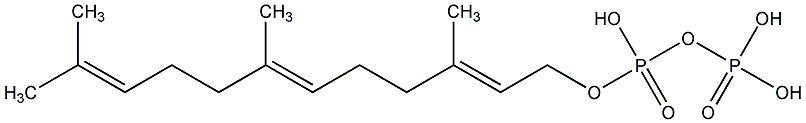
|
|
C00000910
|
Isopentenyl phosphate
|
CHEMBL342070
|
No. 8436 |
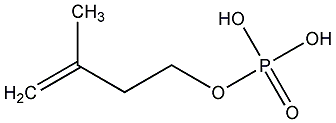
|
Human Protein / Gene in interactions
17 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P53609 | Geranylgeranyl transferase type-1 subunit beta | Enzyme | C00000907 C00000908 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00000911 C00000919 | 11 / 10 |
| P49354 | Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha | Enzyme | C00000907 C00000908 | 0 / 0 |
| P49356 | Protein farnesyltransferase subunit beta | Enzyme | C00000907 C00000908 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00000911 C00000919 | 0 / 0 |
| P14324 | Farnesyl pyrophosphate synthase | Transferase | C00000846 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00000919 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00000911 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00000919 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00000911 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00000911 | 0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00000911 | 2 / 3 |
| O95749 | Geranylgeranyl pyrophosphate synthase | Enzyme | C00000907 | 0 / 0 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00000919 | 0 / 1 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00000919 | 1 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00000919 | 0 / 0 |
| P37268 | Squalene synthase | Enzyme | C00000907 | 0 / 0 |
204 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00000907
C00000908
C00000911
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00000907
C00000908
C00000911
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00000908
C00000911
C00000919
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00000907
C00000908
C00000919
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00000908
C00000911
C00000919
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00000907
C00000908
C00000919
|
| 3156 | HMGCR, LDLCQ3 | 3-hydroxy-3-methylglutaryl-CoA reductase (EC:1.1.1.34) |
C00000846
C00000907
C00000908
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00000908
C00000911
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00000911
C00000919
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00000911
C00000919
|
| 3713 | IVL | involucrin |
C00000911
C00000919
|
| 3265 | HRAS, C-BAS/HAS, C-H-RAS, C-HA-RAS1, CTLO, H-RASIDX, HAMSV, HRAS1, K-RAS, N-RAS, RASH1 | Harvey rat sarcoma viral oncogene homolog |
C00000907
C00000908
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00000911
C00000919
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00000907
C00000911
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00000908
C00000919
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00000908
C00000919
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00000908
C00000919
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00000908
C00000919
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00000908
C00000911
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00000907
C00000908
|
| 4842 | NOS1, IHPS1, N-NOS, NC-NOS, NOS, bNOS, nNOS | nitric oxide synthase 1 (neuronal) (EC:1.14.13.39) |
C00000911
C00000919
|
| 387 | RHOA, ARH12, ARHA, RHO12, RHOH12 | ras homolog family member A |
C00000907
C00000908
|
| 5879 | RAC1, Rac-1, TC-25, p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
C00000907
C00000908
|
| 5444 | PON1, ESA, MVCD5, PON | paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81) |
C00000907
C00000908
|
| 4255 | MGMT | O-6-methylguanine-DNA methyltransferase (EC:2.1.1.63) |
C00000919
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00000908
|
| 8013 | NR4A3, CHN, CSMF, MINOR, NOR1, TEC | nuclear receptor subfamily 4, group A, member 3 |
C00000908
|
| 56729 | RETN, ADSF, FIZZ3, RETN1, RSTN, XCP1 | resistin |
C00000908
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00000908
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00000908
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00000908
|
| 4312 | MMP1, CLG, CLGN | matrix metallopeptidase 1 (interstitial collagenase) (EC:3.4.24.7) |
C00000908
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00000908
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00000908
|
| 3552 | IL1A, IL-1A, IL1, IL1-ALPHA, IL1F1 | interleukin 1, alpha |
C00000908
|
| 5906 | RAP1A, KREV-1, KREV1, RAP1, SMGP21 | RAP1A, member of RAS oncogene family |
C00000846
|
| 118 | ADD1, ADDA | adducin 1 (alpha) |
C00000911
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00000911
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00000911
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00000911
|
| 571 | BACH1, BACH-1 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
C00000911
|
| 8314 | BAP1, HUCEP-13, UCHL2 | BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (EC:3.4.19.12) |
C00000911
|
| 673 | BRAF, B-RAF1, BRAF1, NS7, RAFB1 | v-raf murine sarcoma viral oncogene homolog B (EC:2.7.11.1) |
C00000911
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00000911
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00000911
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00000911
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00000911
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00000911
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00000911
|
| 1020 | CDK5, PSSALRE | cyclin-dependent kinase 5 (EC:2.7.11.22) |
C00000911
|
| 1022 | CDK7, CAK1, CDKN7, HCAK, MO15, STK1, p39MO15 | cyclin-dependent kinase 7 (EC:2.7.11.23 2.7.11.22) |
C00000911
|
| 1025 | CDK9, C-2k, CDC2L4, CTK1, PITALRE, TAK | cyclin-dependent kinase 9 (EC:2.7.11.23 2.7.11.22) |
C00000911
|
| 1113 | CHGA, CGA | chromogranin A (parathyroid secretory protein 1) |
C00000911
|
| 1114 | CHGB, SCG1 | chromogranin B (secretogranin 1) |
C00000911
|
| 23418 | CRB1, LCA8, RP12 | crumbs homolog 1 (Drosophila) |
C00000911
|
| 9453 | GGPS1, GGPPS, GGPPS1 | geranylgeranyl diphosphate synthase 1 (EC:2.5.1.1 2.5.1.10 2.5.1.29) |
C00000908
|
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00000911
|
| 1981 | EIF4G1, EIF-4G1, EIF4F, EIF4G, EIF4GI, P220, PARK18 | eukaryotic translation initiation factor 4 gamma, 1 |
C00000911
|
| 2152 | F3, CD142, TF, TFA | coagulation factor III (thromboplastin, tissue factor) |
C00000908
|
| 1490 | CTGF, CCN2, HCS24, IGFBP8, NOV2 | connective tissue growth factor |
C00000908
|
| 2176 | FANCC, FA3, FAC, FACC | Fanconi anemia, complementation group C |
C00000911
|
| 2188 | FANCF, FAF | Fanconi anemia, complementation group F |
C00000911
|
| 356 | FASLG, ALPS1B, APT1LG1, APTL, CD178, CD95-L, CD95L, FASL, TNFSF6 | Fas ligand (TNF superfamily, member 6) |
C00000911
|
| 2701 | GJA4, CX37 | gap junction protein, alpha 4, 37kDa |
C00000911
|
| 2702 | GJA5, ATFB11, CX40 | gap junction protein, alpha 5, 40kDa |
C00000911
|
| 2879 | GPX4, GPx-4, GSHPx-4, MCSP, PHGPx, snGPx, snPHGPx | glutathione peroxidase 4 (EC:1.11.1.12) |
C00000911
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00000911
|
| 3479 | IGF1, IGF-I, IGF1A, IGFI | insulin-like growth factor 1 (somatomedin C) |
C00000911
|
| 3480 | IGF1R, CD221, IGFIR, IGFR, JTK13 | insulin-like growth factor 1 receptor (EC:2.7.10.1) |
C00000911
|
| 3485 | IGFBP2, IBP2, IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa |
C00000911
|
| 1401 | CRP, PTX1 | C-reactive protein, pentraxin-related |
C00000908
|
| 3716 | JAK1, JAK1A, JAK1B, JTK3 | Janus kinase 1 (EC:2.7.10.2) |
C00000911
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00000911
|
| 3796 | KIF2A, CDCBM3, HK2, KIF2 | kinesin heavy chain member 2A |
C00000911
|
| 3838 | KPNA2, IPOA1, QIP2, RCH1, SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
C00000911
|
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00000911
|
| 3988 | LIPA, CESD, LAL | lipase A, lysosomal acid, cholesterol esterase (EC:3.1.1.13) |
C00000911
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00000911
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00000911
|
| 6416 | MAP2K4, JNKK, JNKK1, MAPKK4, MEK4, MKK4, PRKMK4, SAPKK-1, SAPKK1, SEK1, SERK1, SKK1 | mitogen-activated protein kinase kinase 4 (EC:2.7.12.2) |
C00000911
|
| 5608 | MAP2K6, MAPKK6, MEK6, MKK6, PRKMK6, SAPKK-3, SAPKK3 | mitogen-activated protein kinase kinase 6 (EC:2.7.12.2) |
C00000911
|
| 1432 | MAPK14, CSBP, CSBP1, CSBP2, CSPB1, EXIP, Mxi2, PRKM14, PRKM15, RK, SAPK2A, p38, p38ALPHA | mitogen-activated protein kinase 14 (EC:2.7.11.24) |
C00000911
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00000911
|
| 4247 | MGAT2, CDG2A, CDGS2, GLCNACTII, GNT-II, GNT2 | mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (EC:2.4.1.143) |
C00000911
|
| 100507436 | MICA, MIC-A, PERB11.1 | MHC class I polypeptide-related sequence A |
C00000911
|
| 4292 | MLH1, COCA2, FCC2, HNPCC, HNPCC2, hMLH1 | mutL homolog 1 |
C00000911
|
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00000911
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00000911
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00000908
|
| 8021 | NUP214, CAIN, CAN, D9S46E, N214, p250 | nucleoporin 214kDa |
C00000911
|
| 5320 | PLA2G2A, MOM1, PLA2, PLA2B, PLA2L, PLA2S, PLAS1, sPLA2 | phospholipase A2, group IIA (platelets, synovial fluid) (EC:3.1.1.4) |
C00000911
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00000911
|
| 10111 | RAD50, NBSLD, RAD502, hRad50 | RAD50 homolog (S. cerevisiae) |
C00000911
|
| 8045 | RASSF7, C11orf13, HRAS1, HRC1 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
C00000911
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00000911
|
| 5962 | RDX, DFNB24 | radixin |
C00000911
|
| 6195 | RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1) |
C00000911
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00000911
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00000911
|
| 6414 | SEPP1, SELP, SeP | selenoprotein P, plasma, 1 |
C00000911
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00000911
|
| 8243 | SMC1A, CDLS2, DXS423E, SB1.8, SMC1, SMC1L1, SMC1alpha, SMCB | structural maintenance of chromosomes 1A |
C00000911
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00000911
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00000911
|
| 7980 | TFPI2, PP5, REF1, TFPI-2 | tissue factor pathway inhibitor 2 |
C00000911
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00000911
|
| 7097 | TLR2, CD282, TIL4 | toll-like receptor 2 |
C00000911
|
| 7099 | TLR4, ARMD10, CD284, TLR-4, TOLL | toll-like receptor 4 |
C00000911
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00000911
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00000911
|
| 7415 | VCP, ALS14, IBMPFD, IBMPFD1, TERA, p97 | valosin containing protein (EC:3.6.4.6) |
C00000911
|
| 7485 | WRB, CHD5 | tryptophan rich basic protein |
C00000911
|
| 7697 | ZNF138, pHZ-32 | zinc finger protein 138 |
C00000911
|
| 124 | ADH1A, ADH1 | alcohol dehydrogenase 1A (class I), alpha polypeptide (EC:1.1.1.1) |
C00000919
|
| 128 | ADH5, ADH-3, ADHX, FALDH, FDH, GSH-FDH, GSNOR | alcohol dehydrogenase 5 (class III), chi polypeptide (EC:1.1.1.1 1.1.1.284) |
C00000919
|
| 221 | ALDH3B1, ALDH4, ALDH7 | aldehyde dehydrogenase 3 family, member B1 (EC:1.2.1.5) |
C00000919
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00000919
|
| 287 | ANK2, ANK-2, LQT4, brank-2 | ankyrin 2, neuronal |
C00000919
|
| 288 | ANK3, ANKYRIN-G, MRT37 | ankyrin 3, node of Ranvier (ankyrin G) |
C00000919
|
| 427 | ASAH1, AC, ACDase, ASAH, PHP, PHP32, SMAPME | N-acylsphingosine amidohydrolase (acid ceramidase) 1 (EC:3.5.1.23) |
C00000919
|
| 51665 | ASB1, ASB-1 | ankyrin repeat and SOCS box containing 1 |
C00000919
|
| 140462 | ASB9 | ankyrin repeat and SOCS box containing 9 |
C00000919
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00000919
|
| 9530 | BAG4, BAG-4, SODD | BCL2-associated athanogene 4 |
C00000919
|
| 9529 | BAG5, BAG-5 | BCL2-associated athanogene 5 |
C00000919
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00000919
|
| 53630 | BCMO1, BCDO, BCDO1, BCMO, BCO, BCO1 | beta-carotene 15,15'-monooxygenase 1 (EC:1.14.99.36) |
C00000919
|
| 999 | CDH1, Arc-1, CD324, CDHE, ECAD, LCAM, UVO | cadherin 1, type 1, E-cadherin (epithelial) |
C00000919
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00000919
|
| 9071 | CLDN10, CPETRL3, OSP-L | claudin 10 |
C00000919
|
| 51208 | CLDN18, SFTA5, SFTPJ | claudin 18 |
C00000919
|
| 1282 | COL4A1, HANAC, ICH, POREN1, arresten | collagen, type IV, alpha 1 |
C00000919
|
| 1284 | COL4A2, ICH, POREN2 | collagen, type IV, alpha 2 |
C00000919
|
| 8738 | CRADD, MRT34, RAIDD | CASP2 and RIPK1 domain containing adaptor with death domain |
C00000919
|
| 1508 | CTSB, APPS, CPSB | cathepsin B (EC:3.4.22.1) |
C00000919
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00000919
|
| 6372 | CXCL6, CKA-3, GCP-2, GCP2, SCYB6 | chemokine (C-X-C motif) ligand 6 |
C00000919
|
| 4283 | CXCL9, CMK, Humig, MIG, SCYB9, crg-10 | chemokine (C-X-C motif) ligand 9 |
C00000919
|
| 7852 | CXCR4, CD184, D2S201E, FB22, HM89, HSY3RR, LAP3, LCR1, LESTR, NPY3R, NPYR, NPYRL, NPYY3R, WHIM | chemokine (C-X-C motif) receptor 4 |
C00000919
|
| 1592 | CYP26A1, CP26, CYP26, P450RAI, P450RAI1 | cytochrome P450, family 26, subfamily A, polypeptide 1 (EC:1.14.-.-) |
C00000919
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00000919
|
| 64816 | CYP3A43 | cytochrome P450, family 3, subfamily A, polypeptide 43 (EC:1.14.14.1) |
C00000919
|
| 1746 | DLX2, TES-1, TES1 | distal-less homeobox 2 |
C00000919
|
| 1748 | DLX4, BP1, DLX7, DLX8, DLX9 | distal-less homeobox 4 |
C00000919
|
| 2068 | ERCC2, COFS2, EM9, TTD, XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 (EC:3.6.4.12) |
C00000919
|
| 23017 | FAIM2, LFG, LFG2, NGP35, NMP35, TMBIM2 | Fas apoptotic inhibitory molecule 2 |
C00000919
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00000919
|
| 3169 | FOXA1, HNF3A, TCF3A | forkhead box A1 |
C00000919
|
| 2627 | GATA6 | GATA binding protein 6 |
C00000919
|
| 2633 | GBP1 | guanylate binding protein 1, interferon-inducible |
C00000919
|
| 81025 | GJA9, CX58, CX59, GJA10 | gap junction protein, alpha 9, 59kDa |
C00000919
|
| 373156 | GSTK1, GST, GST_13-13, GST13, GST13-13, GSTK1-1, hGSTK1 | glutathione S-transferase kappa 1 (EC:2.5.1.18) |
C00000919
|
| 2946 | GSTM2, GST4, GSTM, GSTM2-2, GTHMUS | glutathione S-transferase mu 2 (muscle) (EC:2.5.1.18) |
C00000919
|
| 2948 | GSTM4, GSTM4-4, GTM4 | glutathione S-transferase mu 4 (EC:2.5.1.18) |
C00000919
|
| 2949 | GSTM5, GSTM5-5, GTM5 | glutathione S-transferase mu 5 (EC:2.5.1.18) |
C00000919
|
| 2954 | GSTZ1, GSTZ1-1, MAAI, MAI | glutathione S-transferase zeta 1 (EC:2.5.1.18 5.2.1.2) |
C00000919
|
| 3204 | HOXA7, ANTP, HOX1, HOX1.1, HOX1A | homeobox A7 |
C00000919
|
| 3216 | HOXB6, HOX2, HOX2B, HU-2, Hox-2.2 | homeobox B6 |
C00000919
|
| 3226 | HOXC10, HOX3I | homeobox C10 |
C00000919
|
| 3448 | IFNA14, IFN-alphaH, LEIF2H | interferon, alpha 14 |
C00000919
|
| 3449 | IFNA16, IFN-alphaO | interferon, alpha 16 |
C00000919
|
| 3451 | IFNA17, IFN-alphaI, IFNA, INFA, LEIF2C1 | interferon, alpha 17 |
C00000919
|
| 3442 | IFNA5, IFN-alpha-5, IFN-alphaG, INA5, INFA5, leIF_G | interferon, alpha 5 |
C00000919
|
| 3394 | IRF8, H-ICSBP, ICSBP, ICSBP1, IRF-8 | interferon regulatory factor 8 |
C00000919
|
| 50805 | IRX4, IRXA3 | iroquois homeobox 4 |
C00000919
|
| 3688 | ITGB1, CD29, FNRB, GPIIA, MDF2, MSK12, VLA-BETA, VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
C00000919
|
| 3691 | ITGB4, CD104 | integrin, beta 4 |
C00000919
|
| 3875 | KRT18, CYK18, K18 | keratin 18 |
C00000919
|
| 3880 | KRT19, CK19, K19, K1CS | keratin 19 |
C00000919
|
| 3849 | KRT2, CK-2e, K2e, KRT2A, KRT2E, KRTE | keratin 2 |
C00000919
|
| 3855 | KRT7, CK7, K2C7, K7, SCL | keratin 7 |
C00000919
|
| 3856 | KRT8, CARD2, CK-8, CK8, CYK8, K2C8, K8, KO | keratin 8 |
C00000919
|
| 8022 | LHX3, CPHD3, LIM3, M2-LHX3 | LIM homeobox 3 |
C00000919
|
| 4153 | MBL2, COLEC1, HSMBPC, MBL, MBL2D, MBP, MBP-C, MBP1, MBPD | mannose-binding lectin (protein C) 2, soluble |
C00000919
|
| 4170 | MCL1, BCL2L3, EAT, MCL1-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT | myeloid cell leukemia sequence 1 (BCL2-related) |
C00000919
|
| 4212 | MEIS2, HsT18361, MRG1 | Meis homeobox 2 |
C00000919
|
| 4222 | MEOX1, KFS2, MOX1 | mesenchyme homeobox 1 |
C00000919
|
| 5155 | PDGFB, IBGC5, PDGF-2, PDGF2, SIS, SSV, c-sis | platelet-derived growth factor beta polypeptide |
C00000908
|
| 3110 | MNX1, HB9, HLXB9, HOXHB9, SCRA1 | motor neuron and pancreas homeobox 1 |
C00000919
|
| 4439 | MSH5, G7, MUTSH5, NG23 | mutS homolog 5 |
C00000919
|
| 4586 | MUC5AC, MUC5, TBM, leB | mucin 5AC, oligomeric mucus/gel-forming |
C00000919
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00000919
|
| 4973 | OLR1, CLEC8A, LOX1, LOXIN, SCARE1, SLOX1 | oxidized low density lipoprotein (lectin-like) receptor 1 |
C00000919
|
| 5098 | PCDHGC3, PC43, PCDH-GAMMA-C3, PCDH2 | protocadherin gamma subfamily C, 3 |
C00000919
|
| 5359 | PLSCR1, MMTRA1B | phospholipid scramblase 1 |
C00000919
|
| 57048 | PLSCR3 | phospholipid scramblase 3 |
C00000919
|
| 57088 | PLSCR4, TRA1 | phospholipid scramblase 4 |
C00000919
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00000919
|
| 5892 | RAD51D, BROVCA4, R51H3, RAD51L3, TRAD | RAD51 paralog D |
C00000919
|
| 5914 | RARA, NR1B1, RAR | retinoic acid receptor, alpha |
C00000919
|
| 5915 | RARB, HAP, NR1B2, RRB2 | retinoic acid receptor, beta |
C00000919
|
| 5918 | RARRES1, LXNL, TIG1 | retinoic acid receptor responder (tazarotene induced) 1 |
C00000919
|
| 5950 | RBP4, RDCCAS | retinol binding protein 4, plasma |
C00000919
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00000919
|
| 949 | SCARB1, CD36L1, CLA-1, CLA1, HDLQTL6, SR-BI, SRB1 | scavenger receptor class B, member 1 |
C00000919
|
| 4990 | SIX6, MCOPCT2, OPTX2, Six9 | SIX homeobox 6 |
C00000919
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00000919
|
| 9517 | SPTLC2, HSN1C, LCB2, LCB2A, NSAN1C, SPT2, hLCB2a | serine palmitoyltransferase, long chain base subunit 2 (EC:2.3.1.50) |
C00000919
|
| 6720 | SREBF1, SREBP-1c, SREBP1, bHLHd1 | sterol regulatory element binding transcription factor 1 |
C00000919
|
| 7052 | TGM2, G-ALPHA-h, GNAH, TG2, TGC | transglutaminase 2 (EC:2.3.2.13) |
C00000919
|
| 7047 | TGM4, TGP, hTGP | transglutaminase 4 (EC:2.3.2.13) |
C00000919
|
| 3604 | TNFRSF9, 4-1BB, CD137, CDw137, ILA | tumor necrosis factor receptor superfamily, member 9 |
C00000919
|
| 10190 | TXNDC9, APACD, PHLP3 | thioredoxin domain containing 9 |
C00000919
|
| 7517 | XRCC3, CMM6 | X-ray repair complementing defective repair in Chinese hamster cells 3 |
C00000919
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (14)
| OMIM | preferred title | UniProt |
|---|---|---|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (14)
| KEGG | name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
Diseases related to CTD interactions
31 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D056486 | Drug-Induced Liver Injury |
C00000911
C00000919 |
| D009369 | Neoplasms |
C00000911
C00000919 |
| D001145 | Arrhythmias, Cardiac |
C00000919
|
| D006965 | Hyperplasia |
C00000911
|
| D007248 | Infertility, Male |
C00000911
|
| D007249 | Inflammation |
C00000911
|
| D007674 | Kidney Diseases |
C00000911
|
| D010146 | Pain |
C00000907
|
| D020258 | Neurotoxicity Syndromes |
C00000911
|
| D011471 | Prostatic Neoplasms |
C00000911
|
| D012878 | Skin Neoplasms |
C00000911
|
| D013733 | Testicular Diseases |
C00000911
|
| D014178 | Translocation, Genetic |
C00000911
|
| D015431 | Weight Loss |
C00000911
|
| D000435 | Alcoholic Intoxication |
C00000919
|
| D006943 | Hyperglycemia |
C00000911
|
| D053627 | Asthenozoospermia |
C00000919
|
| D002545 | Brain Ischemia |
C00000919
|
| D006528 | Carcinoma, Hepatocellular |
C00000919
|
| D002318 | Cardiovascular Diseases |
C00000919
|
| D003930 | Diabetic Retinopathy |
C00000919
|
| D006526 | Hepatitis C |
C00000919
|
| D015658 | HIV Infections |
C00000919
|
| D006986 | Hypervitaminosis A |
C00000919
|
| D008107 | Liver Diseases |
C00000919
|
| D008113 | Liver Neoplasms |
C00000919
|
| D008114 | Liver Neoplasms, Experimental |
C00000919
|
| D008175 | Lung Neoplasms |
C00000919
|
| D009845 | Oligospermia |
C00000919
|
| D013272 | Stomach Diseases |
C00000919
|
| D013276 | Stomach Ulcer |
C00000919
|