Helichrysum forskahlii
Species
KNApSAcK Entry
| Organism name | Helichrysum forskahlii |
|---|---|
| Genus | Helichrysum |
| Family | Asteraceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Helichrysum forskahlii |
|---|---|
| Linked NCBI taxonomy ID | 630302 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Asteraceae |
|---|---|
| ID | 4210 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (20)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
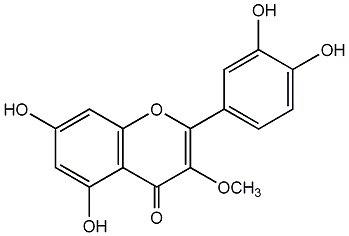
C00004632
|
Quercetin 3-methyl ether
/ Quercetin 3-O-methyl ether |
CHEMBL163316
|
32 / 21 / 21 | No. 3 | No. 15 |
|
||
|
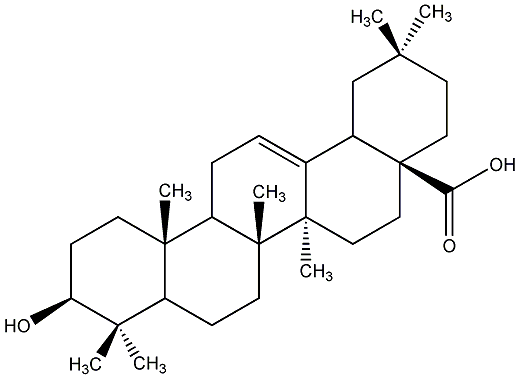
C00019064
|
Oleanolic acid
/ Astrantiagenin C / Virgaureagenin B / 3beta-Hydroxyolean-12-en-28-oic acid |
CHEMBL56615
CHEMBL168 CHEMBL180553 CHEMBL365375 CHEMBL486382 CHEMBL1413646 CHEMBL1436454 |
D009828
|
30 / 8 / 12 | 21 / 15 | No. 13 | No. 51 |
|
|
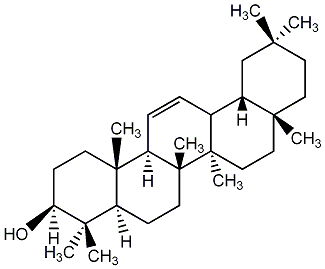
C00003738
|
beta-Amyrin
/ beta-Amirin / beta-Amyrine / beta-Amyrenol |
C036380
|
0 / 4 | No. 23 | No. 51 |
|
||
|
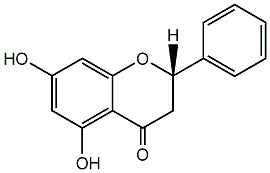
C00000992
|
Pinocembrin
|
CHEMBL70518
CHEMBL399910 CHEMBL399249 |
C016063
|
18 / 23 / 23 | 5 / 0 | No. 25 | No. 14 |
|
|
C00008143
|
Alpinetin
/ (-)-Alpinetin |
CHEMBL427218
CHEMBL254825 |
No. 25 | No. 14 |
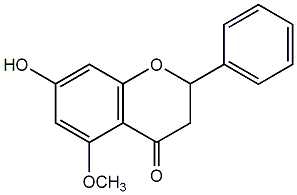
|
|||
|
C00000965
|
Glabranin
|
CHEMBL253998
CHEMBL488888 |
5 / 2 / 3 | No. 28 | No. 14 |

|
||
|
C00033902
|
Helichrysone C
/ (+)-Helichrysone C |
No. 28 | No. 14 |

|
||||
|
C00008189
|
7-O-Prenylpinocembrin
|
No. 28 | No. 14 |
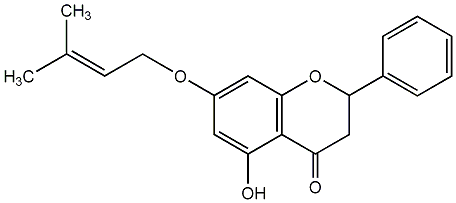
|
||||
|
C00008171
|
Isoglabranin
/ 6-Prenylpinocembrin / 5,7-Dihydroxy-6-C-prenylflavanone |
CHEMBL2143430
CHEMBL2165236 |
1 / 0 / 0 | No. 28 | No. 14 |
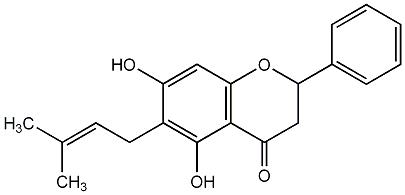
|
||
|
C00033901
|
Helichrysone B
|
No. 90 | No. 13 |

|
||||
|
C00006935
|
Cardamomin
|
CHEMBL378104
|
13 / 18 / 14 | No. 92 | No. 13 |
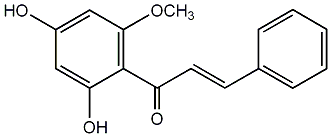
|
||
|
C00006933
|
2',4',6'-Trihydroxychalcone
|
CHEMBL129371
|
2 / 0 / 3 | No. 92 | No. 13 |
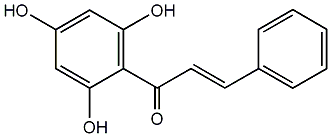
|
||
|
C00007086
|
2',6'-Dihydroxy-4'-prenyloxychalcone
|
No. 133 | No. 13 |

|
||||
|
C00007087
|
2'-Hydroxy-6'-methoxy-4'-prenyloxychalcone
|
No. 133 | No. 13 |

|
||||
|
C00033900
|
Helichrysone A
|
No. 133 | No. 13 |

|
||||
|
C00033565
|
Acetyloleanolic acid
/ Oleanolic acid acetate / 3-O-Acetyloleanolic acid / (+)-Acetyloleanolic acid / (+)-Oleanolic acid acetate / (+)-3-O-Acetyloleanolic acid / 3beta-Acetoxyolean-12-en-28-oic acid |
CHEMBL486822
CHEMBL494653 CHEMBL1315416 |
C052658
|
1 / 0 / 0 | No. 177 |
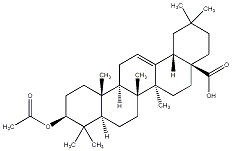
|
||
|
C00033563
|
3'',3''-Dimethyl-5',6'-pyrano-2',4'-dihydroxychalcone
|
No. 337 |

|
|||||
|
C00033772
|
Desmethylhelichromanochalcone
|
No. 337 |

|
|||||
|
C00007983
|
3',4',6'-Trihydroxy-2'-oxo-3',5'-diprenyldihydrochalcone
|
No. 2813 |

|
|||||
|
C00007984
|
Helihumulone
|
No. 2813 |

|
Human Protein / Gene in interactions
70 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00000992 C00004632 C00006935 | 1 / 2 |
| O75496 | Geminin | Unclassified protein | C00004632 C00006935 C00019064 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00000992 C00004632 C00019064 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00004632 C00006935 C00019064 | 0 / 0 |
| O00255 | Menin | Unclassified protein | C00000992 C00004632 C00006935 | 2 / 5 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00000992 C00004632 C00019064 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00000992 C00004632 C00006935 | 4 / 3 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00000965 C00000992 | 1 / 1 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00006933 C00019064 | 0 / 3 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00004632 C00006935 | 0 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00004632 C00006935 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000965 C00000992 | 0 / 1 |
| P06746 | DNA polymerase beta | Enzyme | C00004632 C00019064 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00019064 C00033565 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00004632 C00006935 | 0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00000992 C00004632 | 3 / 3 |
| P11309 | Serine/threonine-protein kinase pim-1 | Pim | C00000965 C00000992 | 0 / 0 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | C00004632 C00006935 | 0 / 0 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00006933 C00019064 | 0 / 0 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00000992 C00006935 | 7 / 3 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00000965 C00000992 | 0 / 1 |
| P15121 | Aldose reductase | Enzyme | C00004632 C00019064 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000965 C00000992 | 1 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00004632 C00006935 | 2 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00000992 C00019064 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00004632 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00019064 | 0 / 0 |
| P42858 | Huntingtin | Unclassified protein | C00004632 | 1 / 1 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00004632 | 1 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00004632 | 1 / 2 |
| P04792 | Heat shock protein beta-1 | Unclassified protein | C00004632 | 2 / 1 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00019064 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00008171 | 0 / 0 |
| P10586 | Receptor-type tyrosine-protein phosphatase F | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00004632 | 1 / 1 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00019064 | 0 / 0 |
| P04798 | Cytochrome P450 1A1 | Cytochrome P450 1A1 | C00000992 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00004632 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00019064 | 2 / 2 |
| Q16678 | Cytochrome P450 1B1 | Cytochrome P450 1B1 | C00000992 | 4 / 4 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00019064 | 0 / 0 |
| P18433 | Receptor-type tyrosine-protein phosphatase alpha | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00019064 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00000992 | 0 / 0 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00004632 | 0 / 1 |
| P08047 | Transcription factor Sp1 | Unclassified protein | C00000992 | 0 / 0 |
| P35228 | Nitric oxide synthase, inducible | Enzyme | C00019064 | 1 / 1 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00019064 | 0 / 0 |
| P24666 | Low molecular weight phosphotyrosine protein phosphatase | Tyr | C00019064 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00000992 | 0 / 0 |
| Q6W5P4 | Neuropeptide S receptor | Neuropeptide receptor | C00006935 | 1 / 0 |
| Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | Tyr | C00019064 | 4 / 2 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00004632 | 1 / 1 |
| P22001 | Potassium voltage-gated channel subfamily A member 3 | KCNA, Kv1.x (Shaker) | C00006935 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00004632 | 0 / 0 |
| P17706 | Tyrosine-protein phosphatase non-receptor type 2 | Tyr | C00019064 | 0 / 1 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | Tyr | C00019064 | 0 / 0 |
| P04054 | Phospholipase A2 | Enzyme | C00019064 | 0 / 0 |
| P23469 | Receptor-type tyrosine-protein phosphatase epsilon | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00004632 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00004632 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00004632 | 0 / 0 |
| Q8IUX4 | DNA dC->dU-editing enzyme APOBEC-3F | Enzyme | C00004632 | 0 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00019064 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00019064 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00019064 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00019064 | 1 / 4 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00004632 | 0 / 0 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00004632 | 0 / 0 |
| Q14191 | Werner syndrome ATP-dependent helicase | Enzyme | C00004632 | 2 / 1 |
24 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00000992
C00019064
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00000992
C00019064
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00019064
|
| 6818 | SULT1A3, HAST, HAST3, M-PST, ST1A3/ST1A4, ST1A5, STM, TL-PST | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (EC:2.8.2.1) |
C00000992
|
| 7052 | TGM2, G-ALPHA-h, GNAH, TG2, TGC | transglutaminase 2 (EC:2.3.2.13) |
C00000992
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00019064
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00019064
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00019064
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00019064
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00019064
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00019064
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00019064
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00000992
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00019064
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00019064
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00019064
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00019064
|
| 5052 | PRDX1, MSP23, NKEF-A, NKEFA, PAG, PAGA, PAGB, PRX1, PRXI, TDPX2 | peroxiredoxin 1 (EC:1.11.1.15) |
C00019064
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00019064
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00019064
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00019064
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00019064
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00019064
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00019064
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (43)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #608584 | Asthma-related traits, susceptibility to, 2 |
Q6W5P4
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #606595 | Charcot-marie-tooth disease, axonal, type 2f; cmt2f |
P04792
|
| #114500 | Colorectal cancer; crc |
P84022
Q14191 |
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #137750 | Glaucoma 1, open angle, a; glc1a |
Q16678
|
| #231300 | Glaucoma 3, primary congenital, a; glc3a |
Q16678
|
| #137760 | Glaucoma, primary open angle; poag |
Q16678
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #143100 | Huntington disease; hd |
P42858
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #607785 | Juvenile myelomonocytic leukemia; jmml |
Q06124
|
| #151100 | Leopard syndrome 1 |
Q06124
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #611162 | Malaria, susceptibility to |
P35228
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #156250 | Metachondromatosis; metcds |
Q06124
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #608634 | Neuronopathy, distal hereditary motor, type iib; hmn2b |
P04792
|
| #163950 | Noonan syndrome 1; ns1 |
Q06124
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #604229 | Peters anomaly |
Q16678
|
| #172700 | Pick disease of brain |
P10636
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #277700 | Werner syndrome; wrn |
Q14191
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (42)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
|
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00856 | Distal hereditary motor neuropathies (dHMN) |
P04792
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00408 | Type I diabetes mellitus |
P17706
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00017 | Esophageal cancer |
P35228
(related)
P35354 (related) |
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00059 | Huntington's disease (HD) |
P42858
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
Q14191 (related) |
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00523 | Noonan syndrome and related disorders |
Q06124
(related)
|
| H01018 | Metachondromatosis |
Q06124
(related)
|
| H00612 | Primary open angle glaucoma |
Q16678
(related)
|
| H01075 | Peters anomaly |
Q16678
(related)
|
| H01159 | Anterior segment dysgenesis (ASD) |
Q16678
(related)
|
| H01203 | Primary congenital glaucoma (PCG) |
Q16678
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
18 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D007249 | Inflammation |
C00003738
C00019064 |
| D007674 | Kidney Diseases |
C00019064
|
| D005157 | Facial Pain |
C00003738
|
| D010146 | Pain |
C00003738
|
| D002252 | Carbon Tetrachloride Poisoning |
C00019064
|
| D056486 | Drug-Induced Liver Injury |
C00019064
|
| D050171 | Dyslipidemias |
C00019064
|
| D018149 | Glucose Intolerance |
C00019064
|
| D006949 | Hyperlipidemias |
C00019064
|
| D006930 | Hyperalgesia |
C00003738
|
| D008103 | Liver Cirrhosis |
C00019064
|
| D008106 | Liver Cirrhosis, Experimental |
C00019064
|
| D008107 | Liver Diseases |
C00019064
|
| D017202 | Myocardial Ischemia |
C00019064
|
| D009369 | Neoplasms |
C00019064
|
| D009765 | Obesity |
C00019064
|
| D011041 | Poisoning |
C00019064
|
| D011230 | Precancerous Conditions |
C00019064
|