Tetracentron sinense
Species
KNApSAcK Entry
| Organism name | Tetracentron sinense |
|---|---|
| Genus | Tetracentron |
| Family | Trochodendraceae / Tetracentraceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Tetracentron sinense |
|---|---|
| Linked NCBI taxonomy ID | 13715 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Trochodendraceae |
|---|---|
| ID | 4405 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | eudicotyledons |
|---|---|
| ID | 71240 |
Metabolite list (12)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
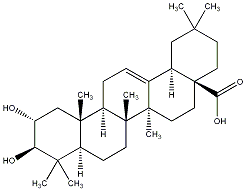
C00030742
|
Maslinic acid
/ Crategolic acid / 2alpha,3beta-Dihydroxyolean-12-en-28-oic acid |
CHEMBL201515
CHEMBL383749 CHEMBL482673 CHEMBL577380 |
C412811
|
8 / 5 / 4 | No. 13 | No. 51 |
|
|
|
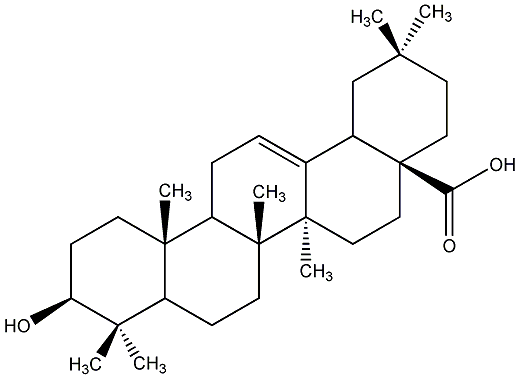
C00019064
|
Oleanolic acid
/ Astrantiagenin C / Virgaureagenin B / 3beta-Hydroxyolean-12-en-28-oic acid |
CHEMBL56615
CHEMBL168 CHEMBL180553 CHEMBL365375 CHEMBL486382 CHEMBL1413646 CHEMBL1436454 |
D009828
|
30 / 8 / 12 | 21 / 15 | No. 13 | No. 51 |
|
|
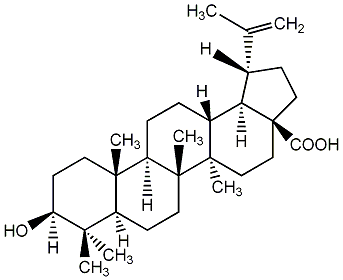
C00003741
|
Betulinic acid
|
CHEMBL269277
CHEMBL71690 CHEMBL519059 CHEMBL1318530 CHEMBL2005635 |
C002070
|
34 / 17 / 14 | 10 / 2 | No. 23 | No. 51 |
|
|
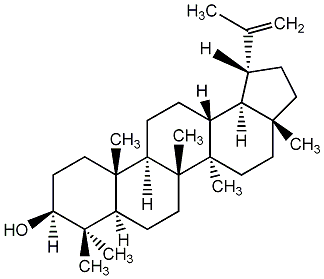
C00003749
|
Lupeol
/ Lupenol / (+)-Lupenol |
CHEMBL289191
CHEMBL459702 |
C010480
|
3 / 0 / 0 | 2 / 6 | No. 23 | No. 51 |
|
|
C00037278
|
Huazhongilexin
|
CHEMBL466534
|
No. 38 | No. 21 |

|
|||
|
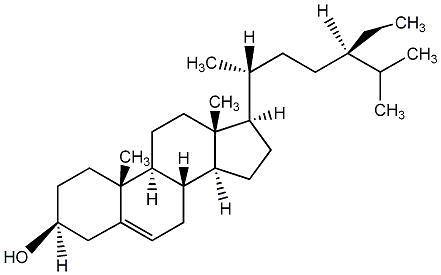
C00003672
|
Sitosterol
/ beta-sitosterl / (-)-beta-Sitosterol / Stigmast-5-en-3beta-ol |
CHEMBL221542
CHEMBL1398443 CHEMBL1875388 |
17 / 19 / 12 | No. 53 | No. 11 |
|
||
|
C00037900
|
Tetracentronside A
/ (-)-Tetracentronside A |
No. 122 | No. 15 |

|
||||
|
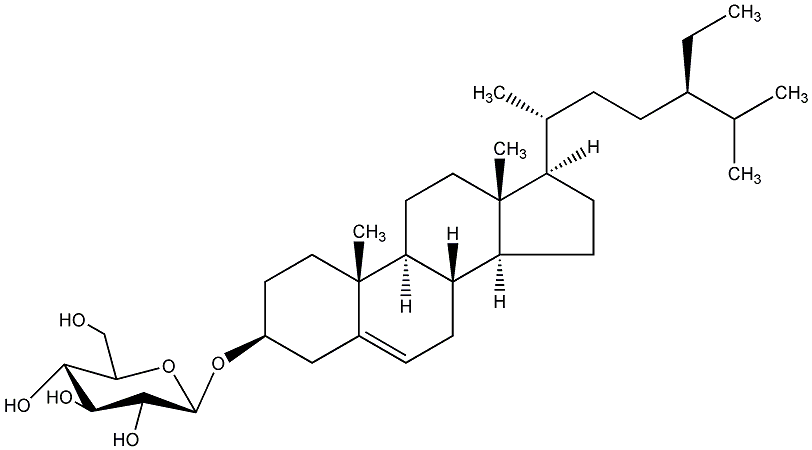
C00019308
|
Doursterol
/ Daucosterin / 3-O-beta-D-Glucopyranosyl sitosterol / beta-Sitosterol 3-O-beta-D-glucopyranoside |
CHEMBL197711
CHEMBL506678 CHEMBL2304043 |
C011015
|
5 / 4 / 2 | 0 / 3 | No. 520 |
|
|
|
C00002599
|
(-)-Dihydrocubebin
|
CHEMBL486597
CHEMBL1477391 |
C011228
|
4 / 8 / 4 | No. 629 | No. 21 |

|
|
|
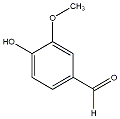
C00029531
|
Vanillic aldehyde
/ 4-Hydroxy-3-methoxybenzaldehyde |
CHEMBL13883
|
C100058
|
18 / 8 / 9 | 33 / 1 | No. 1003 |
|
|
|
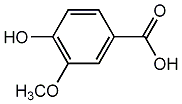
C00002682
|
Vanillic acid
/ 3-Methoxy-4-hydroxybenzoic acid |
CHEMBL120568
|
D014641
|
5 / 3 / 3 | 5 / 0 | No. 1073 |
|
|
|
C00037901
|
Tetracentronside B
/ (-)-Tetracentronside B |
No. 6950 |

|
Human Protein / Gene in interactions
84 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P06746 | DNA polymerase beta | Enzyme | C00003672 C00003741 C00019064 C00019308 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00002682 C00003741 C00019064 C00030742 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00003672 C00019064 C00019308 C00030742 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00003672 C00003741 C00019064 C00019308 | 0 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00003741 C00019064 C00029531 | 0 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00003741 C00019064 C00030742 | 0 / 0 |
| P08047 | Transcription factor Sp1 | Unclassified protein | C00003672 C00003749 C00019308 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002599 C00003672 C00003741 | 0 / 1 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00002599 C00003672 C00003741 | 1 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00003672 C00003741 | 1 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00002682 C00019064 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00003741 C00019064 | 0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00003741 C00019064 | 0 / 0 |
| Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | Tyr | C00019064 C00030742 | 4 / 2 |
| P17706 | Tyrosine-protein phosphatase non-receptor type 2 | Tyr | C00019064 C00030742 | 0 / 1 |
| P03372 | Estrogen receptor | NR3A1 | C00002682 C00003672 | 1 / 1 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00002599 C00019064 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00003672 C00003741 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00003672 C00003741 | 2 / 0 |
| P00734 | Prothrombin | S1A | C00003672 C00019308 | 4 / 2 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | Tyr | C00019064 C00030742 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00003672 C00029531 | 1 / 1 |
| P10586 | Receptor-type tyrosine-protein phosphatase F | Receptor tyrosine-protein phosphatase | C00019064 C00030742 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00003672 C00003741 | 0 / 1 |
| O75496 | Geminin | Unclassified protein | C00003741 C00019064 | 0 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00003741 C00003749 | 0 / 0 |
| P11387 | DNA topoisomerase 1 | Isomerase | C00003741 C00003749 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00003741 | 1 / 1 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00019064 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00029531 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00003741 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00019064 | 0 / 0 |
| P08151 | Zinc finger protein GLI1 | Unclassified protein | C00003741 | 0 / 0 |
| P42858 | Huntingtin | Unclassified protein | C00002682 | 1 / 1 |
| P15559 | NAD(P)H dehydrogenase [quinone] 1 | Enzyme | C00003741 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00029531 | 0 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00003672 | 1 / 0 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00019064 | 0 / 0 |
| O00519 | Fatty-acid amide hydrolase 1 | Enzyme | C00029531 | 0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00003741 | 0 / 0 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00002599 | 7 / 3 |
| Q04206 | Transcription factor p65 | Transcription Factor | C00003741 | 0 / 0 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00029531 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00019064 | 0 / 0 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | C00003741 | 0 / 0 |
| Q02156 | Protein kinase C epsilon type | Eta | C00003741 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00019064 | 2 / 2 |
| P14679 | Tyrosinase | Oxidoreductase | C00003672 | 4 / 2 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00029531 | 1 / 1 |
| P34969 | 5-hydroxytryptamine receptor 7 | Serotonin receptor | C00029531 | 0 / 0 |
| Q13133 | Oxysterols receptor LXR-alpha | NR1H3 | C00003741 | 0 / 0 |
| P18433 | Receptor-type tyrosine-protein phosphatase alpha | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00019064 | 0 / 0 |
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00029531 | 1 / 1 |
| P55055 | Oxysterols receptor LXR-beta | NR1H3 | C00003741 | 0 / 0 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00029531 | 2 / 2 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00029531 | 0 / 0 |
| Q9Y4X1 | UDP-glucuronosyltransferase 2A1 | Enzyme | C00029531 | 0 / 0 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00003672 | 3 / 2 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00003741 | 0 / 0 |
| P05771 | Protein kinase C beta type | Alpha | C00003741 | 0 / 0 |
| P35228 | Nitric oxide synthase, inducible | Enzyme | C00019064 | 1 / 1 |
| P51580 | Thiopurine S-methyltransferase | Enzyme | C00002682 | 1 / 1 |
| P02545 | Prelamin-A/C | Unclassified protein | C00003741 | 11 / 10 |
| P24666 | Low molecular weight phosphotyrosine protein phosphatase | Tyr | C00019064 | 0 / 0 |
| P78527 | DNA-dependent protein kinase catalytic subunit | Atypical serine/threonine protein kinase PIKK subfamily | C00029531 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00019064 | 0 / 0 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00029531 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00029531 | 0 / 0 |
| P10275 | Androgen receptor | NR3C4 | C00029531 | 3 / 4 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00019064 | 0 / 3 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00029531 | 0 / 0 |
| P04054 | Phospholipase A2 | Enzyme | C00019064 | 0 / 0 |
| P80365 | Corticosteroid 11-beta-dehydrogenase isozyme 2 | Enzyme | C00003672 | 1 / 1 |
| P23469 | Receptor-type tyrosine-protein phosphatase epsilon | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00003741 | 0 / 0 |
| P11217 | Glycogen phosphorylase, muscle form | Enzyme | C00030742 | 1 / 1 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00003741 | 1 / 0 |
| Q8IUX4 | DNA dC->dU-editing enzyme APOBEC-3F | Enzyme | C00003741 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00019064 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00019064 | 1 / 4 |
| Q9UBT2 | SUMO-activating enzyme subunit 2 | Enzyme | C00003741 | 0 / 0 |
| Q9UBE0 | SUMO-activating enzyme subunit 1 | Unclassified protein | C00003741 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00029531 | 0 / 0 |
61 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00003741
C00019064
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00002682
C00029531
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00002682
C00029531
|
| 54659 | UGT1A3, UDPGT, UDPGT_1-3, UGT-1C, UGT1-03, UGT1.3, UGT1C | UDP glucuronosyltransferase 1 family, polypeptide A3 (EC:2.4.1.17) |
C00002682
C00029531
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00002682
C00029531
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00003749
C00019064
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00003741
C00019064
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00003741
C00019064
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00003741
C00019064
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00003741
C00019064
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00019064
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00003741
|
| 1646 | AKR1C2, AKR1C-pseudo, BABP, DD, DD2, DDH2, HAKRD, HBAB, MCDR2, SRXY8 | aldo-keto reductase family 1, member C2 (EC:1.3.1.20 1.1.1.357) |
C00029531
|
| 759 | CA1, CA-I, CAB, Car1 | carbonic anhydrase I (EC:4.2.1.1) |
C00029531
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00029531
|
| 1196 | CLK2, hCLK2 | CDC-like kinase 2 (EC:2.7.12.1) |
C00029531
|
| 1490 | CTGF, CCN2, HCS24, IGFBP8, NOV2 | connective tissue growth factor |
C00029531
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00029531
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00029531
|
| 1789 | DNMT3B, ICF, ICF1, M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta (EC:2.1.1.37) |
C00029531
|
| 1848 | DUSP6, HH19, MKP3, PYST1 | dual specificity phosphatase 6 (EC:3.1.3.16 3.1.3.48) |
C00029531
|
| 2263 | FGFR2, BBDS, BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 | fibroblast growth factor receptor 2 (EC:2.7.10.1) |
C00029531
|
| 2535 | FZD2, Fz2, fz-2, fzE2, hFz2 | frizzled family receptor 2 |
C00029531
|
| 2729 | GCLC, GCL, GCS, GLCL, GLCLC | glutamate-cysteine ligase, catalytic subunit (EC:6.3.2.2) |
C00029531
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00029531
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00029531
|
| 3251 | HPRT1, HGPRT, HPRT | hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8) |
C00029531
|
| 3304 | HSPA1B, HSP70-1B, HSP70-2 | heat shock 70kDa protein 1B |
C00029531
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00029531
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00029531
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00029531
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00029531
|
| 57787 | MARK4, MARK4L, MARK4S, MARKL1, MARKL1L, PAR-1D | MAP/microtubule affinity-regulating kinase 4 (EC:2.7.11.1) |
C00029531
|
| 4502 | MT2A, MT2 | metallothionein 2A |
C00029531
|
| 5514 | PPP1R10, CAT53, FB19, PNUTS, PP1R10, R111, p99 | protein phosphatase 1, regulatory subunit 10 |
C00029531
|
| 11046 | SLC35D2, HFRC1, SQV7L, UGTrel8, hfrc | solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
C00029531
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00029531
|
| 7041 | TGFB1I1, ARA55, HIC-5, HIC5, TSC-5 | transforming growth factor beta 1 induced transcript 1 |
C00029531
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00029531
|
| 10626 | TRIM16, EBBP | tripartite motif containing 16 |
C00029531
|
| 7360 | UGP2, UDPG, UDPGP, UDPGP2, UGP1, UGPP1, UGPP2, pHC379 | UDP-glucose pyrophosphorylase 2 (EC:2.7.7.9) |
C00029531
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00003741
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00003741
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00003741
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00003741
|
| 1499 | CTNNB1, CTNNB, MRD19, armadillo | catenin (cadherin-associated protein), beta 1, 88kDa |
C00003749
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00019064
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00019064
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00019064
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00019064
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00019064
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00019064
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00019064
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00019064
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00019064
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00019064
|
| 5052 | PRDX1, MSP23, NKEF-A, NKEFA, PAG, PAGA, PAGB, PRX1, PRXI, TDPX2 | peroxiredoxin 1 (EC:1.11.1.15) |
C00019064
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00019064
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00019064
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00019064
|
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00002682
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (57)
| OMIM | preferred title | UniProt |
|---|---|---|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #218030 | Apparent mineralocorticoid excess; ame |
P80365
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #232600 | Glycogen storage disease v |
P11217
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #143100 | Huntington disease; hd |
P42858
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #607785 | Juvenile myelomonocytic leukemia; jmml |
Q06124
|
| #151100 | Leopard syndrome 1 |
Q06124
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #611162 | Malaria, susceptibility to |
P35228
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #156250 | Metachondromatosis; metcds |
Q06124
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #163950 | Noonan syndrome 1; ns1 |
Q06124
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #601367 | Stroke, ischemic |
P00734
|
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
| #610460 | Thiopurine s-methyltransferase deficiency |
P51580
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (49)
| KEGG | name | UniProt |
|---|---|---|
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00024 | Prostate cancer |
P10275
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00069 | Glycogen storage diseases (GSD) |
P11217
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00038 | Malignant melanoma |
P14679
(marker)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00408 | Type I diabetes mellitus |
P17706
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00017 | Esophageal cancer |
P35228
(related)
P35354 (related) |
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00059 | Huntington's disease (HD) |
P42858
(related)
|
| H00964 | Thiopurine S-methyltransferase deficiency (TPMT deficiency) |
P51580
(related)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H00259 | Apparent mineralocorticoid excess syndrome |
P80365
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00523 | Noonan syndrome and related disorders |
Q06124
(related)
|
| H01018 | Metachondromatosis |
Q06124
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q9NUW8
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
26 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D008103 | Liver Cirrhosis |
C00003741
C00019064 |
| D056486 | Drug-Induced Liver Injury |
C00019064
|
| D006528 | Carcinoma, Hepatocellular |
C00003749
|
| D009202 | Cardiomyopathies |
C00003749
|
| D006937 | Hypercholesterolemia |
C00003749
|
| D009374 | Neoplasms, Experimental |
C00003749
|
| D012878 | Skin Neoplasms |
C00003749
|
| D014947 | Wounds and Injuries |
C00003749
|
| D002493 | Central Nervous System Diseases |
C00019308
|
| D003072 | Cognition Disorders |
C00019308
|
| D013118 | Spinal Cord Diseases |
C00019308
|
| D008113 | Liver Neoplasms |
C00029531
|
| D002252 | Carbon Tetrachloride Poisoning |
C00019064
|
| D008545 | Melanoma |
C00003741
|
| D050171 | Dyslipidemias |
C00019064
|
| D018149 | Glucose Intolerance |
C00019064
|
| D006949 | Hyperlipidemias |
C00019064
|
| D007249 | Inflammation |
C00019064
|
| D007674 | Kidney Diseases |
C00019064
|
| D008106 | Liver Cirrhosis, Experimental |
C00019064
|
| D008107 | Liver Diseases |
C00019064
|
| D017202 | Myocardial Ischemia |
C00019064
|
| D009369 | Neoplasms |
C00019064
|
| D009765 | Obesity |
C00019064
|
| D011041 | Poisoning |
C00019064
|
| D011230 | Precancerous Conditions |
C00019064
|