Linum usitatissimum
Species
KNApSAcK Entry
| Organism name | Linum usitatissimum |
|---|---|
| Genus | Linum |
| Family | Linaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Linum usitatissimum |
|---|---|
| Linked NCBI taxonomy ID | 4006 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Linaceae |
|---|---|
| ID | 4004 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (33)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
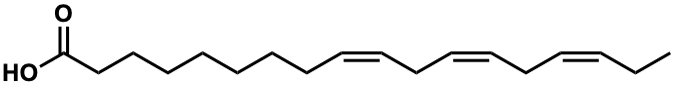
C00007247
|
Linolenic acid
/ alpha-Linolenic acid / (Z,Z,Z)-Octadeca-9,12,15-trienoic acid |
D017962
|
22 / 5 |
|
||||
|
C00006204
|
Orientin 7-O-rhamnoside
|
No. 1 | No. 15 |

|
||||
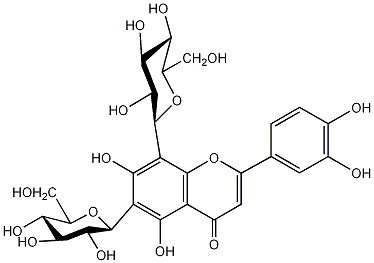
|
C00006231
|
Lucenin 2
/ Luteolin 6,8-C-diglucoside |
No. 1 | No. 15 |
|
||||
|
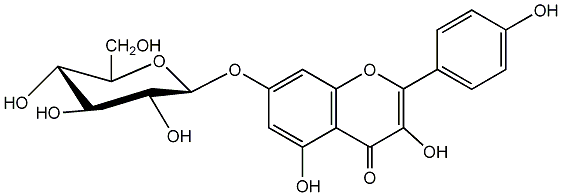
C00005149
|
Populnin
/ Kaempferol 7-glucoside |
CHEMBL469441
CHEMBL1159471 |
No. 2 | No. 15 |
|
|||
|
C00006410
|
Lucenin-2,7-O-rhamnoside
|
No. 5 | No. 15 |

|
||||
|
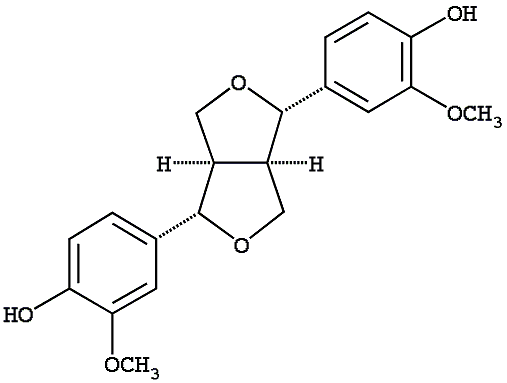
C00007190
|
(+)-Pinoresinol
|
CHEMBL267963
CHEMBL487611 CHEMBL460862 |
C103298
|
7 / 5 / 6 | No. 38 | No. 21 |
|
|
|
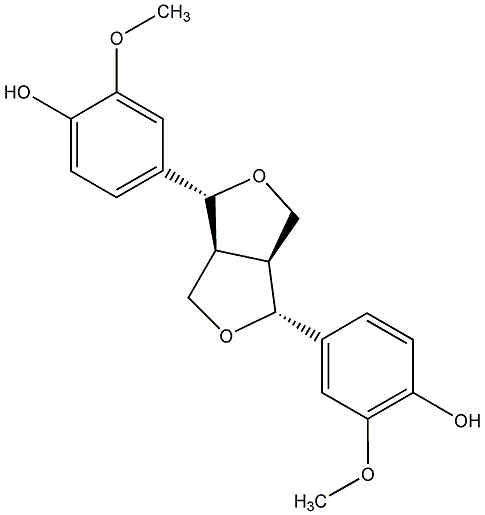
C00000587
|
(-)-Pinoresinol
|
CHEMBL267963
CHEMBL487611 CHEMBL460862 |
7 / 5 / 6 | No. 38 | No. 21 |
|
||
|
C00003650
|
Cycloartenol
|
CHEMBL119938
CHEMBL225815 CHEMBL226036 CHEMBL393145 CHEMBL2007217 |
C100089
|
0 / 1 | No. 129 | No. 11 |

|
|
|
C00000652
|
(+)-Secoisolariciresinol diglucoside
|
No. 152 |

|
|||||
|
C00000606
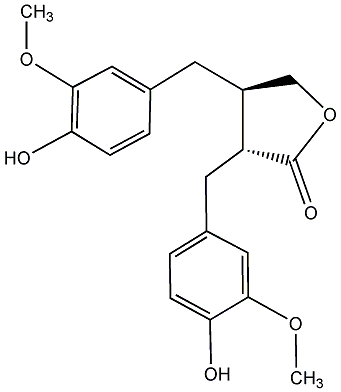
|
(-)-Matairesinol
|
CHEMBL425148
|
C068935
|
1 / 0 / 0 | No. 223 | No. 21 |
|
|
|
C00001448
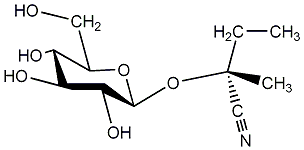
|
Lotaustralin
/ (R)-Lotaustralin |
C001556
|
No. 258 | No. 73 |
|
|||
|
C00001446
|
Linamarin
|
CHEMBL1590222
|
C005091
|
1 / 4 / 3 | No. 258 | No. 73 |

|
|
|
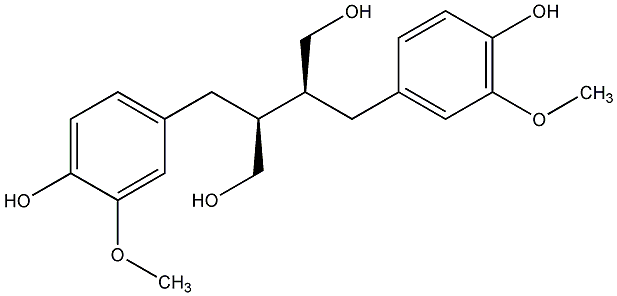
C00000605
|
(+)-Secoisolariciresinol
|
CHEMBL368347
|
2 / 1 / 1 | No. 282 | No. 21 |
|
||
|
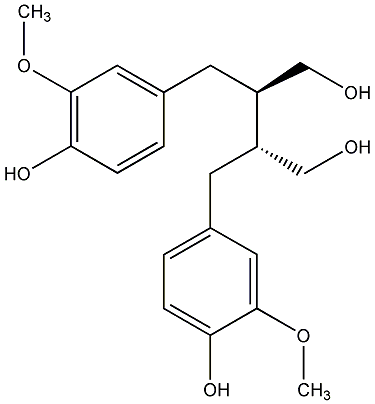
C00000604
|
(-)-Secoisolariciresinol
|
CHEMBL368347
|
C060283
|
2 / 1 / 1 | 1 / 0 | No. 282 | No. 21 |
|
|
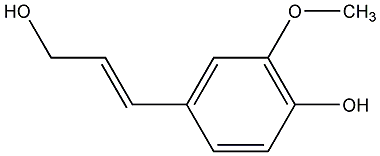
C00000614
|
Coniferyl alcohol
|
CHEMBL501870
CHEMBL2088631 |
C010559
|
No. 310 | No. 6 |
|
||
|
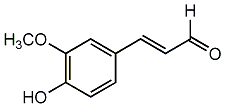
C00002728
|
Coniferaldehyde
/ Coniferyl aldehyde / 4-Hydroxy-3-methoxycinnamaldehyde |
CHEMBL242529
CHEMBL1956165 |
C075384
|
1 / 1 / 1 | No. 310 | No. 6 |
|
|
|
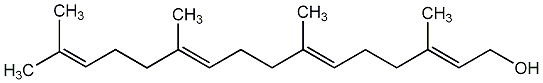
C00003428
|
Geranylgeraniol
|
CHEMBL478589
|
3 / 4 / 3 | No. 369 | No. 38 |
|
||
|
C00001447
|
Linustatin
|
C023022
|
0 / 2 | No. 423 | No. 73 |
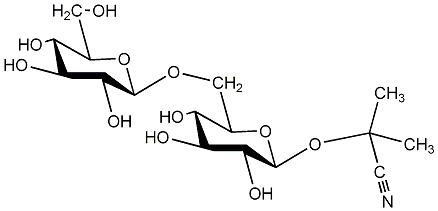
|
||
|
C00001450
|
Neolinustatin
|
C023031
|
0 / 2 | No. 423 | No. 73 |

|
||
|
C00000603
|
(-)-Lariciresinol
|
CHEMBL518421
|
C060282
|
2 / 1 / 1 | No. 700 | No. 21 |
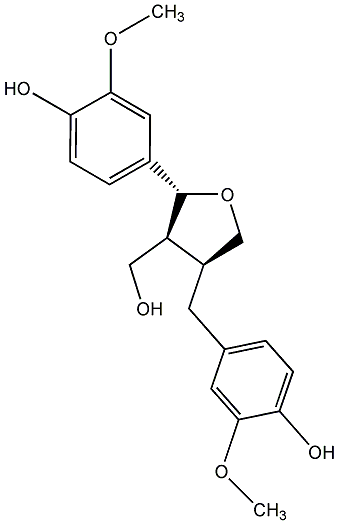
|
|
|
C00000218
|
Jasmonic acid
/ trans-Jasmonic acid / 3-Oxo-2-(2Z-pentenyl)cyclotentylacetic acid |
CHEMBL445499
CHEMBL449572 |
C011006
|
4 / 1 / 0 | No. 707 | No. 70 |
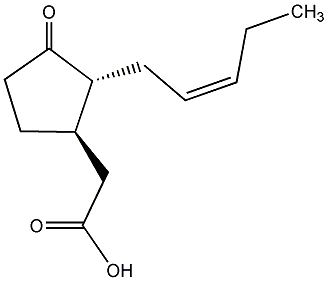
|
|
|
C00000446
|
7-iso-Jasmonic acid
/ (+)-Epijasmonic acid |
CHEMBL445499
CHEMBL449572 |
4 / 1 / 0 | No. 707 | No. 70 |
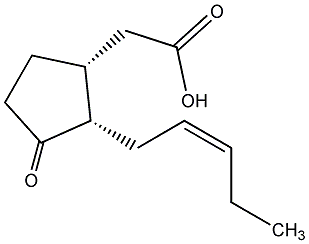
|
||
|
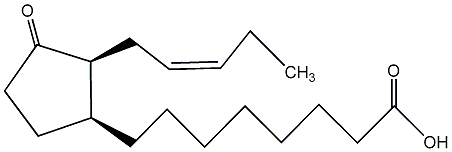
C00000366
|
OPC-8:0
/ 3-Oxo-2-(2-entenyl)cyclopentaneoctanoic acid |
No. 707 | No. 70 |
|
||||
|
C00000361
|
OPC-4:0
|
No. 707 | No. 70 |

|
||||
|
C00027048
|
Cyclolinopeptide I
|
No. 1065 |

|
|||||
|
C00027044
|
Cyclolinopeptide G
|
No. 1065 |

|
|||||
|
C00027046
|
Cyclolinopeptide H
|
No. 1065 |

|
|||||
|
C00027043
|
Cyclolinopeptide F
|
No. 1065 |

|
|||||
|
C00036983
|
Cyclolinopeptide A
|
CHEMBL1793995
|
No. 1065 |

|
||||
|
C00036984
|
Cyclolinopeptide B
/ (-)-Cyclolinopeptide B |
No. 1065 |

|
|||||
|
C00036985
|
Cyclolinopeptide C
|
No. 1065 |

|
|||||
|
C00036986
|
Cyclolinopeptide D
/ (-)-Cyclolinopeptide D |
No. 1065 |

|
|||||
|
C00036987
|
Cyclolinopeptide E
/ (-)-Cyclolinopeptide E |
No. 1065 |

|
Human Protein / Gene in interactions
18 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000603 C00000604 C00000605 | 1 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000603 C00000604 C00000605 | 0 / 1 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00000587 C00007190 | 5 / 3 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00000587 C00007190 | 0 / 3 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00000587 C00007190 | 0 / 0 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00000587 C00007190 | 0 / 0 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | C00000587 C00007190 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00001446 C00003428 | 4 / 3 |
| P42330 | Aldo-keto reductase family 1 member C3 | Enzyme | C00000218 C00000446 | 0 / 0 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00000587 C00007190 | 0 / 0 |
| P52895 | Aldo-keto reductase family 1 member C2 | Enzyme | C00000218 C00000446 | 1 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00000587 C00007190 | 0 / 0 |
| P17516 | Aldo-keto reductase family 1 member C4 | Enzyme | C00000218 C00000446 | 1 / 0 |
| Q04828 | Aldo-keto reductase family 1 member C1 | Enzyme | C00000218 C00000446 | 0 / 0 |
| P04278 | Sex hormone-binding globulin | Secreted protein | C00000606 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00003428 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00003428 | 0 / 0 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00002728 | 1 / 1 |
23 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00000604
|
| 8647 | ABCB11, ABC16, BRIC2, BSEP, PFIC-2, PFIC2, PGY4, SPGP | ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
C00007247
|
| 1374 | CPT1A, CPT1, CPT1-L, L-CPT1 | carnitine palmitoyltransferase 1A (liver) (EC:2.3.1.21) |
C00007247
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00007247
|
| 1591 | CYP24A1, CP24, CYP24, HCAI, P450-CC24 | cytochrome P450, family 24, subfamily A, polypeptide 1 (EC:1.14.13.126) |
C00007247
|
| 1557 | CYP2C19, CPCJ, CYP2C, P450C2C, P450IIC19 | cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00007247
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00007247
|
| 1571 | CYP2E1, CPE1, CYP2E, P450-J, P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) |
C00007247
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00007247
|
| 3992 | FADS1, D5D, FADS6, FADSD5, LLCDL1, TU12 | fatty acid desaturase 1 (EC:1.14.19.-) |
C00007247
|
| 2729 | GCLC, GCL, GCS, GLCL, GLCLC | glutamate-cysteine ligase, catalytic subunit (EC:6.3.2.2) |
C00007247
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00007247
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00007247
|
| 3565 | IL4, BCGF-1, BCGF1, BSF-1, BSF1, IL-4 | interleukin 4 |
C00007247
|
| 3827 | KNG1, BDK, BK, KNG | kininogen 1 |
C00007247
|
| 9971 | NR1H4, BAR, FXR, HRR-1, HRR1, RIP14 | nuclear receptor subfamily 1, group H, member 4 |
C00007247
|
| 5465 | PPARA, NR1C1, PPAR, PPARalpha, hPPAR | peroxisome proliferator-activated receptor alpha |
C00007247
|
| 100136658 |
C00007247
|
||
| 5467 | PPARD, FAAR, NR1C2, NUC1, NUCI, NUCII, PPARB | peroxisome proliferator-activated receptor delta |
C00007247
|
| 5468 | PPARG, CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | peroxisome proliferator-activated receptor gamma |
C00007247
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00007247
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00007247
|
| 7421 | VDR, NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
C00007247
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (12)
| OMIM | preferred title | UniProt |
|---|---|---|
| #614279 | 46,xy sex reversal 8; srxy8 |
P17516
P52895 |
| %606641 | Body mass index; bmi |
P37231
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #137800 | Glioma susceptibility 1; glm1 |
P37231
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #601665 | Obesity |
P37231
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #278300 | Xanthinuria, type i |
P47989
|
KEGG DISEASE (11)
| KEGG | name | UniProt |
|---|---|---|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00017 | Esophageal cancer |
P35354
(related)
|
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00032 | Thyroid cancer |
P37231
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P37231
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
Diseases related to CTD interactions
8 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D011041 | Poisoning |
C00001447
C00001450 |
| D015431 | Weight Loss |
C00001447
C00001450 |
| D003866 | Depressive Disorder |
C00007247
|
| D001930 | Brain Injuries |
C00007247
|
| D012878 | Skin Neoplasms |
C00003650
|
| D064420 | Drug-Related Side Effects and Adverse Reactions |
C00007247
|
| D007249 | Inflammation |
C00007247
|
| D009422 | Nervous System Diseases |
C00007247
|