Pimpinella anisum
Species
KNApSAcK Entry
| Organism name | Pimpinella anisum |
|---|---|
| Genus | Pimpinella |
| Family | Apiaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Pimpinella anisum |
|---|---|
| Linked NCBI taxonomy ID | 271192 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Apiaceae |
|---|---|
| ID | 4037 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (8)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
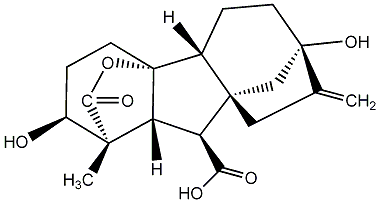
C00000001
|
GA1
/ Gibberellin A1 |
C422660
|
No. 40 | No. 41 |
|
|||
|
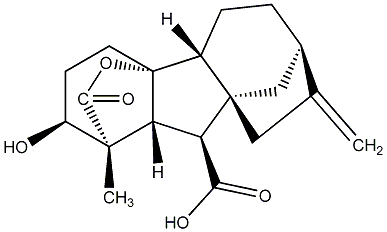
C00000004
|
GA4
/ Gibberellin A4 |
No. 40 | No. 41 |
|
||||
|
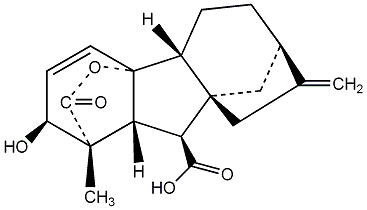
C00000007
|
GA7
/ Gibberellin A7 |
No. 40 | No. 41 |
|
||||
|
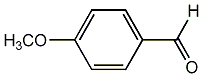
C00002636
|
Anisaldehyde
/ p-Anisaldehyde |
CHEMBL161598
|
C024896
|
11 / 18 / 13 | 1 / 0 | No. 1003 |
|
|
|
C00002645
|
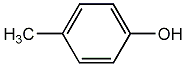
p-Cresol
/ 4-Methylphenol |
CHEMBL16645
|
C032538
|
8 / 4 / 4 | 10 / 4 | No. 1546 |
|
|
|
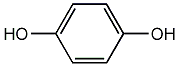
C00002656
|
Hydroquinone
/ 1,4-Benzenediol |
CHEMBL537
|
C031927
|
55 / 48 / 51 | 156 / 14 | No. 1590 | No. 82 |
|
|
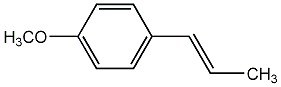
C00002713
|
Anethole
|
CHEMBL452630
CHEMBL1468832 |
C006578
|
6 / 21 / 49 | 16 / 13 | No. 2748 | No. 6 |
|
|
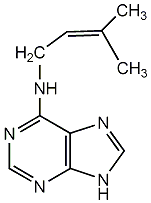
C00000094
|
2iP
/ N6-(delta2-Isopentenyl)adenine |
CHEMBL476189
|
C001478
|
5 / 2 / 3 | 18 / 0 | No. 2879 |
|
Human Protein / Gene in interactions
70 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10275 | Androgen receptor | NR3C4 | C00002636 C00002645 C00002656 | 3 / 4 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00002636 C00002656 C00002713 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000094 C00002656 C00002713 | 0 / 1 |
| P51449 | Nuclear receptor ROR-gamma | Nuclear hormone receptor subfamily 1 group F member 3 | C00002645 C00002656 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00002656 C00002713 | 11 / 10 |
| P03372 | Estrogen receptor | NR3A1 | C00002636 C00002656 | 1 / 1 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00002645 C00002656 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00000094 C00002636 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00002636 C00002656 | 0 / 0 |
| P10145 | Interleukin-8 | Secreted protein | C00002645 C00002656 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00002656 C00002713 | 0 / 0 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00002636 C00002656 | 3 / 1 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00002656 | 0 / 0 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00002713 | 3 / 2 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00002656 | 0 / 0 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00002656 | 0 / 0 |
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00002636 | 1 / 1 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00002656 | 1 / 2 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00000094 | 0 / 1 |
| P54132 | Bloom syndrome protein | Enzyme | C00002656 | 1 / 2 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00002656 | 0 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00002656 | 0 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00002656 | 0 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00002656 | 0 / 0 |
| P68871 | Hemoglobin subunit beta | Secreted protein | C00002656 | 4 / 4 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00002656 | 1 / 2 |
| P11509 | Cytochrome P450 2A6 | Cytochrome P450 2A6 | C00002636 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00002656 | 0 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002656 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00002656 | 0 / 0 |
| P51151 | Ras-related protein Rab-9A | Unclassified protein | C00002656 | 0 / 0 |
| P50225 | Sulfotransferase 1A1 | Enzyme | C00002645 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00002645 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00002656 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00002656 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00002656 | 0 / 0 |
| P04150 | Glucocorticoid receptor | NR3C1 | C00002656 | 0 / 1 |
| P14679 | Tyrosinase | Oxidoreductase | C00002636 | 4 / 2 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00002636 | 5 / 3 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00002636 | 1 / 1 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00002656 | 0 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00002656 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00002656 | 0 / 1 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00002656 | 3 / 3 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00002656 | 2 / 2 |
| O15118 | Niemann-Pick C1 protein | Unclassified protein | C00002656 | 1 / 1 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00002656 | 5 / 2 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00002656 | 0 / 3 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00002656 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00002656 | 2 / 0 |
| P22303 | Acetylcholinesterase | Hydrolase | C00002645 | 1 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00002656 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000094 | 1 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00000094 | 1 / 1 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002656 | 1 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00002656 | 4 / 3 |
| Q99549 | M-phase phosphoprotein 8 | Unclassified protein | C00002656 | 0 / 0 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00002645 | 0 / 0 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00002656 | 0 / 0 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00002713 | 7 / 37 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00002656 | 0 / 0 |
| P35869 | Aryl hydrocarbon receptor | Transcription Factor | C00002656 | 0 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00002656 | 1 / 1 |
| P42345 | Serine/threonine-protein kinase mTOR | Enzyme | C00002656 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00002656 | 0 / 0 |
| P40225 | Thrombopoietin | Unclassified protein | C00002656 | 1 / 1 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00002656 | 0 / 0 |
| O00255 | Menin | Unclassified protein | C00002656 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00002656 | 1 / 2 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002656 | 0 / 0 |
188 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00002645
C00002656
C00002713
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00002656
C00002713
|
| 1565 | CYP2D6, CPD6, CYP2D, CYP2D7AP, CYP2D7BP, CYP2D7P2, CYP2D8P2, CYP2DL1, CYPIID6, P450-DB1, P450C2D, P450DB1 | cytochrome P450, family 2, subfamily D, polypeptide 6 (EC:1.14.14.1) |
C00002645
C00002656
|
| 8989 | TRPA1, ANKTM1, FEPS | transient receptor potential cation channel, subfamily A, member 1 |
C00002636
C00002656
|
| 182 | JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1 | jagged 1 |
C00000094
C00002656
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00002656
C00002713
|
| 1571 | CYP2E1, CPE1, CYP2E, P450-J, P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) |
C00002645
C00002656
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00002656
C00002713
|
| 4313 | MMP2, CLG4, CLG4A, MMP-II, MONA, TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) |
C00002656
C00002713
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00002656
C00002713
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00002656
C00002713
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00000094
C00002656
|
| 4324 | MMP15, MT2-MMP, MTMMP2, SMCP-2 | matrix metallopeptidase 15 (membrane-inserted) (EC:3.4.24.-) |
C00002656
|
| 6286 | S100P, MIG9 | S100 calcium binding protein P |
C00000094
|
| 7029 | TFDP2, DP2 | transcription factor Dp-2 (E2F dimerization partner 2) |
C00000094
|
| 7127 | TNFAIP2, B94, EXOC3L3 | tumor necrosis factor, alpha-induced protein 2 |
C00000094
|
| 6004 | RGS16, A28-RGS14, A28-RGS14P, RGS-R | regulator of G-protein signaling 16 |
C00000094
|
| 7431 | VIM, CTRCT30 | vimentin |
C00000094
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00002713
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00002713
|
| 3551 | IKBKB, IKK-beta, IKK2, IKKB, NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (EC:2.7.11.10) |
C00002713
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00002713
|
| 5774 | PTPN3, PTP-H1, PTPH1 | protein tyrosine phosphatase, non-receptor type 3 (EC:3.1.3.48) |
C00000094
|
| 5782 | PTPN12, PTP-PEST, PTPG1 | protein tyrosine phosphatase, non-receptor type 12 (EC:3.1.3.48) |
C00000094
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00002713
|
| 9360 | PPIG, CARS-Cyp, CYP, SCAF10, SRCyp | peptidylprolyl isomerase G (cyclophilin G) (EC:5.2.1.8) |
C00000094
|
| 5366 | PMAIP1, APR, NOXA | phorbol-12-myristate-13-acetate-induced protein 1 |
C00000094
|
| 4790 | NFKB1, EBP-1, KBF1, NF-kB1, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
C00002713
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00002713
|
| 8544 | PIR | pirin (iron-binding nuclear protein) (EC:1.13.11.24) |
C00000094
|
| 5328 | PLAU, ATF, BDPLT5, QPD, UPA, URK, u-PA | plasminogen activator, urokinase (EC:3.4.21.73) |
C00002713
|
| 5142 | PDE4B, DPDE4, PDE4B5, PDEIVB | phosphodiesterase 4B, cAMP-specific (EC:3.1.4.17) |
C00000094
|
| 7076 | TIMP1, CLGI, EPA, EPO, HCI, TIMP | TIMP metallopeptidase inhibitor 1 |
C00002713
|
| 9818 | NUPL1, PRO2463 | nucleoporin like 1 |
C00000094
|
| 440 | ASNS, TS11 | asparagine synthetase (glutamine-hydrolyzing) (EC:6.3.5.4) |
C00000094
|
| 64782 | AEN, ISG20L1, pp12744 | apoptosis enhancing nuclease |
C00002656
|
| 217 | ALDH2, ALDH-E2, ALDHI, ALDM | aldehyde dehydrogenase 2 family (mitochondrial) (EC:1.2.1.3) |
C00002656
|
| 396 | ARHGDIA, GDIA1, NPHS8, RHOGDI, RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha |
C00002656
|
| 468 | ATF4, CREB-2, CREB2, TAXREB67, TXREB | activating transcription factor 4 |
C00002656
|
| 572 | BAD, BBC2, BCL2L8 | BCL2-associated agonist of cell death |
C00002656
|
| 27113 | BBC3, JFY-1, JFY1, PUMA | BCL2 binding component 3 |
C00002656
|
| 330 | BIRC3, AIP1, API2, CIAP2, HAIP1, HIAP1, MALT2, MIHC, RNF49, c-IAP2 | baculoviral IAP repeat containing 3 |
C00002656
|
| 664 | BNIP3, NIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 |
C00002656
|
| 79866 | BORA, C13orf34 | bora, aurora kinase A activator |
C00002656
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00002656
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00002656
|
| 839 | CASP6, MCH2 | caspase 6, apoptosis-related cysteine peptidase (EC:3.4.22.59) |
C00002656
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00002656
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00002656
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00002656
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00002656
|
| 6356 | CCL11, SCYA11 | chemokine (C-C motif) ligand 11 |
C00002656
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00002656
|
| 6348 | CCL3, G0S19-1, LD78ALPHA, MIP-1-alpha, MIP1A, SCYA3 | chemokine (C-C motif) ligand 3 |
C00002656
|
| 6351 | CCL4, ACT2, AT744.1, G-26, HC21, LAG-1, LAG1, MIP-1-beta, MIP1B, MIP1B1, SCYA2, SCYA4 | chemokine (C-C motif) ligand 4 |
C00002656
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00002656
|
| 9134 | CCNE2, CYCE2 | cyclin E2 |
C00002656
|
| 899 | CCNF, FBX1, FBXO1 | cyclin F |
C00002656
|
| 942 | CD86, B7-2, B7.2, B70, CD28LG2, LAB72 | CD86 molecule |
C00002656
|
| 157313 | CDCA2, Repo-Man | cell division cycle associated 2 |
C00002656
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002656
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00002656
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00002656
|
| 1282 | COL4A1, HANAC, ICH, POREN1, arresten | collagen, type IV, alpha 1 |
C00002656
|
| 1436 | CSF1R, C-FMS, CD115, CSF-1R, CSFR, FIM2, FMS, HDLS, M-CSF-R | colony stimulating factor 1 receptor (EC:2.7.10.1) |
C00002656
|
| 1437 | CSF2, GMCSF | colony stimulating factor 2 (granulocyte-macrophage) |
C00002656
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00002656
|
| 54205 | CYCS, CYC, HCS, THC4 | cytochrome c, somatic |
C00002656
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002656
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00002656
|
| 24300 |
C00002656
|
||
| 10681 | GNB5, GB5 | guanine nucleotide binding protein (G protein), beta 5 |
C00000094
|
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative |
C00000094
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00002656
|
| 1577 | CYP3A5, CP35, CYPIIIA5, P450PCN3, PCN3 | cytochrome P450, family 3, subfamily A, polypeptide 5 (EC:1.14.14.1) |
C00002656
|
| 3491 | CYR61, CCN1, GIG1, IGFBP10 | cysteine-rich, angiogenic inducer, 61 |
C00002656
|
| 1611 | DAP | death-associated protein |
C00002656
|
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00002656
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00002656
|
| 22943 | DKK1, DKK-1, SK | dickkopf WNT signaling pathway inhibitor 1 |
C00002656
|
| 3301 | DNAJA1, DJ-2, DjA1, HDJ2, HSDJ, HSJ2, HSPF4, NEDD7, hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 |
C00002656
|
| 3337 | DNAJB1, HSPF1, Hdj1, Hsp40, RSPH16B, Sis1 | DnaJ (Hsp40) homolog, subfamily B, member 1 |
C00002656
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00002656
|
| 1788 | DNMT3A, DNMT3A2, M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha (EC:2.1.1.37) |
C00002656
|
| 1789 | DNMT3B, ICF, ICF1, M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta (EC:2.1.1.37) |
C00002656
|
| 1999 | ELF3, EPR-1, ERT, ESE-1, ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
C00002656
|
| 8772 | FADD, MORT1 | Fas (TNFRSF6)-associated via death domain |
C00002656
|
| 81610 | FAM83D, C20orf129, CHICA, dJ616B8.3 | family with sequence similarity 83, member D |
C00002656
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00002656
|
| 2335 | FN1, CIG, ED-B, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | fibronectin 1 |
C00002656
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00002656
|
| 2495 | FTH1, FHC, FTH, FTHL6, PIG15, PLIF | ferritin, heavy polypeptide 1 (EC:1.16.3.1) |
C00002656
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00002656
|
| 2623 | GATA1, ERYF1, GATA-1, GF-1, GF1, NF-E1, NFE1, XLANP, XLTDA, XLTT | GATA binding protein 1 (globin transcription factor 1) |
C00002656
|
| 2624 | GATA2, DCML, MONOMAC, NFE1B | GATA binding protein 2 |
C00002656
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00002656
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00002656
|
| 2937 | GSS, GSHS | glutathione synthetase (EC:6.3.2.3) |
C00002656
|
| 2944 | GSTM1, GST1, GSTM1-1, GSTM1a-1a, GSTM1b-1b, GTH4, GTM1, H-B, MU, MU-1 | glutathione S-transferase mu 1 (EC:2.5.1.18) |
C00002656
|
| 2993 | GYPA, CD235a, GPA, GPErik, GPSAT, HGpMiV, HGpMiXI, HGpSta(C), MN, MNS, PAS-2 | glycophorin A (MNS blood group) |
C00002656
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00002656
|
| 3043 | HBB, CD113t-C, beta-globin | hemoglobin, beta |
C00002656
|
| 3009 | HIST1H1B, H1, H1.5, H1B, H1F5, H1s-3 | histone cluster 1, H1b |
C00002656
|
| 3007 | HIST1H1D, H1.3, H1D, H1F3, H1s-2 | histone cluster 1, H1d |
C00002656
|
| 3008 | HIST1H1E, H1.4, H1E, H1F4, H1s-4, dJ221C16.5 | histone cluster 1, H1e |
C00002656
|
| 85235 | HIST1H2AH, H2A/S, H2AFALii, H2AH, dJ86C11.1 | histone cluster 1, H2ah |
C00002656
|
| 8329 | HIST1H2AI, H2A/c, H2AFC | histone cluster 1, H2ai |
C00002656
|
| 8331 | HIST1H2AJ, H2A/E, H2AFE, dJ160A22.4 | histone cluster 1, H2aj |
C00002656
|
| 8358 | HIST1H3B, H3/l, H3FL | histone cluster 1, H3b |
C00002656
|
| 8352 | HIST1H3C, H3.1, H3/c, H3FC | histone cluster 1, H3c |
C00002656
|
| 8359 | HIST1H4A, H4FA | histone cluster 1, H4a |
C00002656
|
| 8366 | HIST1H4B, H4/I, H4FI | histone cluster 1, H4b |
C00002656
|
| 8364 | HIST1H4C, H4/g, H4FG, dJ221C16.1 | histone cluster 1, H4c |
C00002656
|
| 8367 | HIST1H4E, H4/j, H4FJ | histone cluster 1, H4e |
C00002656
|
| 8361 | HIST1H4F, H4, H4/c, H4FC | histone cluster 1, H4f |
C00002656
|
| 8365 | HIST1H4H, H4/h, H4FH | histone cluster 1, H4h |
C00002656
|
| 8362 | HIST1H4K, H4/d, H4F2iii, H4FD, dJ160A22.1 | histone cluster 1, H4k |
C00002656
|
| 8337 | HIST2H2AA3, H2A, H2A.2, H2A/O, H2A/q, H2AFO, H2a-615, HIST2H2AA | histone cluster 2, H2aa3 |
C00002656
|
| 317772 | HIST2H2AB, H2AB | histone cluster 2, H2ab |
C00002656
|
| 8338 | HIST2H2AC, H2A, H2A-GL101, H2A/q, H2AFQ | histone cluster 2, H2ac |
C00002656
|
| 8370 | HIST2H4A, FO108, H4, H4/n, H4F2, H4FN, HIST2H4 | histone cluster 2, H4a |
C00002656
|
| 55355 | HJURP, FAKTS, URLC9, hFLEG1 | Holliday junction recognition protein |
C00002656
|
| 3145 | HMBS, PBG-D, PBGD, PORC, UPS | hydroxymethylbilane synthase (EC:2.5.1.61) |
C00002656
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00002656
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00002656
|
| 3306 | HSPA2, HSP70-2, HSP70-3 | heat shock 70kDa protein 2 |
C00002656
|
| 219844 | HYLS1, HLS | hydrolethalus syndrome 1 |
C00002656
|
| 3586 | IL10, CSIF, GVHDS, IL-10, IL10A, TGIF | interleukin 10 |
C00002656
|
| 3558 | IL2, IL-2, TCGF, lymphokine | interleukin 2 |
C00002656
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00002656
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00002656
|
| 3688 | ITGB1, CD29, FNRB, GPIIA, MDF2, MSK12, VLA-BETA, VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
C00002656
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00002656
|
| 81930 | KIF18A, MS-KIF18A | kinesin family member 18A |
C00002656
|
| 4254 | KITLG, FPH2, KL-1, Kitl, MGF, SCF, SF, SHEP7 | KIT ligand |
C00002656
|
| 3910 | LAMA4, CMD1JJ, LAMA3, LAMA4*-1 | laminin, alpha 4 |
C00002656
|
| 11061 | LECT1, BRICD3, CHM-I, CHM1, MYETS1 | leukocyte cell derived chemotaxin 1 |
C00002656
|
| 4000 | LMNA, CDCD1, CDDC, CMD1A, CMT2B1, EMD2, FPL, FPLD, FPLD2, HGPS, IDC, LDP1, LFP, LGMD1B, LMN1, LMNC, LMNL1, PRO1 | lamin A/C |
C00002656
|
| 8932 | MBD2, DMTase, NY-CO-41 | methyl-CpG binding domain protein 2 |
C00002656
|
| 4257 | MGST1, GST12, MGST, MGST-I | microsomal glutathione S-transferase 1 (EC:2.5.1.18) |
C00002656
|
| 4323 | MMP14, MMP-14, MMP-X1, MT-MMP, MT-MMP_1, MT1-MMP, MT1MMP, MTMMP1, WNCHRS | matrix metallopeptidase 14 (membrane-inserted) (EC:3.4.24.80) |
C00002656
|
| 6016 | RIT1, NS8, RIBB, RIT, ROC1 | Ras-like without CAAX 1 |
C00000094
|
| 4314 | MMP3, CHDS6, MMP-3, SL-1, STMY, STMY1, STR1 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) (EC:3.4.24.17) |
C00002656
|
| 4352 | MPL, C-MPL, CD110, MPLV, THCYT2, TPOR | myeloproliferative leukemia virus oncogene |
C00002656
|
| 4353 | MPO | myeloperoxidase (EC:1.11.2.2) |
C00002656
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00002656
|
| 4778 | NFE2, NF-E2, p45 | nuclear factor, erythroid 2 |
C00002656
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00002656
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00002656
|
| 5036 | PA2G4, EBP1, HG4-1, p38-2G4 | proliferation-associated 2G4, 38kDa |
C00002656
|
| 25796 | PGLS, 6PGL | 6-phosphogluconolactonase (EC:3.1.1.31) |
C00002656
|
| 5429 | POLH, RAD30, RAD30A, XP-V, XPV | polymerase (DNA directed), eta (EC:2.7.7.7) |
C00002656
|
| 8493 | PPM1D, PP2C-DELTA, WIP1 | protein phosphatase, Mg2+/Mn2+ dependent, 1D (EC:3.1.3.16) |
C00002656
|
| 5052 | PRDX1, MSP23, NKEF-A, NKEFA, PAG, PAGA, PAGB, PRX1, PRXI, TDPX2 | peroxiredoxin 1 (EC:1.11.1.15) |
C00002656
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00002656
|
| 84722 | PSRC1, DDA3, FP3214 | proline/serine-rich coiled-coil 1 |
C00002656
|
| 9401 | RECQL4, RECQ4 | RecQ protein-like 4 (EC:3.6.4.12) |
C00002656
|
| 860 | RUNX2, AML3, CBF-alpha-1, CBFA1, CCD, CCD1, CLCD, OSF-2, OSF2, PEA2aA, PEBP2aA | runt-related transcription factor 2 |
C00002656
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00002656
|
| 83667 | SESN2, HI95, SES2, SEST2 | sestrin 2 |
C00002656
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00002656
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00002656
|
| 6688 | SPI1, OF, PU.1, SFPI1, SPI-1, SPI-A | spleen focus forming virus (SFFV) proviral integration oncogene |
C00002656
|
| 8614 | STC2, STC-2, STCRP | stanniocalcin 2 |
C00002656
|
| 7037 | TFRC, CD71, T9, TFR, TFR1, TR, TRFR, p90 | transferrin receptor |
C00002656
|
| 7077 | TIMP2, CSC-21K, DDC8 | TIMP metallopeptidase inhibitor 2 |
C00002656
|
| 25976 | TIPARP, ARTD14, PARP7, pART14 | TCDD-inducible poly(ADP-ribose) polymerase (EC:2.4.2.30) |
C00002656
|
| 8795 | TNFRSF10B, CD262, DR5, KILLER, KILLER/DR5, TRAIL-R2, TRAILR2, TRICK2, TRICK2A, TRICK2B, TRICKB, ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b |
C00002656
|
| 8793 | TNFRSF10D, CD264, DCR2, TRAIL-R4, TRAILR4, TRUNDD | tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
C00002656
|
| 8718 | TNFRSF25, APO-3, DDR3, DR3, LARD, TNFRSF12, TR3, TRAMP, WSL-1, WSL-LR | tumor necrosis factor receptor superfamily, member 25 |
C00002656
|
| 3604 | TNFRSF9, 4-1BB, CD137, CDw137, ILA | tumor necrosis factor receptor superfamily, member 9 |
C00002656
|
| 8742 | TNFSF12, APO3L, DR3LG, TWEAK | tumor necrosis factor (ligand) superfamily, member 12 |
C00002656
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00002656
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00002656
|
| 94241 | TP53INP1, SIP, TP53DINP1, TP53INP1A, TP53INP1B, Teap, p53DINP1 | tumor protein p53 inducible nuclear protein 1 |
C00002656
|
| 8717 | TRADD, Hs.89862 | TNFRSF1A-associated via death domain |
C00002656
|
| 9618 | TRAF4, CART1, MLN62, RNF83 | TNF receptor-associated factor 4 |
C00002656
|
| 286827 | TRIM59, MRF1, RNF104, TRIM57, TSBF1 | tripartite motif containing 59 |
C00002656
|
| 7295 | TXN, TRDX, TRX, TRX1 | thioredoxin |
C00002656
|
| 7474 | WNT5A, hWNT5A | wingless-type MMTV integration site family, member 5A |
C00002656
|
| 7486 | WRN, RECQ3, RECQL2, RECQL3 | Werner syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00002656
|
| 213 | ALB, PRO0883, PRO0903, PRO1341 | albumin |
C00002645
|
| 6376 | CX3CL1, ABCD-3, C3Xkine, CXC3, CXC3C, NTN, NTT, SCYD1, fractalkine, neurotactin | chemokine (C-X3-C motif) ligand 1 |
C00002645
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00002645
|
| 50848 | F11R, CD321, JAM, JAM1, JAMA, JCAM, KAT, PAM-1 | F11 receptor |
C00002645
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00002645
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00002645
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002645
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (72)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #613985 | Beta-thalassemia |
P68871
|
| #603902 | Beta-thalassemia, dominant inclusion body type |
P68871
|
| #210900 | Bloom syndrome; blm |
P54132
|
| %606641 | Body mass index; bmi |
P37231
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #133239 | Esophageal cancer |
P04637
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #137800 | Glioma susceptibility 1; glm1 |
P37231
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #140700 | Heinz body anemias |
P68871
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #211980 | Lung cancer |
P04637
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #257220 | Niemann-pick disease, type c1; npc1 |
O15118
|
| #601665 | Obesity |
P37231
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #603903 | Sickle cell anemia |
P68871
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
P04637
|
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #187950 | Thrombocythemia 1; thcyt1 |
P40225
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (87)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00136 | Niemann-Pick disease type C (NPC) |
O15118
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
P04637 (marker) Q16790 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
Q01453 (related) |
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
P37231 (related) |
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04637 (related) |
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P04150
(related)
|
| H00004 | Chronic myeloid leukemia (CML) |
P04637
(related)
|
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
|
| H00006 | Hairy-cell leukemia |
P04637
(related)
|
| H00008 | Burkitt lymphoma |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00010 | Multiple myeloma |
P04637
(related)
|
| H00013 | Small cell lung cancer |
P04637
(related)
|
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00016 | Oral cancer |
P04637
(related)
P04637 (marker) |
| H00017 | Esophageal cancer |
P04637
(related)
P04637 (marker) P35354 (related) |
| H00018 | Gastric cancer |
P04637
(related)
|
| H00019 | Pancreatic cancer |
P04637
(related)
P04637 (marker) |
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) P68871 (marker) |
| H00022 | Bladder cancer |
P04637
(related)
P68871 (marker) |
| H00025 | Penile cancer |
P04637
(related)
P04637 (marker) P35354 (related) |
| H00027 | Ovarian cancer |
P04637
(related)
|
| H00028 | Choriocarcinoma |
P04637
(related)
|
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00031 | Breast cancer |
P04637
(related)
|
| H00032 | Thyroid cancer |
P04637
(related)
P37231 (related) |
| H00036 | Osteosarcoma |
P04637
(related)
P08684 (marker) |
| H00038 | Malignant melanoma |
P04637
(related)
P14679 (marker) |
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00041 | Kaposi's sarcoma |
P04637
(related)
|
| H00042 | Glioma |
P04637
(related)
P04637 (marker) |
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00046 | Cholangiocarcinoma |
P04637
(related)
P35354 (related) |
| H00047 | Gallbladder cancer |
P04637
(related)
|
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00055 | Laryngeal cancer |
P04637
(related)
P04637 (marker) |
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00024 | Prostate cancer |
P10275
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00227 | Congenital amegakaryocytic thrombocytopenia (CAMT) |
P40225
(marker)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H00228 | Thalassemia |
P68871
(related)
|
| H00229 | Sickle cell anemia (SCA) |
P68871
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q03164
(related)
Q03164 (marker) |
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
Diseases related to CTD interactions
29 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D056486 | Drug-Induced Liver Injury |
C00002713
C00002656 |
| D007674 | Kidney Diseases |
C00002656
C00002645 |
| D002869 | Chromosome Aberrations |
C00002656
|
| D001919 | Bradycardia |
C00002713
|
| D005767 | Gastrointestinal Diseases |
C00002713
|
| D007022 | Hypotension |
C00002713
|
| D007249 | Inflammation |
C00002713
|
| D008107 | Liver Diseases |
C00002713
|
| D008113 | Liver Neoplasms |
C00002713
|
| D009362 | Neoplasm Metastasis |
C00002713
|
| D010243 | Paralysis |
C00002713
|
| D013276 | Stomach Ulcer |
C00002713
|
| D013927 | Thrombosis |
C00002713
|
| D001855 | Bone Marrow Diseases |
C00002656
|
| D003877 | Dermatitis, Contact |
C00002713
|
| D019457 | Chromosome Breakage |
C00002656
|
| D004487 | Edema |
C00002656
|
| D002286 | Carcinoma, Ehrlich Tumor |
C00002713
|
| D007680 | Kidney Neoplasms |
C00002656
|
| D008548 | Melanosis |
C00002656
|
| D048629 | Micronuclei, Chromosome-Defective |
C00002656
|
| D009784 | Occupational Diseases |
C00002656
|
| D011041 | Poisoning |
C00002656
|
| D011123 | Polyploidy |
C00002656
|
| D012140 | Respiratory Tract Diseases |
C00002656
|
| D012772 | Shock, Septic |
C00002656
|
| D002318 | Cardiovascular Diseases |
C00002645
|
| D003873 | Dermatitis, Exfoliative |
C00002645
|
| D006965 | Hyperplasia |
C00002645
|