Alpinia blepharocalyx
Species
KNApSAcK Entry
| Organism name | Alpinia blepharocalyx |
|---|---|
| Genus | Alpinia |
| Family | Zingiberaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Alpinia blepharocalyx |
|---|---|
| Linked NCBI taxonomy ID | 105668 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Zingiberaceae |
|---|---|
| ID | 4642 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | Liliopsida |
|---|---|
| ID | 4447 |
Metabolite list (36)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00006919
|
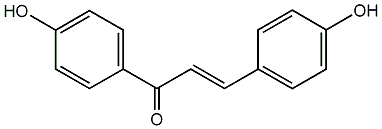
4,4'-Dihydroxychalcone
|
CHEMBL145927
|
C007423
|
No. 92 | No. 13 |
|
||
|
C00006954
|
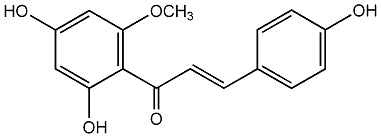
Helichrysetin
/ 4,2',4'-Trihydroxy-6'-methoxychalcone |
CHEMBL507998
|
C061023
|
No. 92 | No. 13 |
|
||
|
C00014289
|
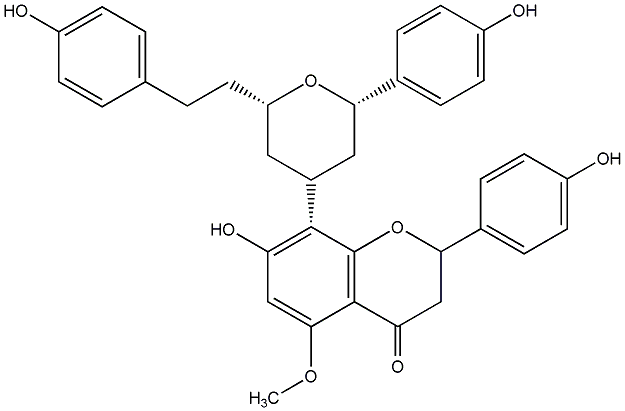
Calyxin K
|
CHEMBL455334
CHEMBL450782 |
No. 200 |
|
||||
|
C00014576
|
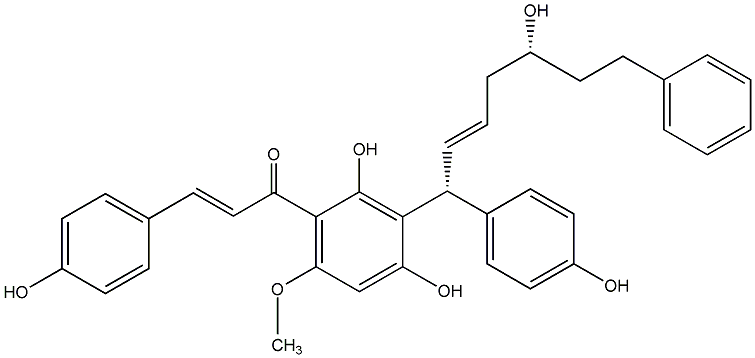
Epicalyxin H
|
CHEMBL1926699
|
No. 200 |
|
||||
|
C00008123
|
Calyxin A
|
No. 200 |
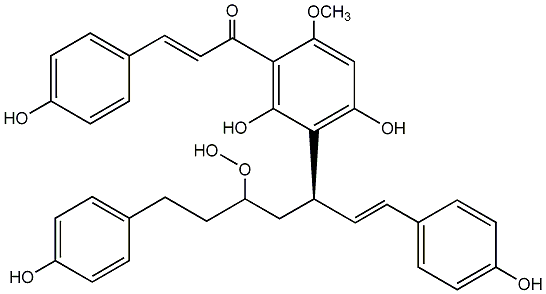
|
|||||
|
C00008124
|
Calyxin B
|
No. 200 |
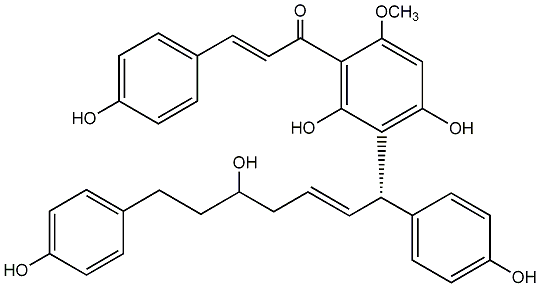
|
|||||
|
C00014285
|
Calyxin C
/ 2,3-Dihydro-7-hydroxy-8-[(1S,2E,5S)-5-hydroxy-1,7-bis(4-hydroxyphenyl)-2-heptenyl]-2-(4-hydroxyphenyl)-5-methoxy-4H-1-benzopyran-4-one |
No. 200 |
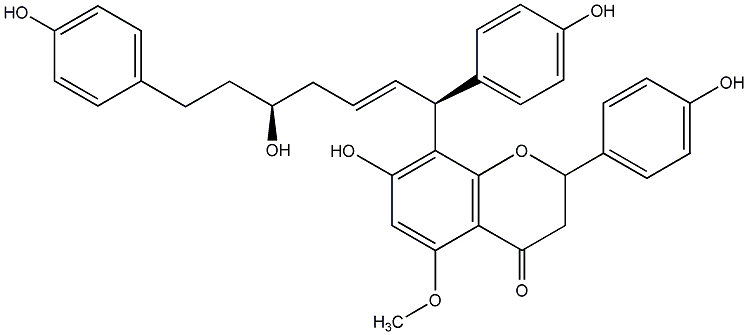
|
|||||
|
C00014286
|
Calyxin D
/ 2,3-Dihydro-7-hydroxy-8-[(1R,2E,5S)-5-hydroxy-1,7-bis(4-hydroxyphenyl)-2-heptenyl]-2-(4-hydroxyphenyl)-5-methoxy-4H-1-benzopyran-4-one |
No. 200 |
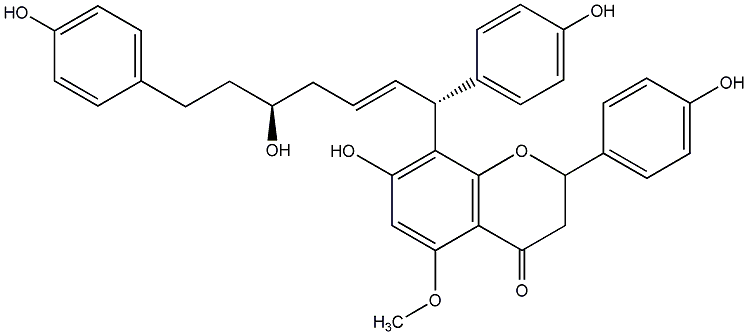
|
|||||
|
C00014287
|
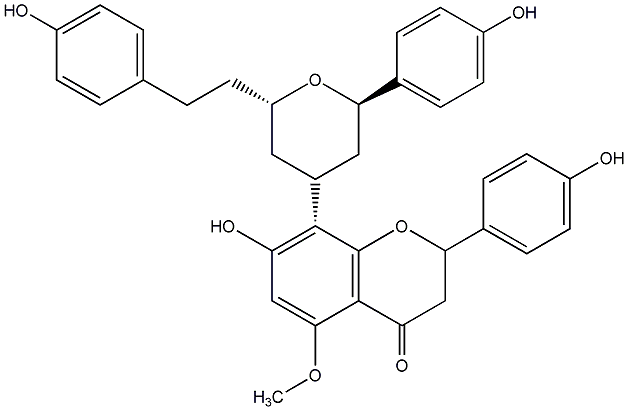
Calyxin G
|
CHEMBL455334
CHEMBL450782 |
No. 200 |
|
||||
|
C00014575
|
Calyxin H
|
CHEMBL1926699
|
No. 200 |

|
||||
|
C00014571
|
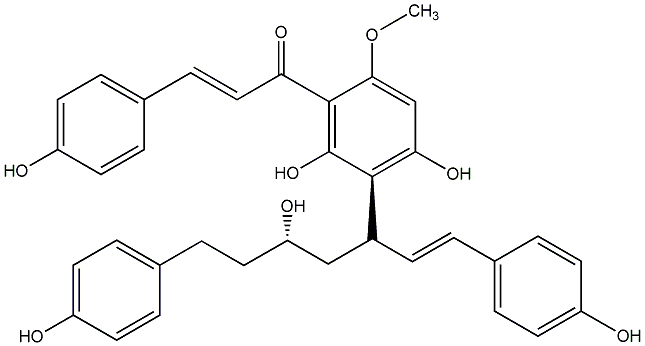
Deoxycalyxin A
|
CHEMBL504530
|
No. 200 |
|
||||
|
C00014290
|
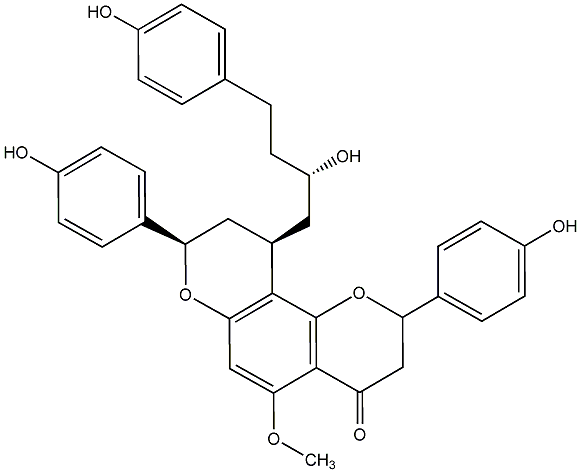
Calyxin M
|
CHEMBL457496
|
No. 200 |
|
||||
|
C00014291
|
Epicalyxin C
|
No. 200 |
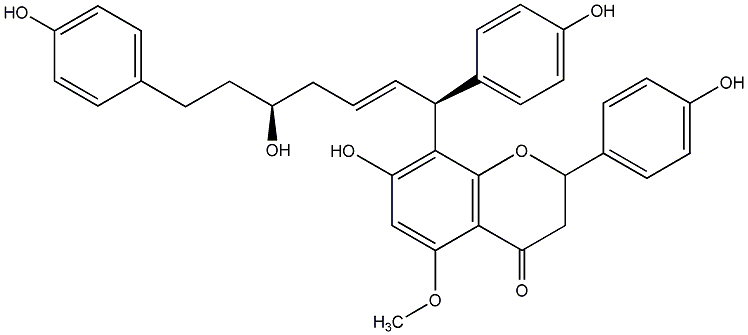
|
|||||
|
C00014292
|
Epicalyxin D
|
No. 200 |
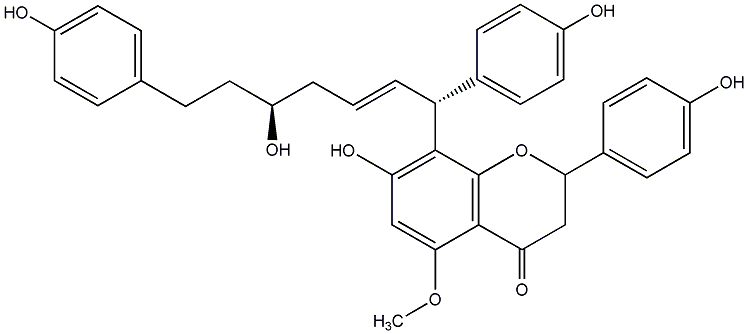
|
|||||
|
C00014293
|
Epicalyxin G
|
CHEMBL455334
CHEMBL450782 |
No. 200 |
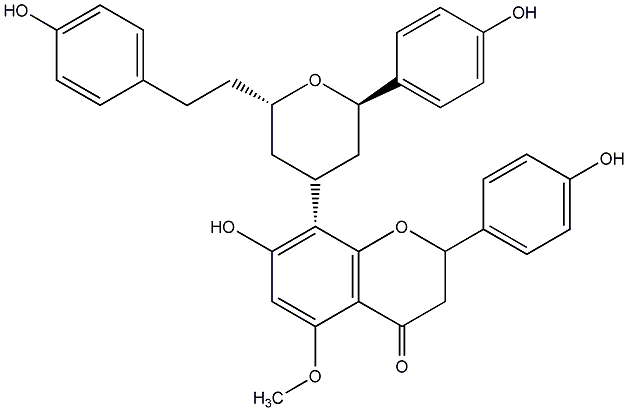
|
||||
|
C00014573
|
Calyxin F
|
No. 200 |
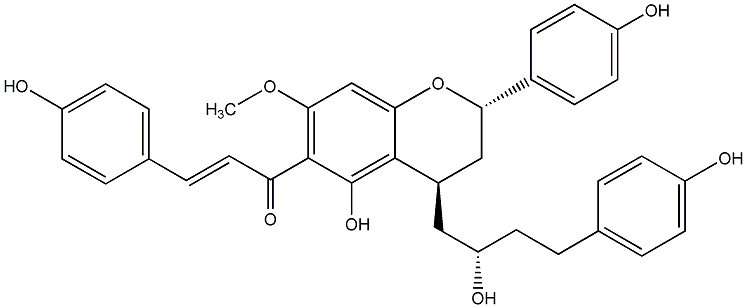
|
|||||
|
C00014295
|
Epicalyxin K
|
CHEMBL455334
CHEMBL450782 |
No. 200 |
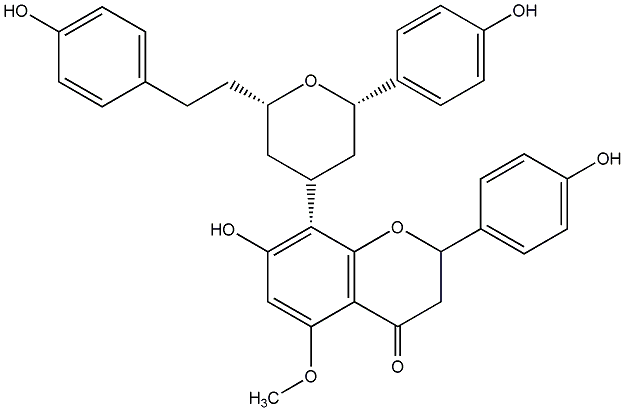
|
||||
|
C00014296
|
Epicalyxin M
|
CHEMBL457496
|
No. 200 |
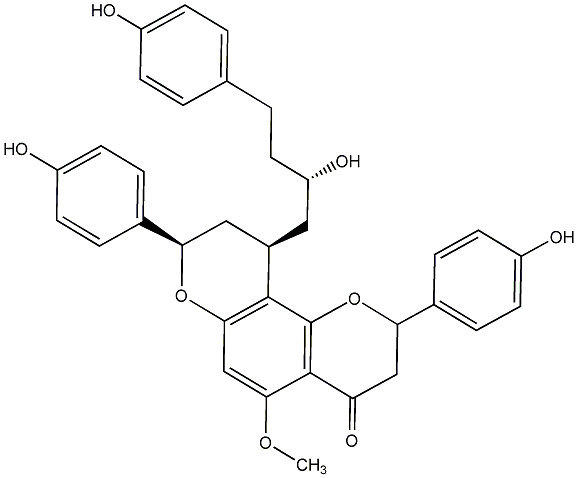
|
||||
|
C00014574
|
Calyxin L
/ Epicalyxin F |
CHEMBL451268
CHEMBL447717 |
No. 200 |
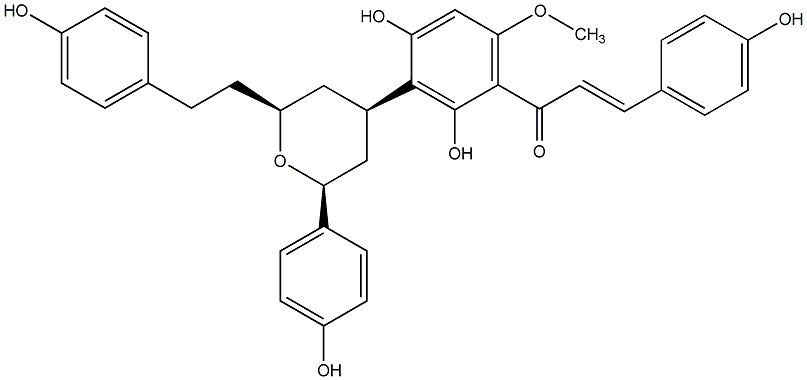
|
||||
|
C00014572
|
Epicalyxin B
/ 3-epi-Calyxin B |
No. 200 |
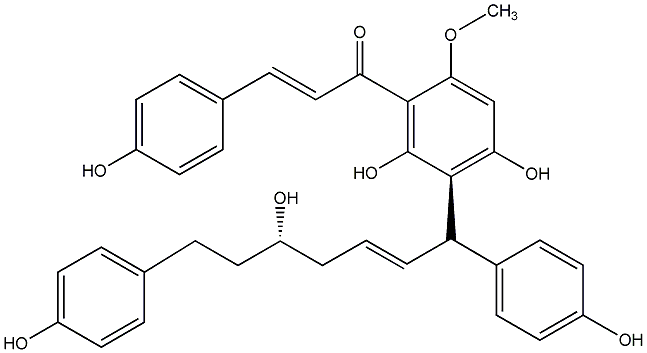
|
|||||
|
C00019308
|
Doursterol
/ Daucosterin / 3-O-beta-D-Glucopyranosyl sitosterol / beta-Sitosterol 3-O-beta-D-glucopyranoside |
CHEMBL197711
CHEMBL506678 CHEMBL2304043 |
C011015
|
5 / 4 / 2 | 0 / 3 | No. 520 |
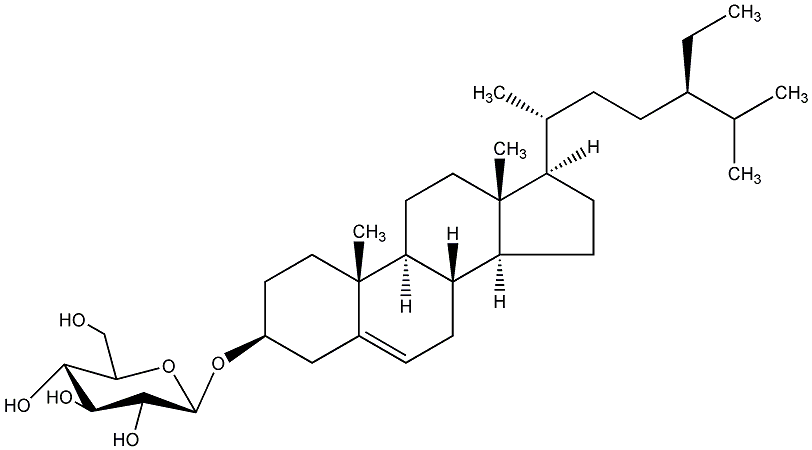
|
|
|
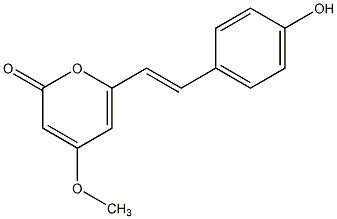
C00045571
|
p-Hydroxy-5,6-dehydrokawain
/ 4'-Hydroxy-5,6-dehydrokawain |
CHEMBL464111
|
No. 1312 | No. 63 |
|
|||
|
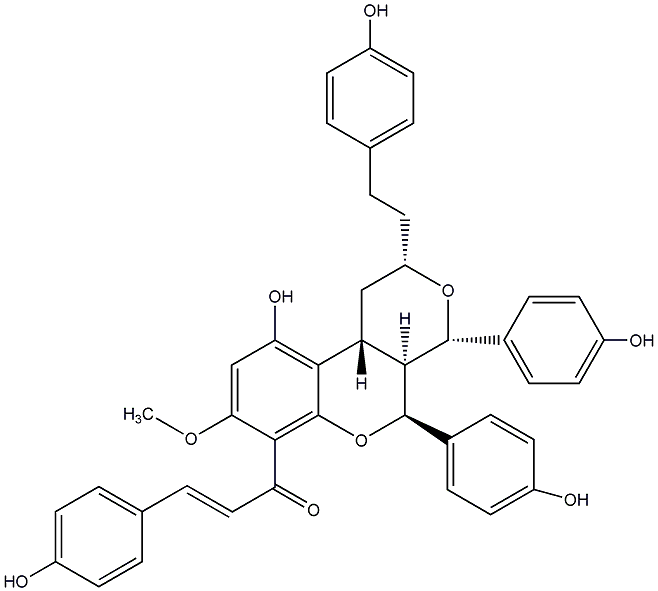
C00014578
|
Epicalyxin I
|
CHEMBL455049
CHEMBL503028 |
No. 1371 |
|
||||
|
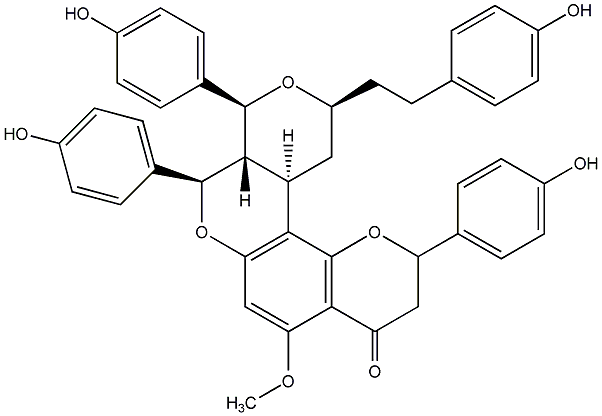
C00014294
|
Epicalyxin J
|
CHEMBL510115
|
No. 1371 |
|
||||
|
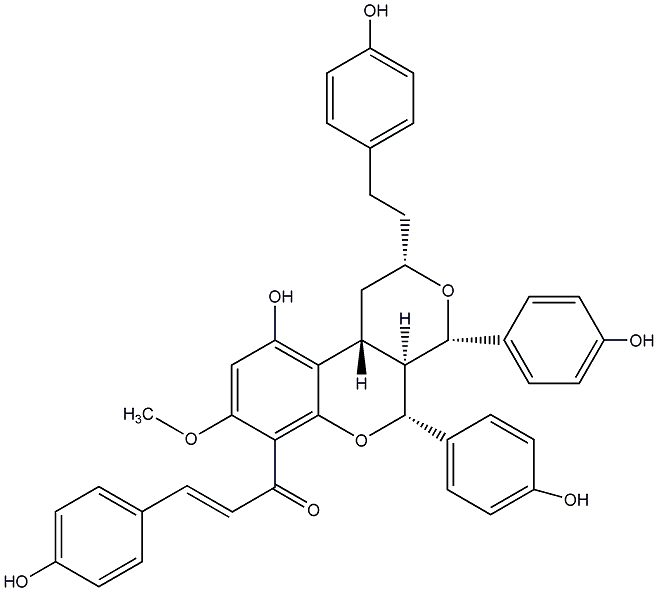
C00014577
|
Calyxin I
|
CHEMBL455049
CHEMBL503028 |
No. 1371 |
|
||||
|
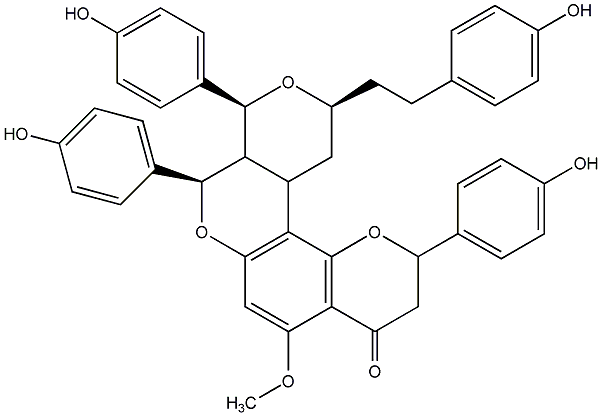
C00014288
|
Calyxin J
|
CHEMBL510115
|
No. 1371 |
|
||||
|
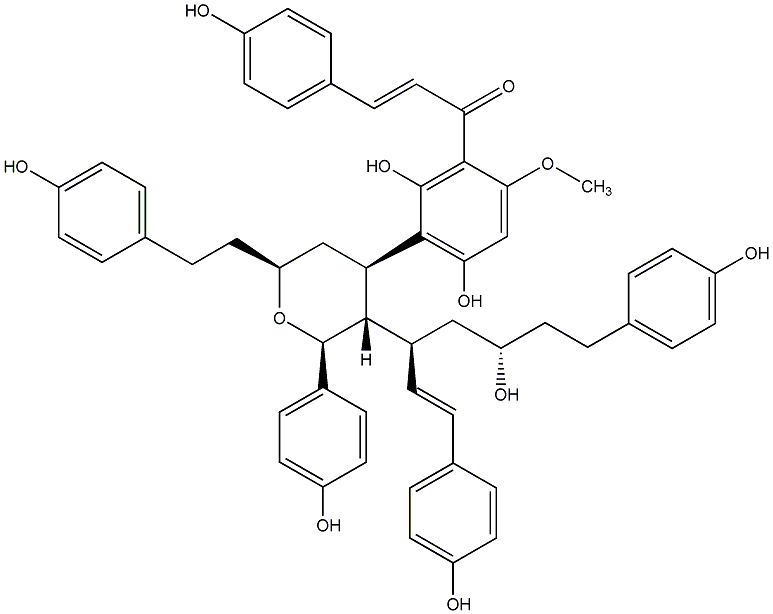
C00014581
|
Blepharocalyxin E
|
No. 1371 |
|
|||||
|
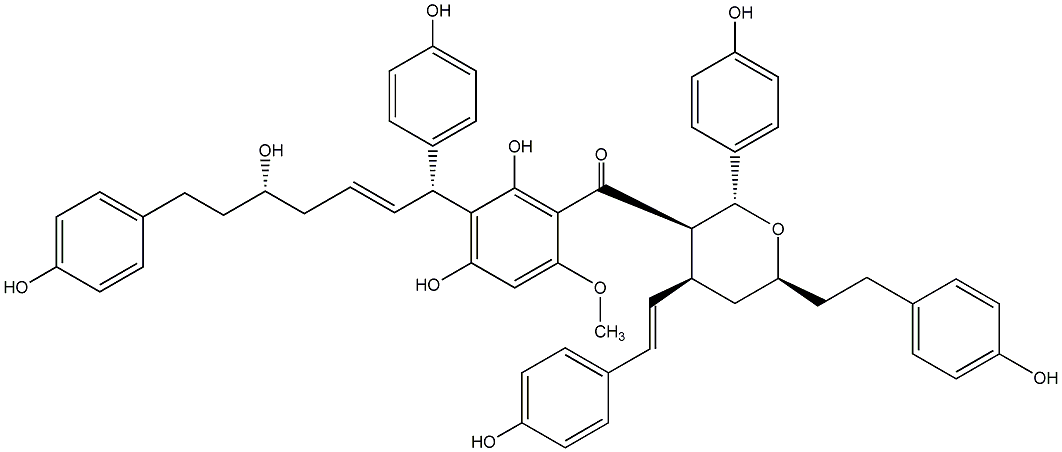
C00014580
|
Blepharocalyxin B
/ (-)-Blepharocalyxin B |
No. 1371 |
|
|||||
|
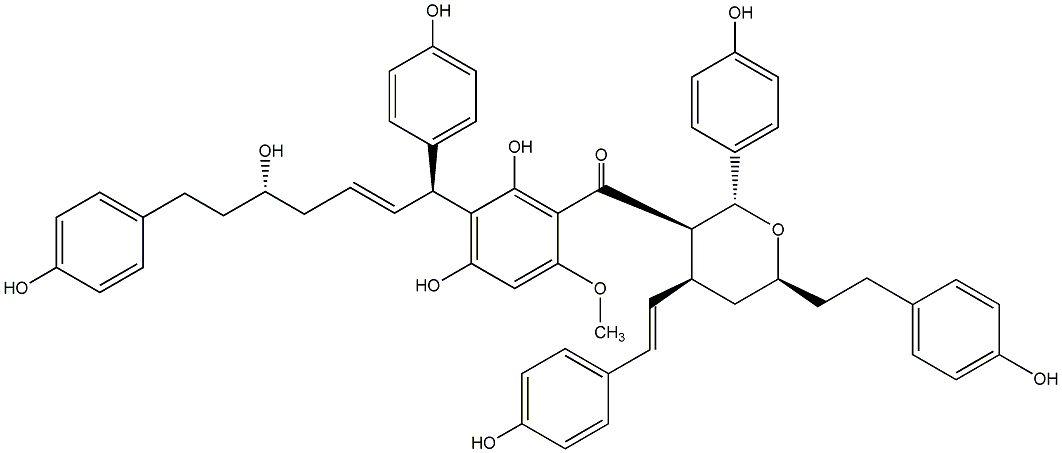
C00014579
|
Blepharocalyxin A
/ (-)-Blepharocalyxin A |
No. 1371 |
|
|||||
|
C00002665
|
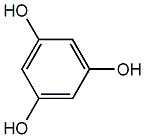
Phloroglucinol
|
CHEMBL473159
|
D010696
|
3 / 4 / 2 | 8 / 2 | No. 1590 | No. 82 |
|
|
C00002657
|
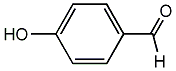
p-Formylphenol
/ 4-Hydroxybenzaldehyde / p-Hydroxybenzaldehyde |
CHEMBL14193
|
C011483
|
3 / 2 / 2 | No. 2076 |
|
||
|
C00032012
|
Methyl p-coumarate
/ Methyl p-hydroxycinnamate / Methyl 4-hydroxy cinnamate / 4-Hydroxycinnamic acid methyl ester |
CHEMBL146816
|
C057918
|
6 / 2 / 4 | No. 2416 |

|
||
|
C00046202
|
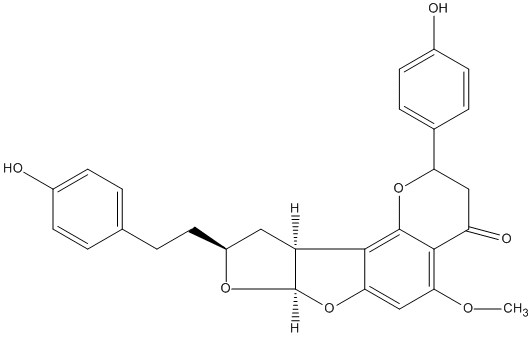
Neocalyxin A
|
CHEMBL518161
|
No. 4904 |
|
||||
|
C00046203
|
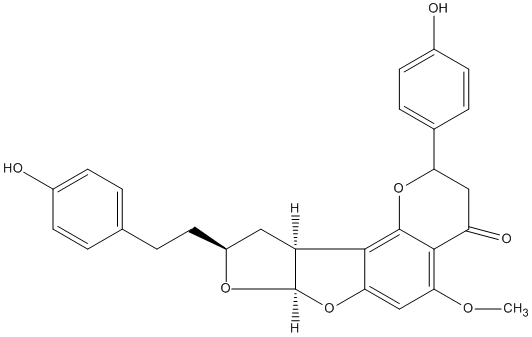
Neocalyxin B
/ (-)-Neocalyxin B |
CHEMBL518161
|
No. 4904 |
|
||||
|
C00045687
|
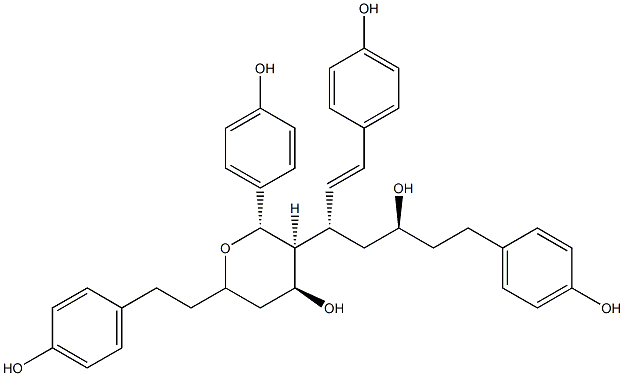
Blepharocalyxin C
/ (+)-Blepharocalyxin C |
CHEMBL448211
|
No. 5225 |
|
||||
|
C00045688
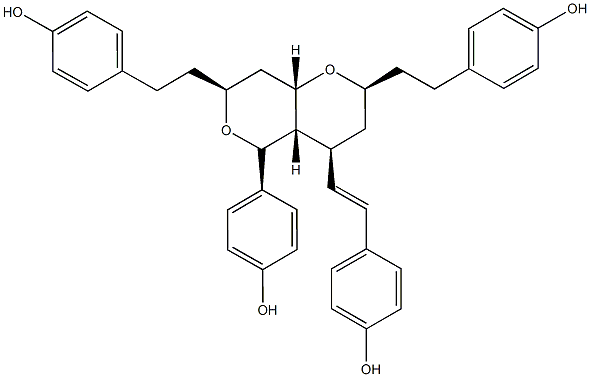
|
Blepharocalyxin D
/ (+)-Blepharocalyxin D |
CHEMBL446847
|
No. 5225 |
|
Human Protein / Gene in interactions
17 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00019308 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00019308 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00032012 | 0 / 0 |
| P08047 | Transcription factor Sp1 | Unclassified protein | C00019308 | 0 / 0 |
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00002657 | 1 / 1 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00032012 | 1 / 2 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00032012 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00032012 | 1 / 2 |
| P14902 | Indoleamine 2,3-dioxygenase 1 | Enzyme | C00002657 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00032012 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00019308 | 0 / 0 |
| P00734 | Prothrombin | S1A | C00019308 | 4 / 2 |
| P14679 | Tyrosinase | Oxidoreductase | C00002665 | 4 / 2 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00002657 | 1 / 1 |
| P56817 | Beta-secretase 1 | A1A | C00002665 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00032012 | 0 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00002665 | 0 / 0 |
8 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00002665
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00002665
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00002665
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00002665
|
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00002665
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00002665
|
| 2932 | GSK3B | glycogen synthase kinase 3 beta (EC:2.7.11.1 2.7.11.26) |
C00002665
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00002665
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (12)
| OMIM | preferred title | UniProt |
|---|---|---|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #601367 | Stroke, ischemic |
P00734
|
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
KEGG DISEASE (10)
| KEGG | name | UniProt |
|---|---|---|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00038 | Malignant melanoma |
P14679
(marker)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|