Polygala caudata
Species
KNApSAcK Entry
| Organism name | Polygala caudata |
|---|---|
| Genus | Polygala |
| Family | Polygalaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Polygala |
|---|---|
| Linked NCBI taxonomy ID | 4275 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Polygalaceae |
|---|---|
| ID | 4274 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (15)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00040043
|
Polycaudoside A
|
No. 1 | No. 15 |

|
||||
|
C00040666
|
Wubangziside A
|
C046985
|
No. 1 | No. 15 |

|
|||
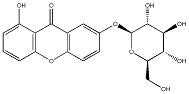
|
C00040667
|
Wubangziside B
|
C046986
|
No. 2 | No. 15 |
|
|||
|
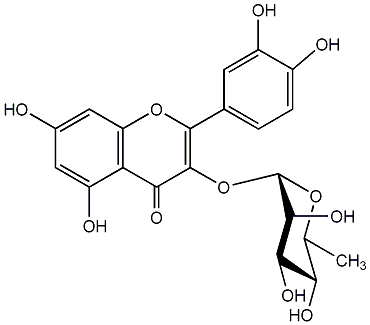
C00005374
|
Quercetin
|
CHEMBL82242
CHEMBL479232 CHEMBL1437696 |
C012526
|
14 / 2 / 2 | 2 / 1 | No. 2 | No. 15 |
|
|
C00034991
|
1,3-Dihydroxy-2-methoxyxanthone
|
No. 3 | No. 15 |

|
||||
|
C00002960
|
Lancerin
|
No. 22 | No. 15 |

|
||||
|
C00032065
|
Neolancerin
|
No. 22 | No. 15 |

|
||||
|
C00038223
|
2'-Benzoylmangiferin
|
CHEMBL466996
|
No. 30 | No. 15 |

|
|||
|
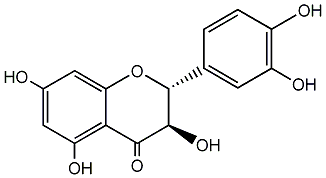
C00000677
|
Distylin
/ Dihydroquercetin / (2R,3R)-Taxifolin |
CHEMBL66
CHEMBL9249 CHEMBL337309 CHEMBL1492383 |
C003377
|
65 / 41 / 37 | 29 / 1 | No. 42 | No. 14 |
|
|
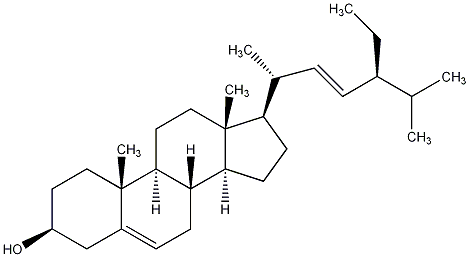
C00023774
|
Fucostanol
/ Stigmasterol / Dihydro-beta-sitosterol / (24S)24-Ethylcholestain-3beta-ol |
CHEMBL66943
CHEMBL186373 CHEMBL400247 CHEMBL1568947 |
D013265
|
5 / 0 / 0 | 1 / 0 | No. 53 | No. 11 |
|
|
C00038109
|
1,2,8-Trihydroxyxanthone
|
No. 71 | No. 15 |

|
||||
|
C00002953
|
Gentisein
|
CHEMBL361025
|
2 / 1 / 1 | No. 71 | No. 15 |

|
||
|
C00029364
|
Euxanthone
/ 1,7-Dihydroxyxanthone |
CHEMBL389166
|
C404221
|
1 / 0 / 0 | No. 76 | No. 15 |

|
|
|
C00029509
|
Stigmasterol-3-O-beta-D-glucopyranoside
/ beta-Stigmasteryl 3-O-beta-D-glucopyranoside |
CHEMBL447335
|
C054293
|
2 / 0 / 0 | No. 520 |

|
||
|
C00000856
|
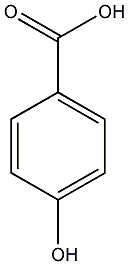
4-Hydroxybenzoic acid
/ p-Hydroxybenzoic acid |
CHEMBL441343
|
C038193
|
21 / 7 / 16 | 2 / 1 | No. 817 | No. 81 |
|
Human Protein / Gene in interactions
102 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P06746 | DNA polymerase beta | Enzyme | C00000677 C00023774 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00000677 C00005374 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00000677 C00005374 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00000677 C00005374 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00000677 C00005374 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00000677 C00005374 | 1 / 1 |
| P39748 | Flap endonuclease 1 | Enzyme | C00000677 C00005374 | 0 / 0 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00000677 C00005374 | 1 / 1 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00000856 | 0 / 0 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00000677 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000677 | 1 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00000677 | 1 / 1 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00000856 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00000856 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00000856 | 0 / 0 |
| P39900 | Macrophage metalloelastase | M10A | C00000677 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00000677 | 2 / 2 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00000677 | 0 / 0 |
| P28907 | ADP-ribosyl cyclase 1 | Enzyme | C00000677 | 0 / 1 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00000677 | 0 / 0 |
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00000856 | 1 / 1 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00000856 | 1 / 2 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00002953 | 1 / 1 |
| P34969 | 5-hydroxytryptamine receptor 7 | Serotonin receptor | C00023774 | 0 / 0 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00029509 | 0 / 0 |
| P49327 | Fatty acid synthase | Transferase | C00002953 | 0 / 0 |
| Q9UQ49 | Sialidase-3 | Enzyme | C00000856 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00000677 | 0 / 1 |
| P07237 | Protein disulfide-isomerase | Enzyme | C00005374 | 0 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00000677 | 1 / 2 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00000856 | 0 / 0 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00000856 | 1 / 8 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00000677 | 0 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00000856 | 0 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00000856 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00005374 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00000856 | 1 / 2 |
| P36888 | Receptor-type tyrosine-protein kinase FLT3 | Pdgfr | C00000677 | 1 / 1 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00000677 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00000677 | 2 / 0 |
| O75496 | Geminin | Unclassified protein | C00000677 | 0 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00000856 | 0 / 0 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00000677 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00005374 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00000856 | 0 / 0 |
| P03956 | Interstitial collagenase | M10A | C00000677 | 0 / 1 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00000856 | 0 / 0 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00005374 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00000677 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00000677 | 0 / 0 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00000856 | 1 / 1 |
| P45452 | Collagenase 3 | M10A | C00000677 | 1 / 1 |
| P18054 | Arachidonate 12-lipoxygenase, 12S-type | Enzyme | C00000677 | 2 / 0 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00029509 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00000677 | 0 / 0 |
| P56817 | Beta-secretase 1 | A1A | C00000677 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00000677 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00000856 | 0 / 1 |
| P08253 | 72 kDa type IV collagenase | M10A | C00000677 | 1 / 3 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00000677 | 3 / 3 |
| Q92887 | Canalicular multispecific organic anion transporter 1 | Unclassified protein | C00000677 | 1 / 1 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00000677 | 2 / 2 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00000677 | 5 / 2 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00000856 | 0 / 0 |
| P09884 | DNA polymerase alpha catalytic subunit | Transferase | C00023774 | 0 / 0 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00000677 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00000677 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00000677 | 0 / 0 |
| P51580 | Thiopurine S-methyltransferase | Enzyme | C00000856 | 1 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00000677 | 1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000677 | 0 / 1 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00000677 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00005374 | 0 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00000677 | 4 / 1 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00000677 | 4 / 3 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00000856 | 0 / 0 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00000677 | 0 / 0 |
| P08254 | Stromelysin-1 | M10A | C00000677 | 1 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00000856 | 1 / 1 |
| Q9P1W9 | Serine/threonine-protein kinase pim-2 | Pim | C00005374 | 0 / 0 |
| P24298 | Alanine aminotransferase 1 | Enzyme | C00000677 | 0 / 0 |
| P35610 | Sterol O-acyltransferase 1 | Enzyme | C00029364 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00000677 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00000677 | 0 / 0 |
| O00255 | Menin | Unclassified protein | C00000677 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00000677 | 1 / 2 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00000677 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00000677 | 1 / 4 |
| Q05513 | Protein kinase C zeta type | Iota | C00000677 | 0 / 0 |
| Q04759 | Protein kinase C theta type | Delta | C00000677 | 0 / 1 |
| Q02156 | Protein kinase C epsilon type | Eta | C00000677 | 0 / 0 |
| O94806 | Serine/threonine-protein kinase D3 | Pkd | C00000677 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00000677 | 0 / 0 |
| Q05655 | Protein kinase C delta type | Delta | C00000677 | 0 / 0 |
| P05129 | Protein kinase C gamma type | Alpha | C00000677 | 1 / 1 |
| P05771 | Protein kinase C beta type | Alpha | C00000677 | 0 / 0 |
| P24723 | Protein kinase C eta type | Eta | C00000677 | 1 / 0 |
| P41743 | Protein kinase C iota type | Iota | C00000677 | 0 / 0 |
| Q15139 | Serine/threonine-protein kinase D1 | Pkd | C00000677 | 0 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00023774 | 0 / 0 |
| Q02880 | DNA topoisomerase 2-beta | Isomerase | C00023774 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00005374 | 0 / 0 |
34 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00000677
|
| 335 | APOA1 | apolipoprotein A-I |
C00000677
|
| 338 | APOB, FLDB, LDLCQ4 | apolipoprotein B |
C00000677
|
| 8900 | CCNA1 | cyclin A1 |
C00000677
|
| 993 | CDC25A, CDC25A2 | cell division cycle 25A (EC:3.1.3.48) |
C00000677
|
| 1571 | CYP2E1, CPE1, CYP2E, P450-J, P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) |
C00000677
|
| 1630 | DCC, CRC18, CRCR1, IGDCC1, MRMV1 | deleted in colorectal carcinoma |
C00000677
|
| 1950 | EGF, HOMG4, URG | epidermal growth factor |
C00000677
|
| 8817 | FGF18, FGF-18, ZFGF5 | fibroblast growth factor 18 |
C00000677
|
| 2248 | FGF3, HBGF-3, INT2 | fibroblast growth factor 3 |
C00000677
|
| 2621 | GAS6, AXLLG, AXSF | growth arrest-specific 6 |
C00000677
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00000677
|
| 2944 | GSTM1, GST1, GSTM1-1, GSTM1a-1a, GSTM1b-1b, GTH4, GTM1, H-B, MU, MU-1 | glutathione S-transferase mu 1 (EC:2.5.1.18) |
C00000677
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00000677
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00000677
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00000677
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00000677
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00000677
|
| 4547 | MTTP, ABL, MTP | microsomal triglyceride transfer protein |
C00000677
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00000677
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00000677
|
| 5155 | PDGFB, IBGC5, PDGF-2, PDGF2, SIS, SSV, c-sis | platelet-derived growth factor beta polypeptide |
C00000677
|
| 5921 | RASA1, CM-AVM, CMAVM, GAP, PKWS, RASA, RASGAP, p120GAP, p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 |
C00000677
|
| 6772 | STAT1, CANDF7, ISGF-3, STAT91 | signal transducer and activator of transcription 1, 91kDa |
C00000677
|
| 7187 | TRAF3, CAP-1, CAP1, CD40bp, CRAF1, IIAE5, LAP1 | TNF receptor-associated factor 3 |
C00000677
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00000677
|
| 7498 | XDH, XO, XOR | xanthine dehydrogenase (EC:1.17.1.4 1.17.3.2) |
C00000677
|
| 7507 | XPA, XP1, XPAC | xeroderma pigmentosum, complementation group A |
C00000677
|
| 7518 | XRCC4 | X-ray repair complementing defective repair in Chinese hamster cells 4 |
C00000677
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00005374
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00005374
|
| 3952 | LEP, LEPD, OB, OBS | leptin |
C00000856
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00000856
|
| 10599 | SLCO1B1, HBLRR, LST-1, LST1, OATP-C, OATP1B1, OATP2, OATPC, SLC21A6 | solute carrier organic anion transporter family, member 1B1 |
C00023774
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (49)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #300615 | Brunner syndrome |
P21397
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #114500 | Colorectal cancer; crc |
P18054
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #237500 | Dubin-johnson syndrome; djs |
Q92887
|
| #133239 | Esophageal cancer |
P18054
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #601626 | Leukemia, acute myeloid; aml |
P36888
|
| #211980 | Lung cancer |
P00533
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #605361 | Spinocerebellar ataxia 14; sca14 |
P05129
|
| #602111 | Spondyloepimetaphyseal dysplasia, missouri type |
P45452
|
| #601367 | Stroke, ischemic |
P24723
|
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #610460 | Thiopurine s-methyltransferase deficiency |
P51580
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (53)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
|
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
|
| H00018 | Gastric cancer |
P00533
(related)
|
| H00022 | Bladder cancer |
P00533
(related)
|
| H00028 | Choriocarcinoma |
P00533
(related)
P03956 (related) P08253 (related) |
| H00030 | Cervical cancer |
P00533
(related)
|
| H00042 | Glioma |
P00533
(related)
P00533 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
P05129
(related)
|
| H00025 | Penile cancer |
P08253
(related)
P14780 (related) |
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
P45452 (related) |
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H00005 | Chronic lymphocytic leukemia (CLL) |
P28907
(marker)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00003 | Acute myeloid leukemia (AML) |
P36888
(related)
Q01196 (related) Q01196 (marker) Q13951 (marker) |
| H00192 | Xanthinuria |
P47989
(related)
|
| H00964 | Thiopurine S-methyltransferase deficiency (TPMT deficiency) |
P51580
(related)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
Q01453
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00408 | Type I diabetes mellitus |
Q04759
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00208 | Hyperbilirubinemia |
Q92887
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|