Balanophora abbreviata B1
Species
KNApSAcK Entry
| Organism name | Balanophora abbreviata B1 |
|---|---|
| Genus | Balanophora |
| Family | Balanophoraceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Balanophora |
|---|---|
| Linked NCBI taxonomy ID | 25674 |
| Linked level | genus |
Family
| Family in NCBI taxonomy | Balanophoraceae |
|---|---|
| ID | 25673 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | eudicotyledons |
|---|---|
| ID | 71240 |
Metabolite list (9)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
C00007190
|
(+)-Pinoresinol
|
CHEMBL267963
CHEMBL487611 CHEMBL460862 |
C103298
|
7 / 5 / 6 | No. 38 | No. 21 |
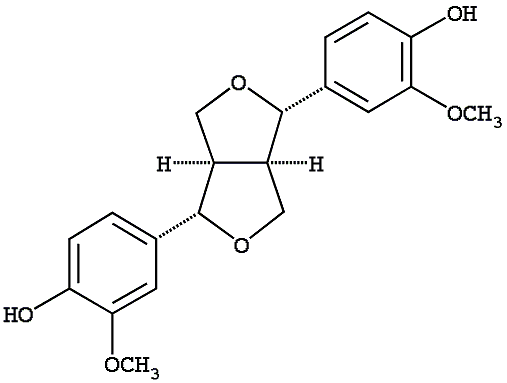
|
|
|
C00031451
|
(-)-Pinoresinol O-beta-D-glucopyranoside
/ (-)-Pinoresinol 4-O-beta-D-glucopyranoside |
CHEMBL573336
CHEMBL1083218 CHEMBL1302893 |
9 / 6 / 4 | No. 174 | No. 22 |
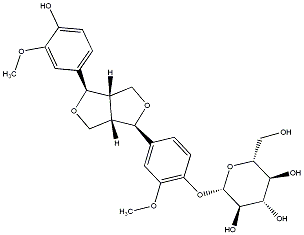
|
||
|
C00002728
|
Coniferaldehyde
/ Coniferyl aldehyde / 4-Hydroxy-3-methoxycinnamaldehyde |
CHEMBL242529
CHEMBL1956165 |
C075384
|
1 / 1 / 1 | No. 310 | No. 6 |
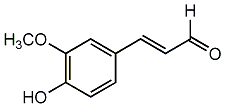
|
|
|
C00031446
|
(-)-ent-Isolariciresinol
|
CHEMBL399512
CHEMBL1760593 |
2 / 1 / 1 | No. 406 |

|
|||
|
C00019308
|
Doursterol
/ Daucosterin / 3-O-beta-D-Glucopyranosyl sitosterol / beta-Sitosterol 3-O-beta-D-glucopyranoside |
CHEMBL197711
CHEMBL506678 CHEMBL2304043 |
C011015
|
5 / 4 / 2 | 0 / 3 | No. 520 |
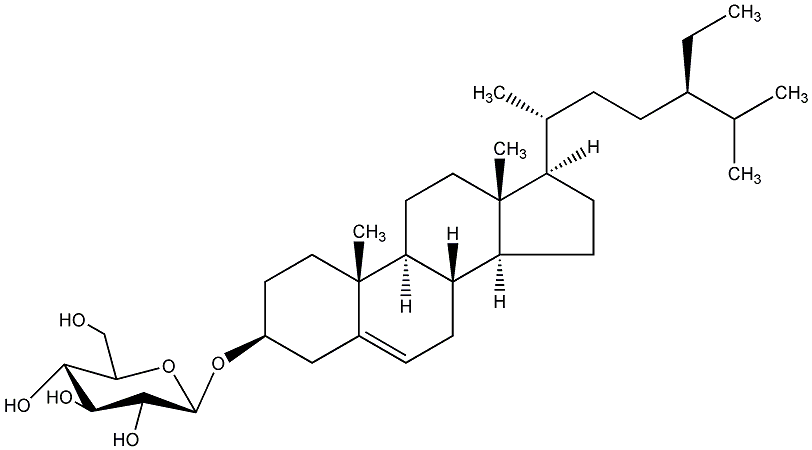
|
|
|
C00002727
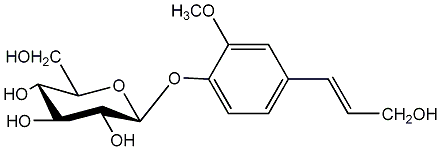
|
Coniferin
/ Coniferoside |
CHEMBL459056
|
C016316
|
2 / 0 / 0 | No. 678 | No. 6 |
|
|
|
C00000603
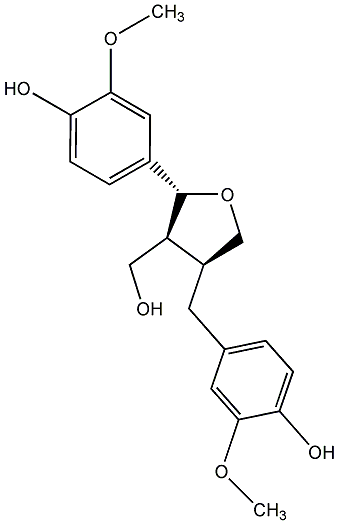
|
(-)-Lariciresinol
|
CHEMBL518421
|
C060282
|
2 / 1 / 1 | No. 700 | No. 21 |
|
|
|
C00029961
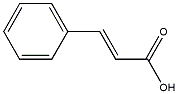
|
Cinnamic acid
/ .beta-Phenylacrylic acid |
CHEMBL27246
|
C029010
|
5 / 2 / 2 | 16 / 2 | No. 904 | No. 6 |
|
|
C00000152
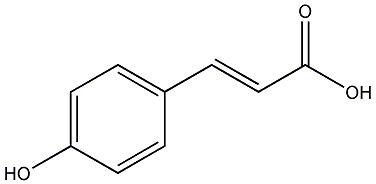
|
p-Coumaric acid
|
CHEMBL66879
CHEMBL2336752 |
C032171
|
23 / 13 / 18 | 6 / 1 | No. 904 | No. 6 |
|
Human Protein / Gene in interactions
47 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000603 C00031446 | 1 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00019308 C00029961 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000603 C00031446 | 0 / 1 |
| P03372 | Estrogen receptor | NR3A1 | C00000152 C00029961 | 1 / 1 |
| P15121 | Aldose reductase | Enzyme | C00000152 C00029961 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00000152 C00007190 | 0 / 3 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00002728 C00029961 | 1 / 1 |
| P06746 | DNA polymerase beta | Enzyme | C00019308 C00031451 | 0 / 0 |
| Q8TDS4 | Hydroxycarboxylic acid receptor 2 | Hydroxycarboxylic acid receptor | C00000152 C00029961 | 0 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00000152 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00000152 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00000152 | 0 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00007190 | 0 / 0 |
| P08047 | Transcription factor Sp1 | Unclassified protein | C00019308 | 0 / 0 |
| P37840 | Alpha-synuclein | Unclassified protein | C00031451 | 4 / 2 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00031451 | 1 / 1 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00000152 | 1 / 2 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00000152 | 1 / 8 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00002727 | 0 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00000152 | 0 / 0 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00000152 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00000152 | 1 / 2 |
| P42330 | Aldo-keto reductase family 1 member C3 | Enzyme | C00000152 | 0 / 0 |
| Q13093 | Platelet-activating factor acetylhydrolase | Enzyme | C00000152 | 3 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00002727 | 0 / 0 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00007190 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00000152 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00019308 | 0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00031451 | 1 / 1 |
| P00734 | Prothrombin | S1A | C00019308 | 4 / 2 |
| P14679 | Tyrosinase | Oxidoreductase | C00000152 | 4 / 2 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00007190 | 5 / 3 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00000152 | 0 / 1 |
| Q04828 | Aldo-keto reductase family 1 member C1 | Enzyme | C00000152 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00007190 | 0 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00000152 | 0 / 0 |
| P17516 | Aldo-keto reductase family 1 member C4 | Enzyme | C00000152 | 1 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00031451 | 0 / 0 |
| P52895 | Aldo-keto reductase family 1 member C2 | Enzyme | C00000152 | 1 / 0 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | C00007190 | 0 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00000152 | 0 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00000152 | 1 / 1 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00031451 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00031451 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00031451 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00031451 | 0 / 0 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00007190 | 0 / 0 |
20 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00000152
C00029961
|
| 7299 | TYR, ATN, CMM8, OCA1, OCA1A, OCAIA, SHEP3 | tyrosinase (EC:1.14.18.1) |
C00000152
C00029961
|
| 1586 | CYP17A1, CPT7, CYP17, P450C17, S17AH | cytochrome P450, family 17, subfamily A, polypeptide 1 (EC:1.14.99.9 4.1.2.30) |
C00029961
|
| 1558 | CYP2C8, CPC8, CYPIIC8, MP-12/MP-20 | cytochrome P450, family 2, subfamily C, polypeptide 8 (EC:1.14.14.1) |
C00029961
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00029961
|
| 1565 | CYP2D6, CPD6, CYP2D, CYP2D7AP, CYP2D7BP, CYP2D7P2, CYP2D8P2, CYP2DL1, CYPIID6, P450-DB1, P450C2D, P450DB1 | cytochrome P450, family 2, subfamily D, polypeptide 6 (EC:1.14.14.1) |
C00029961
|
| 1571 | CYP2E1, CPE1, CYP2E, P450-J, P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) |
C00029961
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00029961
|
| 338442 | HCAR2, GPR109A, HCA2, HM74a, HM74b, NIACR1, PUMAG, Puma-g | hydroxycarboxylic acid receptor 2 |
C00029961
|
| 3284 | HSD3B2, HSD3B, HSDB, SDR11E2 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 (EC:1.1.1.145 5.3.3.1) |
C00029961
|
| 1557 | CYP2C19, CPCJ, CYP2C, P450C2C, P450IIC19 | cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00029961
|
| 1548 | CYP2A6, CPA6, CYP2A, CYP2A3, CYPIIA6, P450C2A, P450PB | cytochrome P450, family 2, subfamily A, polypeptide 6 (EC:1.14.14.1) |
C00029961
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00029961
|
| 54659 | UGT1A3, UDPGT, UDPGT_1-3, UGT-1C, UGT1-03, UGT1.3, UGT1C | UDP glucuronosyltransferase 1 family, polypeptide A3 (EC:2.4.1.17) |
C00029961
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00029961
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00029961
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00000152
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00000152
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00000152
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00000152
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (30)
| OMIM | preferred title | UniProt |
|---|---|---|
| #614279 | 46,xy sex reversal 8; srxy8 |
P17516
P52895 |
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #600807 | Asthma, susceptibility to |
Q13093
|
| %606641 | Body mass index; bmi |
P37231
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #127750 | Dementia, lewy body; dlb |
P37840
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #137800 | Glioma susceptibility 1; glm1 |
P37231
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #147050 | Ige responsiveness, atopic; iger |
Q13093
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #211980 | Lung cancer |
P00533
|
| #601665 | Obesity |
P37231
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #168601 | Parkinson disease 1, autosomal dominant; park1 |
P37840
|
| #605543 | Parkinson disease 4, autosomal dominant; park4 |
P37840
|
| #168600 | Parkinson disease, late-onset; pd |
P37840
|
| #614278 | Platelet-activating factor acetylhydrolase deficiency; pafad |
Q13093
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #601367 | Stroke, ischemic |
P00734
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (29)
| KEGG | name | UniProt |
|---|---|---|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
Q16790 (marker) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
|
| H00022 | Bladder cancer |
P00533
(related)
|
| H00028 | Choriocarcinoma |
P00533
(related)
|
| H00030 | Cervical cancer |
P00533
(related)
|
| H00042 | Glioma |
P00533
(related)
P00533 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) |
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00038 | Malignant melanoma |
P14679
(marker)
|
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00032 | Thyroid cancer |
P37231
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P37231
(related)
|
| H00057 | Parkinson's disease (PD) |
P37840
(related)
|
| H00066 | Lewy body dementia (LBD) |
P37840
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|