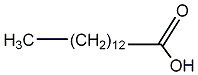Metabolite
KNApSAcK Entry
| id | C00001228 |
|---|---|
| Name | Myristic acid / Tetradecanoic acid / n-Tetradecanoic acid |
| CAS RN | 544-63-8 |
| Standard InChI | InChI=1S/C14H28O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14(15)16/h2-13H2,1H3,(H,15,16) |
| Standard InChI (Main Layer) | InChI=1S/C14H28O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14(15)16/h2-13H2,1H3,(H,15,16) |
Cluster
| Phytochemical cluster | No. 68 |
|---|---|
| KCF-S cluster | No. 1141 |
Link
ChEMBL
| By standard InChI | CHEMBL111077 |
|---|---|
| By standard InChI Main Layer | CHEMBL111077 |
KEGG
| By LinkDB | C06424 |
|---|
CTD
| By CAS RN | D019814 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| Embryophyta | 6 |
| rosids | 6 |
| eudicotyledons | 3 |
| Magnoliophyta | 3 |
| asterids | 2 |
| Liliopsida | 1 |
Family (Top 10)
| family name | count |
|---|---|
| Frullaniaceae | 6 |
| Brassicaceae | 2 |
| Calophyllaceae | 2 |
| Saururaceae | 2 |
| Myristicaceae | 1 |
| Cistaceae | 1 |
| Iridaceae | 1 |
| Crassulaceae | 1 |
| Saxifragaceae | 1 |
| Malvaceae | 1 |
List (28)
* NCBI
Human Protein / Gene in interaction
17 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| O60603 | Toll-like receptor 2 | Membrane receptor | CHEMBL111077 |
CHEMBL867222
(2)
|
1 / 1 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | CHEMBL111077 |
CHEMBL1908087
(1)
|
0 / 0 |
| Q05193 | Dynamin-1 | Structural | CHEMBL111077 |
CHEMBL673521
(1)
|
0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | CHEMBL111077 |
CHEMBL1794339
(1)
|
2 / 3 |
| O15296 | Arachidonate 15-lipoxygenase B | Enzyme | CHEMBL111077 |
CHEMBL1613800
(1)
|
0 / 0 |
| P17405 | Sphingomyelin phosphodiesterase | Enzyme | CHEMBL111077 |
CHEMBL1794495
(1)
|
2 / 2 |
| P39748 | Flap endonuclease 1 | Enzyme | CHEMBL111077 |
CHEMBL1794486
(1)
|
0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | CHEMBL111077 |
CHEMBL1738606
(1)
|
0 / 0 |
| O75496 | Geminin | Unclassified protein | CHEMBL111077 |
CHEMBL2114780
(1)
|
0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | CHEMBL111077 |
CHEMBL2114788
(1)
|
0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | CHEMBL111077 |
CHEMBL1794401
(1)
|
0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | CHEMBL111077 |
CHEMBL976708
(1)
|
2 / 2 |
| Q96RI1 | Bile acid receptor | NR1H4 | CHEMBL111077 |
CHEMBL1794415
(1)
|
0 / 0 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | CHEMBL111077 |
CHEMBL1908085
(1)
|
0 / 0 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | CHEMBL111077 |
CHEMBL2114915
(2)
|
0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | CHEMBL111077 |
CHEMBL1614364
(1)
|
1 / 1 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | CHEMBL111077 |
CHEMBL2354287
(1)
|
1 / 1 |
CTD interaction (4)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| D019814 | 199974 |
CYP4Z1
CYP4A20 |
cytochrome P450, family 4, subfamily Z, polypeptide 1 (EC:1.14.14.1) | CYP4Z1 protein results in decreased abundance of Myristic Acid |
decreases abundance
|
protein |
22841774
|
| D019814 | 199974 |
CYP4Z1
CYP4A20 |
cytochrome P450, family 4, subfamily Z, polypeptide 1 (EC:1.14.14.1) | N-hydroxy-N'-(4-butyl-2-methylphenyl)formamidine inhibits the reaction [CYP4Z1 protein results in decreased abundance of Myristic Acid] |
decreases abundance
/ decreases reaction |
protein |
22841774
|
| D019814 | 5465 |
PPARA
NR1C1 PPAR PPARalpha hPPAR |
peroxisome proliferator-activated receptor alpha | Myristic Acid binds to PPARA protein |
affects binding
|
protein |
10403814
|
| D019814 | 10891 |
PPARGC1A
LEM6 PGC-1(alpha) PGC-1v PGC1 PGC1A PPARGC1 |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | Myristic Acid results in increased expression of PPARGC1A mRNA |
increases expression
|
mRNA |
23056435
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (9)
| OMIM | preferred title | UniProt |
|---|---|---|
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #246300 | Leprosy, susceptibility to, 3; lprs3 |
O60603
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #257200 | Niemann-pick disease, type a |
P17405
|
| #607616 | Niemann-pick disease, type b |
P17405
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
KEGG DISEASE (10)
| KEGG | disease name | UniProt |
|---|---|---|
| H00344 | Leprosy |
O60603
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H00137 | Niemann-Pick disease (NPD) typeA and B |
P17405
(related)
|
| H00424 | Defects in the degradation of sphingomyelin |
P17405
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
Q13148
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q9NUW8
(related)
|