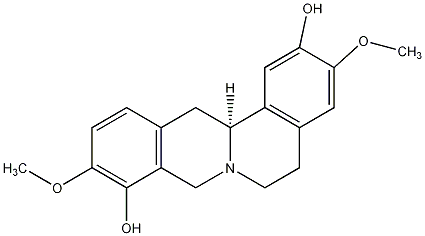
Metabolite
KNApSAcK Entry
| id | C00026092 |
|---|---|
| Name | Scoulerine / (-)-Scoulerine |
| CAS RN | 6451-73-6 |
| Standard InChI | InChI=1S/C19H21NO4/c1-23-17-4-3-11-7-15-13-9-16(21)18(24-2)8-12(13)5-6-20(15)10-14(11)19(17)22/h3-4,8-9,15,21-22H,5-7,10H2,1-2H3/t15-/m0/s1 |
| Standard InChI (Main Layer) | InChI=1S/C19H21NO4/c1-23-17-4-3-11-7-15-13-9-16(21)18(24-2)8-12(13)5-6-20(15)10-14(11)19(17)22/h3-4,8-9,15,21-22H,5-7,10H2,1-2H3 |
Cluster
| Phytochemical cluster | No. 4 |
|---|---|
| KCF-S cluster | No. 37 |
Link
ChEMBL
| By standard InChI | CHEMBL1235966 |
|---|---|
| By standard InChI Main Layer | CHEMBL191133 CHEMBL1235966 CHEMBL1395394 |
KEGG
| By LinkDB | C02106 |
|---|
CTD
| By CAS RN |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| eudicotyledons | 31 |
| Magnoliophyta | 2 |
| rosids | 2 |
Family
| family name | count |
|---|---|
| Papaveraceae | 30 |
| Rutaceae | 1 |
| Menispermaceae | 1 |
| Lauraceae | 1 |
| Annonaceae | 1 |
| Fabaceae | 1 |
List (36)
* NCBI
Human Protein / Gene in interaction
27 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | CHEMBL1395394 |
CHEMBL1614110
(1)
CHEMBL1741321
(1)
|
1 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | CHEMBL1235966 CHEMBL1395394 |
CHEMBL2114784
(2)
|
1 / 1 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | CHEMBL1235966 CHEMBL1395394 |
CHEMBL2343280
(2)
CHEMBL2343284
(2)
|
0 / 0 |
| P29466 | Caspase-1 | C14 | CHEMBL1395394 |
CHEMBL1614158
(1)
|
0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | CHEMBL1395394 |
CHEMBL1614544
(1)
|
11 / 10 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | CHEMBL1395394 |
CHEMBL1741325
(1)
|
0 / 1 |
| P14416 | D(2) dopamine receptor | Dopamine receptor | CHEMBL1235966 CHEMBL1395394 |
CHEMBL2343279
(2)
CHEMBL2343283
(2)
|
2 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | CHEMBL1235966 |
CHEMBL1794486
(1)
|
0 / 0 |
| O75496 | Geminin | Unclassified protein | CHEMBL1235966 |
CHEMBL2114843
(1)
CHEMBL2114780
(1)
|
0 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | CHEMBL1235966 |
CHEMBL2114788
(1)
|
0 / 0 |
| Q9Y253 | DNA polymerase eta | Enzyme | CHEMBL1235966 |
CHEMBL1794569
(1)
|
1 / 1 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | CHEMBL1235966 |
CHEMBL1614280
(1)
|
0 / 0 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | CHEMBL1235966 |
CHEMBL1963863
(1)
CHEMBL2114934
(1)
|
0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | CHEMBL1395394 |
CHEMBL1794467
(1)
CHEMBL1794492
(1)
|
0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | CHEMBL1395394 |
CHEMBL1741322
(1)
|
0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | CHEMBL1235966 |
CHEMBL1738588
(1)
|
0 / 0 |
| P28223 | 5-hydroxytryptamine receptor 2A | Serotonin receptor | CHEMBL1235966 CHEMBL1395394 |
CHEMBL2343277
(2)
CHEMBL2343281
(2)
|
0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | CHEMBL1395394 |
CHEMBL1613777
(1)
CHEMBL1741323
(1)
|
1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | CHEMBL1395394 |
CHEMBL1614108
(1)
CHEMBL1613886
(1)
CHEMBL1741324 (1) |
0 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | CHEMBL1235966 |
CHEMBL1794483
(1)
|
0 / 0 |
| O14727 | Apoptotic protease-activating factor 1 | Unclassified protein | CHEMBL1235966 |
CHEMBL1794319
(1)
CHEMBL1794383
(1)
|
0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | CHEMBL1235966 |
CHEMBL1738184
(1)
|
0 / 0 |
| P08908 | 5-hydroxytryptamine receptor 1A | Serotonin receptor | CHEMBL1235966 CHEMBL1395394 |
CHEMBL2343278
(2)
CHEMBL2343282
(2)
|
1 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | CHEMBL1395394 |
CHEMBL1613914
(1)
|
0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | CHEMBL1235966 |
CHEMBL1738442
(1)
|
0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | CHEMBL1235966 |
CHEMBL1964002
(1)
CHEMBL2354311
(1)
|
1 / 0 |
| Q8IUX4 | DNA dC->dU-editing enzyme APOBEC-3F | Enzyme | CHEMBL1235966 |
CHEMBL2114859
(1)
CHEMBL2114734
(1)
|
0 / 0 |
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (19)
| OMIM | preferred title | UniProt |
|---|---|---|
| #103780 | Alcohol dependence |
P14416
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #159900 | Myoclonic dystonia |
P14416
|
| #614674 | Periodic fever, menstrual cycle-dependent |
P08908
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (15)
| KEGG | disease name | UniProt |
|---|---|---|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|