Diospyros kaki
Species
KNApSAcK Entry
| Organism name | Diospyros kaki |
|---|---|
| Genus | Diospyros |
| Family | Ebenaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Diospyros kaki |
|---|---|
| Linked NCBI taxonomy ID | 35925 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Ebenaceae |
|---|---|
| ID | 19955 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | asterids |
|---|---|
| ID | 71274 |
Metabolite list (38)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
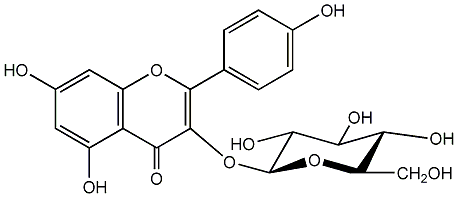
C00005138
|
Astragalin
/ Kaempferol 3-glucoside / Kaempferol 3-O-beta-D-glucoside / Kaempferol 3-O-beta-D-glucopyranoside |
CHEMBL233930
CHEMBL453290 CHEMBL1572115 |
C001579
|
10 / 6 / 7 | 0 / 1 | No. 2 | No. 15 |
|
|
C00009020
|
Leucodelphinidin 3-glucoside
|
No. 12 | No. 14 |

|
||||
|
C00036107
|
Coussaric acid
|
CHEMBL492458
|
1 / 0 / 0 | No. 13 | No. 51 |

|
||
|
C00031074
|
Pomolic acid
|
CHEMBL486986
CHEMBL464567 |
2 / 0 / 0 | No. 13 | No. 51 |

|
||
|
C00029789
|
Barbinervic acid
|
CHEMBL523209
CHEMBL493908 CHEMBL1271052 |
C052363
|
2 / 0 / 0 | No. 13 | No. 51 |

|
|
|
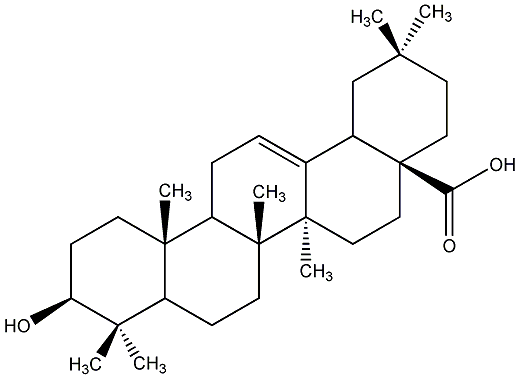
C00019064
|
Oleanolic acid
/ Astrantiagenin C / Virgaureagenin B / 3beta-Hydroxyolean-12-en-28-oic acid |
CHEMBL56615
CHEMBL168 CHEMBL180553 CHEMBL365375 CHEMBL486382 CHEMBL1413646 CHEMBL1436454 |
D009828
|
30 / 8 / 12 | 21 / 15 | No. 13 | No. 51 |
|
|
C00031228
|
Rotungenic acid
/ (+)-Rotungenic acid |
CHEMBL523209
CHEMBL493908 CHEMBL1271052 |
2 / 0 / 0 | No. 13 | No. 51 |

|
||
|
C00032467
|
Uvaol
/ Uvalol / Urs-12-ene-3beta,28-diol / 3beta,28-Dihydroxyurs-12-ene |
CHEMBL58872
CHEMBL399873 |
C013330
|
5 / 2 / 2 | No. 13 | No. 51 |

|
|
|
C00003758
|
Alnulin
/ Tiliadin / Skimmiol / Taraxerol |
CHEMBL511822
CHEMBL470462 |
C005802
|
No. 13 | No. 51 |

|
||
|
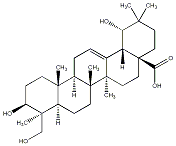
C00040339
|
Spathodic acid
|
CHEMBL493893
|
1 / 0 / 0 | No. 13 | No. 51 |
|
||
|
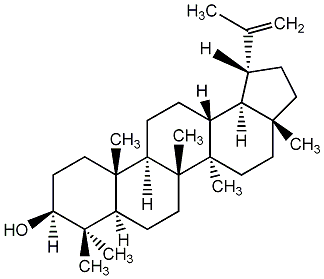
C00003749
|
Lupeol
/ Lupenol / (+)-Lupenol |
CHEMBL289191
CHEMBL459702 |
C010480
|
3 / 0 / 0 | 2 / 6 | No. 23 | No. 51 |
|
|
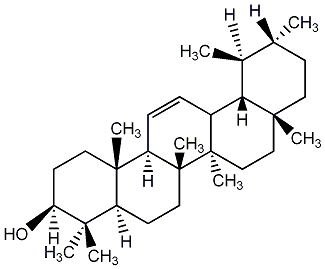
C00003737
|
alpha-Amyrin
/ alpha-Amyrine / alpha-Amyrenol |
No. 23 | No. 51 |
|
||||
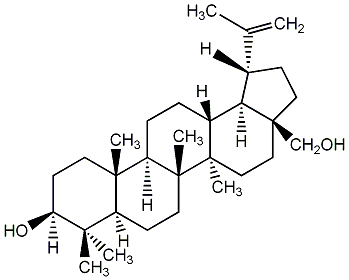
|
C00003740
|
Betulin
|
CHEMBL23236
CHEMBL140040 CHEMBL1610940 CHEMBL2000891 CHEMBL2069124 |
C002503
|
18 / 16 / 17 | 0 / 2 | No. 23 | No. 51 |
|
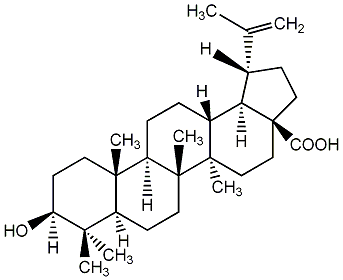
|
C00003741
|
Betulinic acid
|
CHEMBL269277
CHEMBL71690 CHEMBL519059 CHEMBL1318530 CHEMBL2005635 |
C002070
|
34 / 17 / 14 | 10 / 2 | No. 23 | No. 51 |
|
|
C00003765
|
alpha-Carotene
|
No. 26 | No. 59 |

|
||||
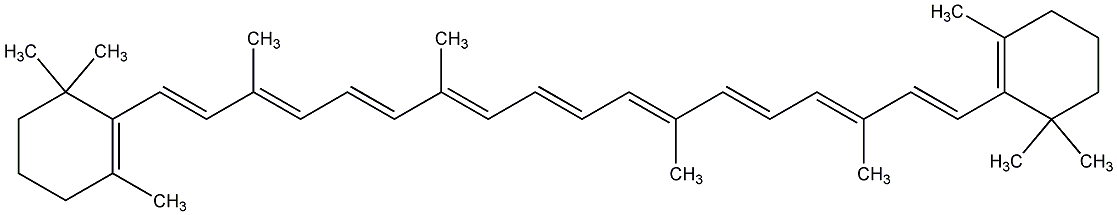
|
C00000919
|
beta-Carotene
|
CHEMBL1293
CHEMBL1966639 |
D019207
|
7 / 12 / 11 | 104 / 19 | No. 26 | No. 59 |
|
|
C00003672
|
Sitosterol
/ beta-sitosterl / (-)-beta-Sitosterol / Stigmast-5-en-3beta-ol |
CHEMBL221542
CHEMBL1398443 CHEMBL1875388 |
17 / 19 / 12 | No. 53 | No. 11 |
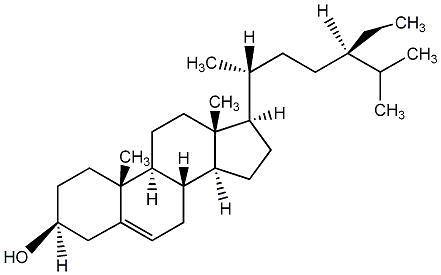
|
||
|
C00023774
|
Fucostanol
/ Stigmasterol / Dihydro-beta-sitosterol / (24S)24-Ethylcholestain-3beta-ol |
CHEMBL66943
CHEMBL186373 CHEMBL400247 CHEMBL1568947 |
D013265
|
5 / 0 / 0 | 1 / 0 | No. 53 | No. 11 |
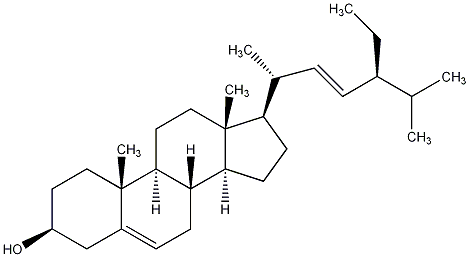
|
|
C00003647
|
Campesterol
/ 24alpha-Methylcholesterol / (24R)24-Methylcholest-5-en-3beta-ol |
CHEMBL520535
CHEMBL485421 CHEMBL1836653 |
C021273
|
No. 53 | No. 11 |
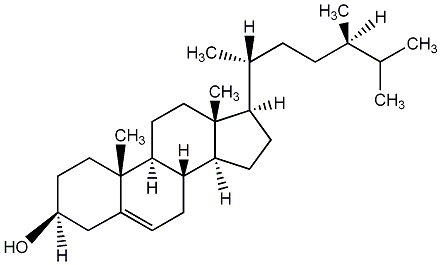
|
||
|
C00000911
|
Lycopene
/ all-trans-Lycopene |
CHEMBL501174
CHEMBL1984187 |
C015329
|
6 / 13 / 13 | 84 / 13 | No. 97 | No. 59 |
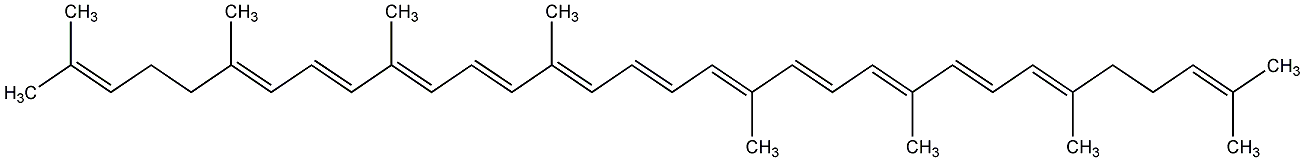
|
|
C00005957
|
Quercetin 3-(2''-galloylglucoside)
|
CHEMBL459260
CHEMBL505880 |
No. 98 |
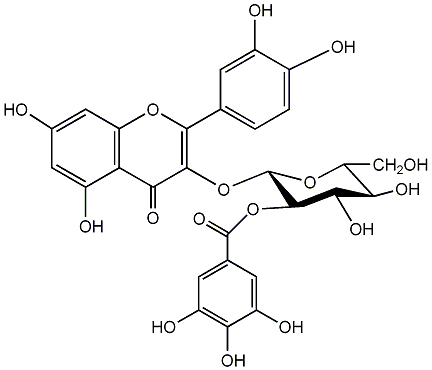
|
||||
|
C00005846
|
Kaempferol 3-(2''-galloylglucoside)
|
CHEMBL444191
|
No. 98 |

|
||||
|
C00000926
|
gamma-Carotene
|
No. 131 | No. 59 |
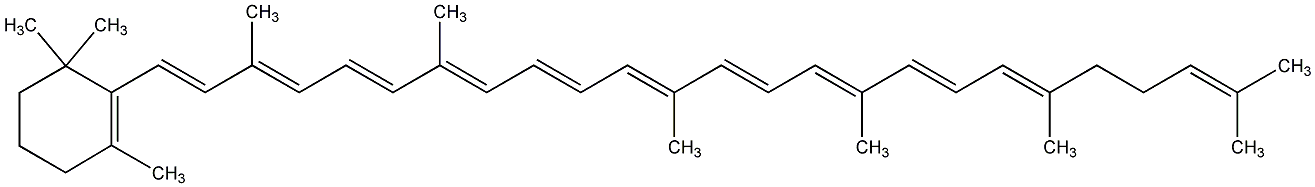
|
||||
|
C00029633
|
Ursolic acid
/ Acetylursolic acid |
CHEMBL55086
CHEMBL410525 |
4 / 2 / 2 | No. 177 |
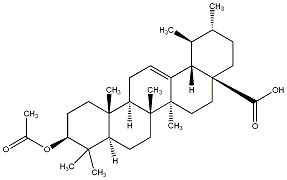
|
|||
|
C00002831
|
Isodiospyrin
|
CHEMBL1992900
|
C409666
|
No. 312 |

|
|||
|
C00033160
|
Maritinone
/ 8,8'-Biplumbagin |
CHEMBL512154
|
C079947
|
No. 312 |

|
|||
|
C00002816
|
Euclein
/ Diospyrin |
CHEMBL242550
|
C043546
|
No. 312 |

|
|||
|
C00043762
|
Neodiospyrin
|
CHEMBL241392
|
No. 312 |

|
||||
|
C00043693
|
Mamegakinone
|
No. 312 |

|
|||||
|
C00000107
|
IAN
/ Indole-3-acetonitrile |
CHEMBL1812654
|
C016516
|
No. 710 | No. 4 |
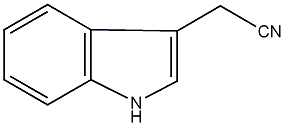
|
||
|
C00000332
|
Methyl pheophorbide a
|
CHEMBL408290
CHEMBL504821 CHEMBL1985396 |
C032625
|
No. 990 |

|
|||
|
C00000323
|
Methyl pheophorbide b
|
No. 990 |

|
|||||
|
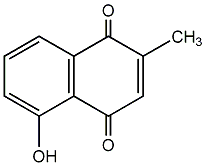
C00002852
|
Plumbagin
/ Plumbagine |
CHEMBL295316
|
C014758
|
28 / 30 / 59 | 14 / 3 | No. 1047 | No. 80 |
|
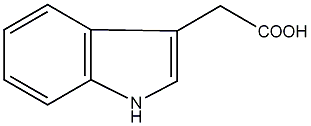
|
C00000100
|
IAA
/ Indole-3-acetic acid |
CHEMBL82411
|
C030737
|
3 / 0 / 0 | 2 / 1 | No. 1432 | No. 4 |
|
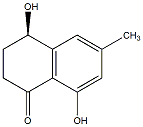
|
C00043916
|
Shinanolone
|
CHEMBL391802
CHEMBL459915 |
No. 1520 |
|
||||
|
C00043192
|
3-Methoxy-7-methyljuglone
|
CHEMBL449429
|
No. 3269 |

|
||||
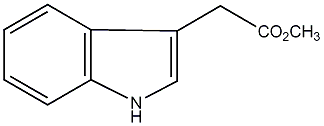
|
C00000101
|
IAA methyl ester
/ Methyl indole-3-acetate |
No. 4225 |
|
|||||
|
C00002859
|
Ramentaceone
/ 7-Methyljuglone |
CHEMBL430853
|
No. 4286 | No. 80 |
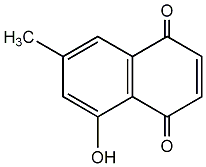
|
Human Protein / Gene in interactions
101 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00003672 C00003740 C00019064 C00029789 C00031074 C00031228 C00032467 C00036107 C00040339 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00003672 C00003741 C00019064 C00023774 C00029633 | 0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | C00000911 C00000919 C00003740 C00003741 | 11 / 10 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00002852 C00005138 C00019064 C00029633 | 0 / 0 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00002852 C00003672 C00003740 C00003741 | 0 / 1 |
| P11387 | DNA topoisomerase 1 | Isomerase | C00002852 C00003740 C00003741 C00003749 | 0 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00003740 C00003741 C00003749 C00023774 | 0 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00003740 C00003741 C00019064 C00032467 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00000911 C00000919 C00003741 C00019064 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00003740 C00003741 C00019064 C00032467 | 0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | C00003672 C00003741 C00019064 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00003741 C00005138 C00019064 | 0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00003672 C00003740 C00003741 | 0 / 1 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00000919 C00002852 C00019064 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00003672 C00003740 C00003741 | 0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00003672 C00003740 C00003741 | 1 / 1 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00003740 C00003741 C00019064 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00003672 C00003740 C00003741 | 1 / 0 |
| P08047 | Transcription factor Sp1 | Unclassified protein | C00003672 C00003749 C00031074 | 0 / 0 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00000911 C00002852 C00003741 | 0 / 0 |
| P15559 | NAD(P)H dehydrogenase [quinone] 1 | Enzyme | C00003741 C00029789 C00031228 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00000911 C00002852 C00003740 | 0 / 0 |
| Q02156 | Protein kinase C epsilon type | Eta | C00003740 C00003741 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00002852 C00003672 | 1 / 1 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00002852 C00029633 | 0 / 0 |
| P80365 | Corticosteroid 11-beta-dehydrogenase isozyme 2 | Enzyme | C00003672 C00032467 | 1 / 1 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00005138 C00019064 | 0 / 0 |
| P05771 | Protein kinase C beta type | Alpha | C00003740 C00003741 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00005138 C00019064 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00003672 C00003741 | 2 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00003741 C00019064 | 0 / 0 |
| P14679 | Tyrosinase | Oxidoreductase | C00003672 C00005138 | 4 / 2 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00019064 C00029633 | 2 / 2 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00003740 C00019064 | 0 / 3 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00000911 C00002852 | 2 / 3 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00003741 C00005138 | 1 / 1 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00000919 C00002852 | 0 / 0 |
| Q04206 | Transcription factor p65 | Transcription Factor | C00002852 C00003741 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00003740 C00003741 | 0 / 0 |
| P04054 | Phospholipase A2 | Enzyme | C00019064 | 0 / 0 |
| P08151 | Zinc finger protein GLI1 | Unclassified protein | C00003741 | 0 / 0 |
| P29466 | Caspase-1 | C14 | C00002852 | 0 / 0 |
| Q16773 | Kynurenine--oxoglutarate transaminase 1 | Enzyme | C00000100 | 0 / 0 |
| Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | Tyr | C00019064 | 4 / 2 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00003741 | 0 / 0 |
| P28845 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | Enzyme | C00032467 | 1 / 1 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00019064 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00019064 | 0 / 0 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | C00003741 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00002852 | 0 / 0 |
| P24666 | Low molecular weight phosphotyrosine protein phosphatase | Tyr | C00019064 | 0 / 0 |
| P00734 | Prothrombin | S1A | C00003672 | 4 / 2 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00003672 | 3 / 2 |
| P54132 | Bloom syndrome protein | Enzyme | C00002852 | 1 / 2 |
| Q9HBH1 | Peptide deformylase, mitochondrial | Enzyme | C00000100 | 0 / 0 |
| Q99558 | Mitogen-activated protein kinase kinase kinase 14 | Unique STE serine/threonine protein kinase | C00002852 | 0 / 0 |
| Q13133 | Oxysterols receptor LXR-alpha | NR1H3 | C00003741 | 0 / 0 |
| P18433 | Receptor-type tyrosine-protein phosphatase alpha | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00019064 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00002852 | 0 / 0 |
| P07237 | Protein disulfide-isomerase | Enzyme | C00005138 | 0 / 0 |
| P55055 | Oxysterols receptor LXR-beta | NR1H3 | C00003741 | 0 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00002852 | 3 / 3 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00002852 | 2 / 2 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00003740 | 3 / 1 |
| P03372 | Estrogen receptor | NR3A1 | C00003672 | 1 / 1 |
| P09884 | DNA polymerase alpha catalytic subunit | Transferase | C00023774 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00003741 | 0 / 0 |
| P34969 | 5-hydroxytryptamine receptor 7 | Serotonin receptor | C00023774 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00002852 | 0 / 0 |
| P55210 | Caspase-7 | C14 | C00002852 | 0 / 0 |
| P35228 | Nitric oxide synthase, inducible | Enzyme | C00019064 | 1 / 1 |
| Q99700 | Ataxin-2 | Unclassified protein | C00005138 | 1 / 1 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00002852 | 4 / 1 |
| P10586 | Receptor-type tyrosine-protein phosphatase F | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00002852 | 7 / 37 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00002852 | 4 / 3 |
| P17706 | Tyrosine-protein phosphatase non-receptor type 2 | Tyr | C00019064 | 0 / 1 |
| P04049 | RAF proto-oncogene serine/threonine-protein kinase | Raf | C00002852 | 2 / 0 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | Tyr | C00019064 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00005138 | 0 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00003672 | 1 / 0 |
| P23469 | Receptor-type tyrosine-protein phosphatase epsilon | Receptor tyrosine-protein phosphatase | C00019064 | 0 / 0 |
| Q02750 | Dual specificity mitogen-activated protein kinase kinase 1 | Ste7 | C00002852 | 1 / 1 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00002852 | 0 / 0 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00019064 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00000911 | 0 / 0 |
| P02768 | Serum albumin | Secreted protein | C00000100 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00003741 | 1 / 0 |
| Q8IUX4 | DNA dC->dU-editing enzyme APOBEC-3F | Enzyme | C00003741 | 0 / 0 |
| O00255 | Menin | Unclassified protein | C00002852 | 2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00002852 | 1 / 2 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00019064 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00019064 | 1 / 4 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00000919 | 0 / 1 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00000919 | 1 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00000919 | 0 / 0 |
| Q02880 | DNA topoisomerase 2-beta | Isomerase | C00023774 | 0 / 0 |
| Q9UBT2 | SUMO-activating enzyme subunit 2 | Enzyme | C00003741 | 0 / 0 |
| Q9UBE0 | SUMO-activating enzyme subunit 1 | Unclassified protein | C00003741 | 0 / 0 |
| P01215 | Glycoprotein hormones alpha chain | Unclassified protein | C00005138 | 0 / 3 |
205 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00000911
C00000919
C00002852
C00003741
C00019064 |
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00000919
C00002852
C00003741
C00019064
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00000919
C00003741
C00019064
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00000911
C00000919
C00019064
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00000911
C00000919
C00019064
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00000911
C00002852
C00003741
|
| 3713 | IVL | involucrin |
C00000911
C00000919
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00000911
C00019064
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00003741
C00019064
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00003741
C00019064
|
| 4842 | NOS1, IHPS1, N-NOS, NC-NOS, NOS, bNOS, nNOS | nitric oxide synthase 1 (neuronal) (EC:1.14.13.39) |
C00000911
C00000919
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00003749
C00019064
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00000911
C00019064
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00000911
C00019064
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00000911
C00019064
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00002852
C00019064
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00000911
C00000919
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00000911
C00000919
|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00000919
C00003741
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00000911
C00019064
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00000911
C00019064
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00000911
C00019064
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00000100
C00000911
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00000911
C00019064
|
| 4255 | MGMT | O-6-methylguanine-DNA methyltransferase (EC:2.1.1.63) |
C00000919
|
| 54600 | UGT1A9, HLUGP4, LUGP4, UDPGT, UDPGT_1-9, UGT-1I, UGT1-09, UGT1-9, UGT1.9, UGT1AI, UGT1I | UDP glucuronosyltransferase 1 family, polypeptide A9 (EC:2.4.1.17) |
C00002852
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00000911
|
| 571 | BACH1, BACH-1 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
C00000911
|
| 8314 | BAP1, HUCEP-13, UCHL2 | BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (EC:3.4.19.12) |
C00000911
|
| 673 | BRAF, B-RAF1, BRAF1, NS7, RAFB1 | v-raf murine sarcoma viral oncogene homolog B (EC:2.7.11.1) |
C00000911
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00002852
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00002852
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00002852
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00000911
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00000911
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00000911
|
| 1020 | CDK5, PSSALRE | cyclin-dependent kinase 5 (EC:2.7.11.22) |
C00000911
|
| 1022 | CDK7, CAK1, CDKN7, HCAK, MO15, STK1, p39MO15 | cyclin-dependent kinase 7 (EC:2.7.11.23 2.7.11.22) |
C00000911
|
| 1025 | CDK9, C-2k, CDC2L4, CTK1, PITALRE, TAK | cyclin-dependent kinase 9 (EC:2.7.11.23 2.7.11.22) |
C00000911
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00000911
|
| 1113 | CHGA, CGA | chromogranin A (parathyroid secretory protein 1) |
C00000911
|
| 1114 | CHGB, SCG1 | chromogranin B (secretogranin 1) |
C00000911
|
| 23418 | CRB1, LCA8, RP12 | crumbs homolog 1 (Drosophila) |
C00000911
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00002852
|
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00000911
|
| 1981 | EIF4G1, EIF-4G1, EIF4F, EIF4G, EIF4GI, P220, PARK18 | eukaryotic translation initiation factor 4 gamma, 1 |
C00000911
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00002852
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00002852
|
| 2176 | FANCC, FA3, FAC, FACC | Fanconi anemia, complementation group C |
C00000911
|
| 2188 | FANCF, FAF | Fanconi anemia, complementation group F |
C00000911
|
| 356 | FASLG, ALPS1B, APT1LG1, APTL, CD178, CD95-L, CD95L, FASL, TNFSF6 | Fas ligand (TNF superfamily, member 6) |
C00000911
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00000911
|
| 2701 | GJA4, CX37 | gap junction protein, alpha 4, 37kDa |
C00000911
|
| 2702 | GJA5, ATFB11, CX40 | gap junction protein, alpha 5, 40kDa |
C00000911
|
| 2879 | GPX4, GPx-4, GSHPx-4, MCSP, PHGPx, snGPx, snPHGPx | glutathione peroxidase 4 (EC:1.11.1.12) |
C00000911
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00002852
|
| 2950 | GSTP1, DFN7, FAEES3, GST3, GSTP, PI | glutathione S-transferase pi 1 (EC:2.5.1.18) |
C00002852
|
| 3479 | IGF1, IGF-I, IGF1A, IGFI | insulin-like growth factor 1 (somatomedin C) |
C00000911
|
| 3480 | IGF1R, CD221, IGFIR, IGFR, JTK13 | insulin-like growth factor 1 receptor (EC:2.7.10.1) |
C00000911
|
| 3485 | IGFBP2, IBP2, IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa |
C00000911
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00002852
|
| 3716 | JAK1, JAK1A, JAK1B, JTK3 | Janus kinase 1 (EC:2.7.10.2) |
C00000911
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00000911
|
| 3796 | KIF2A, CDCBM3, HK2, KIF2 | kinesin heavy chain member 2A |
C00000911
|
| 3838 | KPNA2, IPOA1, QIP2, RCH1, SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
C00000911
|
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00000911
|
| 3988 | LIPA, CESD, LAL | lipase A, lysosomal acid, cholesterol esterase (EC:3.1.1.13) |
C00000911
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00000911
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00000911
|
| 6416 | MAP2K4, JNKK, JNKK1, MAPKK4, MEK4, MKK4, PRKMK4, SAPKK-1, SAPKK1, SEK1, SERK1, SKK1 | mitogen-activated protein kinase kinase 4 (EC:2.7.12.2) |
C00000911
|
| 5608 | MAP2K6, MAPKK6, MEK6, MKK6, PRKMK6, SAPKK-3, SAPKK3 | mitogen-activated protein kinase kinase 6 (EC:2.7.12.2) |
C00000911
|
| 1432 | MAPK14, CSBP, CSBP1, CSBP2, CSPB1, EXIP, Mxi2, PRKM14, PRKM15, RK, SAPK2A, p38, p38ALPHA | mitogen-activated protein kinase 14 (EC:2.7.11.24) |
C00000911
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00000911
|
| 4247 | MGAT2, CDG2A, CDGS2, GLCNACTII, GNT-II, GNT2 | mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (EC:2.4.1.143) |
C00000911
|
| 100507436 | MICA, MIC-A, PERB11.1 | MHC class I polypeptide-related sequence A |
C00000911
|
| 4292 | MLH1, COCA2, FCC2, HNPCC, HNPCC2, hMLH1 | mutL homolog 1 |
C00000911
|
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00000911
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00000911
|
| 1499 | CTNNB1, CTNNB, MRD19, armadillo | catenin (cadherin-associated protein), beta 1, 88kDa |
C00003749
|
| 8021 | NUP214, CAIN, CAN, D9S46E, N214, p250 | nucleoporin 214kDa |
C00000911
|
| 5320 | PLA2G2A, MOM1, PLA2, PLA2B, PLA2L, PLA2S, PLAS1, sPLA2 | phospholipase A2, group IIA (platelets, synovial fluid) (EC:3.1.1.4) |
C00000911
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00000911
|
| 10111 | RAD50, NBSLD, RAD502, hRad50 | RAD50 homolog (S. cerevisiae) |
C00000911
|
| 8045 | RASSF7, C11orf13, HRAS1, HRC1 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
C00000911
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00000911
|
| 5962 | RDX, DFNB24 | radixin |
C00000911
|
| 6195 | RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1) |
C00000911
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00000911
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00000911
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00003741
|
| 6414 | SEPP1, SELP, SeP | selenoprotein P, plasma, 1 |
C00000911
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00000911
|
| 8243 | SMC1A, CDLS2, DXS423E, SB1.8, SMC1, SMC1L1, SMC1alpha, SMCB | structural maintenance of chromosomes 1A |
C00000911
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00000911
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00000911
|
| 7980 | TFPI2, PP5, REF1, TFPI-2 | tissue factor pathway inhibitor 2 |
C00000911
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00000911
|
| 7097 | TLR2, CD282, TIL4 | toll-like receptor 2 |
C00000911
|
| 7099 | TLR4, ARMD10, CD284, TLR-4, TOLL | toll-like receptor 4 |
C00000911
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00003741
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00000911
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00003741
|
| 7415 | VCP, ALS14, IBMPFD, IBMPFD1, TERA, p97 | valosin containing protein (EC:3.6.4.6) |
C00000911
|
| 7485 | WRB, CHD5 | tryptophan rich basic protein |
C00000911
|
| 7697 | ZNF138, pHZ-32 | zinc finger protein 138 |
C00000911
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00000100
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00019064
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00019064
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00019064
|
| 5052 | PRDX1, MSP23, NKEF-A, NKEFA, PAG, PAGA, PAGB, PRX1, PRXI, TDPX2 | peroxiredoxin 1 (EC:1.11.1.15) |
C00019064
|
| 10599 | SLCO1B1, HBLRR, LST-1, LST1, OATP-C, OATP1B1, OATP2, OATPC, SLC21A6 | solute carrier organic anion transporter family, member 1B1 |
C00023774
|
| 124 | ADH1A, ADH1 | alcohol dehydrogenase 1A (class I), alpha polypeptide (EC:1.1.1.1) |
C00000919
|
| 128 | ADH5, ADH-3, ADHX, FALDH, FDH, GSH-FDH, GSNOR | alcohol dehydrogenase 5 (class III), chi polypeptide (EC:1.1.1.1 1.1.1.284) |
C00000919
|
| 221 | ALDH3B1, ALDH4, ALDH7 | aldehyde dehydrogenase 3 family, member B1 (EC:1.2.1.5) |
C00000919
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00000919
|
| 287 | ANK2, ANK-2, LQT4, brank-2 | ankyrin 2, neuronal |
C00000919
|
| 288 | ANK3, ANKYRIN-G, MRT37 | ankyrin 3, node of Ranvier (ankyrin G) |
C00000919
|
| 427 | ASAH1, AC, ACDase, ASAH, PHP, PHP32, SMAPME | N-acylsphingosine amidohydrolase (acid ceramidase) 1 (EC:3.5.1.23) |
C00000919
|
| 51665 | ASB1, ASB-1 | ankyrin repeat and SOCS box containing 1 |
C00000919
|
| 140462 | ASB9 | ankyrin repeat and SOCS box containing 9 |
C00000919
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00000919
|
| 9530 | BAG4, BAG-4, SODD | BCL2-associated athanogene 4 |
C00000919
|
| 9529 | BAG5, BAG-5 | BCL2-associated athanogene 5 |
C00000919
|
| 53630 | BCMO1, BCDO, BCDO1, BCMO, BCO, BCO1 | beta-carotene 15,15'-monooxygenase 1 (EC:1.14.99.36) |
C00000919
|
| 999 | CDH1, Arc-1, CD324, CDHE, ECAD, LCAM, UVO | cadherin 1, type 1, E-cadherin (epithelial) |
C00000919
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00000919
|
| 9071 | CLDN10, CPETRL3, OSP-L | claudin 10 |
C00000919
|
| 51208 | CLDN18, SFTA5, SFTPJ | claudin 18 |
C00000919
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00000919
|
| 1282 | COL4A1, HANAC, ICH, POREN1, arresten | collagen, type IV, alpha 1 |
C00000919
|
| 1284 | COL4A2, ICH, POREN2 | collagen, type IV, alpha 2 |
C00000919
|
| 8738 | CRADD, MRT34, RAIDD | CASP2 and RIPK1 domain containing adaptor with death domain |
C00000919
|
| 1508 | CTSB, APPS, CPSB | cathepsin B (EC:3.4.22.1) |
C00000919
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00000919
|
| 6372 | CXCL6, CKA-3, GCP-2, GCP2, SCYB6 | chemokine (C-X-C motif) ligand 6 |
C00000919
|
| 4283 | CXCL9, CMK, Humig, MIG, SCYB9, crg-10 | chemokine (C-X-C motif) ligand 9 |
C00000919
|
| 7852 | CXCR4, CD184, D2S201E, FB22, HM89, HSY3RR, LAP3, LCR1, LESTR, NPY3R, NPYR, NPYRL, NPYY3R, WHIM | chemokine (C-X-C motif) receptor 4 |
C00000919
|
| 1592 | CYP26A1, CP26, CYP26, P450RAI, P450RAI1 | cytochrome P450, family 26, subfamily A, polypeptide 1 (EC:1.14.-.-) |
C00000919
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00000919
|
| 64816 | CYP3A43 | cytochrome P450, family 3, subfamily A, polypeptide 43 (EC:1.14.14.1) |
C00000919
|
| 1746 | DLX2, TES-1, TES1 | distal-less homeobox 2 |
C00000919
|
| 1748 | DLX4, BP1, DLX7, DLX8, DLX9 | distal-less homeobox 4 |
C00000919
|
| 2068 | ERCC2, COFS2, EM9, TTD, XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 (EC:3.6.4.12) |
C00000919
|
| 23017 | FAIM2, LFG, LFG2, NGP35, NMP35, TMBIM2 | Fas apoptotic inhibitory molecule 2 |
C00000919
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00000919
|
| 3169 | FOXA1, HNF3A, TCF3A | forkhead box A1 |
C00000919
|
| 2627 | GATA6 | GATA binding protein 6 |
C00000919
|
| 2633 | GBP1 | guanylate binding protein 1, interferon-inducible |
C00000919
|
| 81025 | GJA9, CX58, CX59, GJA10 | gap junction protein, alpha 9, 59kDa |
C00000919
|
| 373156 | GSTK1, GST, GST_13-13, GST13, GST13-13, GSTK1-1, hGSTK1 | glutathione S-transferase kappa 1 (EC:2.5.1.18) |
C00000919
|
| 2946 | GSTM2, GST4, GSTM, GSTM2-2, GTHMUS | glutathione S-transferase mu 2 (muscle) (EC:2.5.1.18) |
C00000919
|
| 2948 | GSTM4, GSTM4-4, GTM4 | glutathione S-transferase mu 4 (EC:2.5.1.18) |
C00000919
|
| 2949 | GSTM5, GSTM5-5, GTM5 | glutathione S-transferase mu 5 (EC:2.5.1.18) |
C00000919
|
| 2954 | GSTZ1, GSTZ1-1, MAAI, MAI | glutathione S-transferase zeta 1 (EC:2.5.1.18 5.2.1.2) |
C00000919
|
| 3204 | HOXA7, ANTP, HOX1, HOX1.1, HOX1A | homeobox A7 |
C00000919
|
| 3216 | HOXB6, HOX2, HOX2B, HU-2, Hox-2.2 | homeobox B6 |
C00000919
|
| 3226 | HOXC10, HOX3I | homeobox C10 |
C00000919
|
| 3448 | IFNA14, IFN-alphaH, LEIF2H | interferon, alpha 14 |
C00000919
|
| 3449 | IFNA16, IFN-alphaO | interferon, alpha 16 |
C00000919
|
| 3451 | IFNA17, IFN-alphaI, IFNA, INFA, LEIF2C1 | interferon, alpha 17 |
C00000919
|
| 3442 | IFNA5, IFN-alpha-5, IFN-alphaG, INA5, INFA5, leIF_G | interferon, alpha 5 |
C00000919
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00000919
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00000919
|
| 3394 | IRF8, H-ICSBP, ICSBP, ICSBP1, IRF-8 | interferon regulatory factor 8 |
C00000919
|
| 50805 | IRX4, IRXA3 | iroquois homeobox 4 |
C00000919
|
| 3688 | ITGB1, CD29, FNRB, GPIIA, MDF2, MSK12, VLA-BETA, VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
C00000919
|
| 3691 | ITGB4, CD104 | integrin, beta 4 |
C00000919
|
| 3875 | KRT18, CYK18, K18 | keratin 18 |
C00000919
|
| 3880 | KRT19, CK19, K19, K1CS | keratin 19 |
C00000919
|
| 3849 | KRT2, CK-2e, K2e, KRT2A, KRT2E, KRTE | keratin 2 |
C00000919
|
| 3855 | KRT7, CK7, K2C7, K7, SCL | keratin 7 |
C00000919
|
| 3856 | KRT8, CARD2, CK-8, CK8, CYK8, K2C8, K8, KO | keratin 8 |
C00000919
|
| 8022 | LHX3, CPHD3, LIM3, M2-LHX3 | LIM homeobox 3 |
C00000919
|
| 4153 | MBL2, COLEC1, HSMBPC, MBL, MBL2D, MBP, MBP-C, MBP1, MBPD | mannose-binding lectin (protein C) 2, soluble |
C00000919
|
| 4170 | MCL1, BCL2L3, EAT, MCL1-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT | myeloid cell leukemia sequence 1 (BCL2-related) |
C00000919
|
| 4212 | MEIS2, HsT18361, MRG1 | Meis homeobox 2 |
C00000919
|
| 4222 | MEOX1, KFS2, MOX1 | mesenchyme homeobox 1 |
C00000919
|
| 118 | ADD1, ADDA | adducin 1 (alpha) |
C00000911
|
| 3110 | MNX1, HB9, HLXB9, HOXHB9, SCRA1 | motor neuron and pancreas homeobox 1 |
C00000919
|
| 4439 | MSH5, G7, MUTSH5, NG23 | mutS homolog 5 |
C00000919
|
| 4586 | MUC5AC, MUC5, TBM, leB | mucin 5AC, oligomeric mucus/gel-forming |
C00000919
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00000919
|
| 4973 | OLR1, CLEC8A, LOX1, LOXIN, SCARE1, SLOX1 | oxidized low density lipoprotein (lectin-like) receptor 1 |
C00000919
|
| 5098 | PCDHGC3, PC43, PCDH-GAMMA-C3, PCDH2 | protocadherin gamma subfamily C, 3 |
C00000919
|
| 5359 | PLSCR1, MMTRA1B | phospholipid scramblase 1 |
C00000919
|
| 57048 | PLSCR3 | phospholipid scramblase 3 |
C00000919
|
| 57088 | PLSCR4, TRA1 | phospholipid scramblase 4 |
C00000919
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00000919
|
| 5892 | RAD51D, BROVCA4, R51H3, RAD51L3, TRAD | RAD51 paralog D |
C00000919
|
| 5914 | RARA, NR1B1, RAR | retinoic acid receptor, alpha |
C00000919
|
| 5915 | RARB, HAP, NR1B2, RRB2 | retinoic acid receptor, beta |
C00000919
|
| 5918 | RARRES1, LXNL, TIG1 | retinoic acid receptor responder (tazarotene induced) 1 |
C00000919
|
| 5950 | RBP4, RDCCAS | retinol binding protein 4, plasma |
C00000919
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00000919
|
| 949 | SCARB1, CD36L1, CLA-1, CLA1, HDLQTL6, SR-BI, SRB1 | scavenger receptor class B, member 1 |
C00000919
|
| 4990 | SIX6, MCOPCT2, OPTX2, Six9 | SIX homeobox 6 |
C00000919
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00000919
|
| 9517 | SPTLC2, HSN1C, LCB2, LCB2A, NSAN1C, SPT2, hLCB2a | serine palmitoyltransferase, long chain base subunit 2 (EC:2.3.1.50) |
C00000919
|
| 6720 | SREBF1, SREBP-1c, SREBP1, bHLHd1 | sterol regulatory element binding transcription factor 1 |
C00000919
|
| 7052 | TGM2, G-ALPHA-h, GNAH, TG2, TGC | transglutaminase 2 (EC:2.3.2.13) |
C00000919
|
| 7047 | TGM4, TGP, hTGP | transglutaminase 4 (EC:2.3.2.13) |
C00000919
|
| 3604 | TNFRSF9, 4-1BB, CD137, CDw137, ILA | tumor necrosis factor receptor superfamily, member 9 |
C00000919
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00000919
|
| 10190 | TXNDC9, APACD, PHLP3 | thioredoxin domain containing 9 |
C00000919
|
| 7517 | XRCC3, CMM6 | X-ray repair complementing defective repair in Chinese hamster cells 3 |
C00000919
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (75)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #218030 | Apparent mineralocorticoid excess; ame |
P80365
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #615279 | Cardiofaciocutaneous syndrome 3; cfc3 |
Q02750
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #604931 | Cortisone reductase deficiency 1; cortrd1 |
P28845
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #133239 | Esophageal cancer |
P04637
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #607785 | Juvenile myelomonocytic leukemia; jmml |
Q06124
|
| #151100 | Leopard syndrome 1 |
Q06124
|
| #611554 | Leopard syndrome 2 |
P04049
|
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #211980 | Lung cancer |
P04637
|
| #611162 | Malaria, susceptibility to |
P35228
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #156250 | Metachondromatosis; metcds |
Q06124
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #163950 | Noonan syndrome 1; ns1 |
Q06124
|
| #611553 | Noonan syndrome 5; ns5 |
P04049
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #172700 | Pick disease of brain |
P10636
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
P04637
|
| #601367 | Stroke, ischemic |
P00734
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
KEGG DISEASE (89)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00081 | Hashimoto's thyroiditis |
P01215
(marker)
|
| H00082 | Graves' disease |
P01215
(marker)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P01215
(marker)
P16473 (related) |
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04637 (related) |
| H00004 | Chronic myeloid leukemia (CML) |
P04637
(related)
Q01196 (related) |
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
|
| H00006 | Hairy-cell leukemia |
P04637
(related)
|
| H00008 | Burkitt lymphoma |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00010 | Multiple myeloma |
P04637
(related)
|
| H00013 | Small cell lung cancer |
P04637
(related)
|
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00016 | Oral cancer |
P04637
(related)
P04637 (marker) |
| H00017 | Esophageal cancer |
P04637
(related)
P04637 (marker) P35228 (related) P35354 (related) |
| H00018 | Gastric cancer |
P04637
(related)
|
| H00019 | Pancreatic cancer |
P04637
(related)
P04637 (marker) |
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) |
| H00022 | Bladder cancer |
P04637
(related)
|
| H00025 | Penile cancer |
P04637
(related)
P04637 (marker) P35354 (related) |
| H00027 | Ovarian cancer |
P04637
(related)
|
| H00028 | Choriocarcinoma |
P04637
(related)
|
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00031 | Breast cancer |
P04637
(related)
|
| H00032 | Thyroid cancer |
P04637
(related)
|
| H00036 | Osteosarcoma |
P04637
(related)
P08684 (marker) |
| H00038 | Malignant melanoma |
P04637
(related)
P14679 (marker) |
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00041 | Kaposi's sarcoma |
P04637
(related)
|
| H00042 | Glioma |
P04637
(related)
P04637 (marker) |
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00046 | Cholangiocarcinoma |
P04637
(related)
P35354 (related) |
| H00047 | Gallbladder cancer |
P04637
(related)
|
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00055 | Laryngeal cancer |
P04637
(related)
P04637 (marker) |
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00021 | Renal cell carcinoma |
P04637
(marker)
|
| H00079 | Asthma |
P07550
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
|
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00408 | Type I diabetes mellitus |
P17706
(related)
|
| H01111 | Cortisone reductase deficiency (CRD) |
P28845
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H00259 | Apparent mineralocorticoid excess syndrome |
P80365
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00523 | Noonan syndrome and related disorders |
Q02750
(related)
Q06124 (related) |
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H01018 | Metachondromatosis |
Q06124
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
Q9NUW8 (related) |
| H00480 | Non-syndromic X-linked mental retardation |
Q99714
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
51 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D056486 | Drug-Induced Liver Injury |
C00000911
C00000100 C00019064 C00000919 |
| D009369 | Neoplasms |
C00000911
C00019064 C00000919 |
| D007249 | Inflammation |
C00000911
C00019064 |
| D007674 | Kidney Diseases |
C00000911
C00019064 |
| D008103 | Liver Cirrhosis |
C00003741
C00019064 |
| D006528 | Carcinoma, Hepatocellular |
C00003749
C00000919 |
| D008107 | Liver Diseases |
C00019064
C00000919 |
| D012878 | Skin Neoplasms |
C00003749
C00000911 |
| D015431 | Weight Loss |
C00000911
|
| D006937 | Hypercholesterolemia |
C00003749
|
| D014947 | Wounds and Injuries |
C00003749
|
| D001284 | Atrophy |
C00002852
|
| D006968 | Hypersensitivity, Delayed |
C00002852
|
| D020244 | Infarction, Middle Cerebral Artery |
C00002852
|
| D009202 | Cardiomyopathies |
C00003749
|
| D006943 | Hyperglycemia |
C00000911
|
| D006965 | Hyperplasia |
C00000911
|
| D007248 | Infertility, Male |
C00000911
|
| D064420 | Drug-Related Side Effects and Adverse Reactions |
C00003740
|
| D003110 | Colonic Neoplasms |
C00003740
|
| D008545 | Melanoma |
C00003741
|
| D020258 | Neurotoxicity Syndromes |
C00000911
|
| D011471 | Prostatic Neoplasms |
C00000911
|
| D013733 | Testicular Diseases |
C00000911
|
| D014178 | Translocation, Genetic |
C00000911
|
| D009374 | Neoplasms, Experimental |
C00003749
|
| D002252 | Carbon Tetrachloride Poisoning |
C00019064
|
| D050171 | Dyslipidemias |
C00019064
|
| D018149 | Glucose Intolerance |
C00019064
|
| D006949 | Hyperlipidemias |
C00019064
|
| D008106 | Liver Cirrhosis, Experimental |
C00019064
|
| D003876 | Dermatitis, Atopic |
C00005138
|
| D017202 | Myocardial Ischemia |
C00019064
|
| D009765 | Obesity |
C00019064
|
| D011041 | Poisoning |
C00019064
|
| D011230 | Precancerous Conditions |
C00019064
|
| D000435 | Alcoholic Intoxication |
C00000919
|
| D001145 | Arrhythmias, Cardiac |
C00000919
|
| D053627 | Asthenozoospermia |
C00000919
|
| D002545 | Brain Ischemia |
C00000919
|
| D002318 | Cardiovascular Diseases |
C00000919
|
| D003930 | Diabetic Retinopathy |
C00000919
|
| D006526 | Hepatitis C |
C00000919
|
| D015658 | HIV Infections |
C00000919
|
| D006986 | Hypervitaminosis A |
C00000919
|
| D008113 | Liver Neoplasms |
C00000919
|
| D008114 | Liver Neoplasms, Experimental |
C00000919
|
| D008175 | Lung Neoplasms |
C00000919
|
| D009845 | Oligospermia |
C00000919
|
| D013272 | Stomach Diseases |
C00000919
|
| D013276 | Stomach Ulcer |
C00000919
|