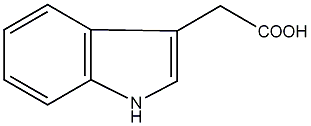
Metabolite
KNApSAcK Entry
| id | C00000100 |
|---|---|
| Name | IAA / Indole-3-acetic acid |
| CAS RN | 87-51-4 |
| Standard InChI | InChI=1S/C10H9NO2/c12-10(13)5-7-6-11-9-4-2-1-3-8(7)9/h1-4,6,11H,5H2,(H,12,13) |
| Standard InChI (Main Layer) | InChI=1S/C10H9NO2/c12-10(13)5-7-6-11-9-4-2-1-3-8(7)9/h1-4,6,11H,5H2,(H,12,13) |
Cluster
| Phytochemical cluster | No. 4 |
|---|---|
| KCF-S cluster | No. 1432 |
Link
ChEMBL
| By standard InChI | CHEMBL82411 |
|---|---|
| By standard InChI Main Layer | CHEMBL82411 |
KEGG
| By LinkDB | C00954 |
|---|
CTD
| By CAS RN | C030737 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| rosids | 24 |
| Spermatophyta | 6 |
| asterids | 4 |
| Liliopsida | 1 |
| Euphyllophyta | 1 |
Family (Top 10)
| family name | count |
|---|---|
| Fabaceae | 9 |
| Pinaceae | 5 |
| Brassicaceae | 3 |
| Rosaceae | 3 |
| Fagaceae | 2 |
| Rutaceae | 2 |
| Solanaceae | 2 |
| Euphorbiaceae | 2 |
| Hominidae | 2 |
| Rhizopodaceae | 1 |
List (39)
* NCBI
Human Protein / Gene in interaction
3 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| Q16773 | Kynurenine--oxoglutarate transaminase 1 | Enzyme | CHEMBL82411 |
CHEMBL1002982
(1)
|
0 / 0 |
| P02768 | Serum albumin | Secreted protein | CHEMBL82411 |
CHEMBL894775
(1)
|
0 / 0 |
| Q9HBH1 | Peptide deformylase, mitochondrial | Enzyme | CHEMBL82411 |
CHEMBL912248
(1)
|
0 / 0 |
CTD interaction (2)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| C030737 | 196 |
AHR
bHLHe76 |
aryl hydrocarbon receptor | indoleacetic acid results in increased activity of [AHR protein binds to ARNT protein] |
affects binding
/ increases activity |
protein |
9407059
|
| C030737 | 405 |
ARNT
HIF-1-beta HIF-1beta HIF1-beta HIF1B HIF1BETA TANGO bHLHe2 |
aryl hydrocarbon receptor nuclear translocator | indoleacetic acid results in increased activity of [AHR protein binds to ARNT protein] |
affects binding
/ increases activity |
protein |
9407059
|