Pisum sativum
Species
KNApSAcK Entry
| Organism name | Pisum sativum |
|---|---|
| Genus | Pisum |
| Family | Fabaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Pisum sativum |
|---|---|
| Linked NCBI taxonomy ID | 3888 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Fabaceae |
|---|---|
| ID | 3803 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (108)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
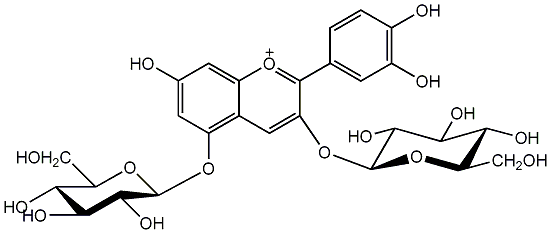
C00002378
|
Cyanin
/ Cyanidin 3,5-diglucoside |
No. 1 | No. 15 |
|
||||
|
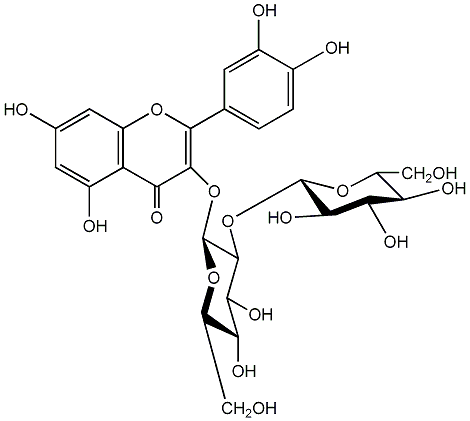
C00005409
|
Quercetin 3-O-sophoroside
|
CHEMBL452919
|
C055545
|
No. 1 | No. 15 |
|
||
|
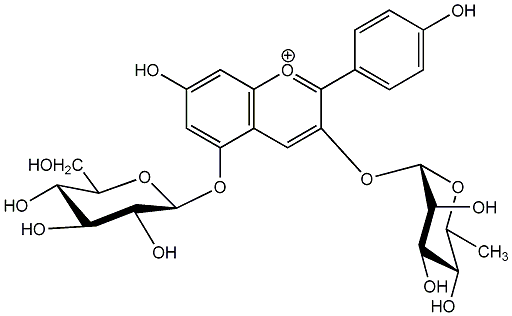
C00006641
|
Pelargonidin 3-rhamnoside-5-glucoside
|
No. 1 | No. 15 |
|
||||
|
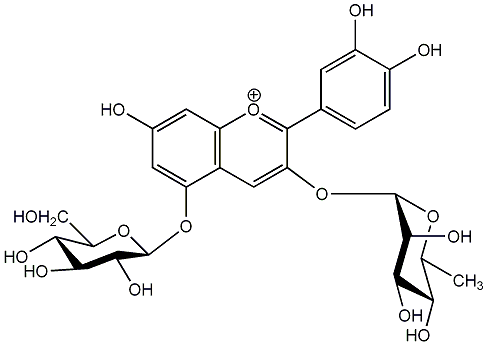
C00006665
|
Cyanidin 3-rhamnoside-5-glucoside
|
No. 1 | No. 15 |
|
||||
|
C00006689
|
Peonidin 3-rhamnoside-5-glucoside
|
No. 1 | No. 15 |

|
||||
|
C00006704
|
Delphinidin 3-sambubioside
|
No. 1 | No. 15 |
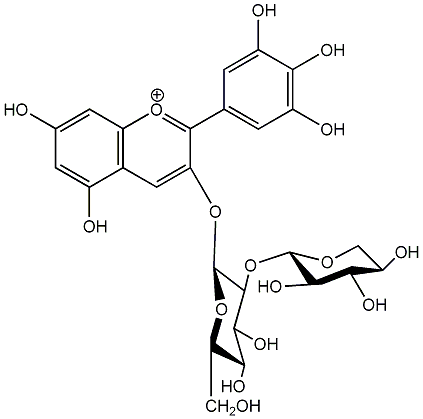
|
||||
|
C00006705
|
Delphinidin 3-sophoroside
|
No. 1 | No. 15 |

|
||||
|
C00006709
|
Delphin
/ Delphinidin 3,5-diglucoside |
No. 1 | No. 15 |
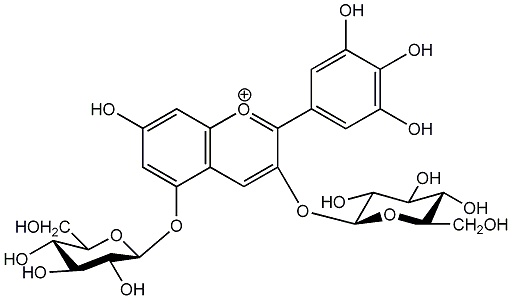
|
||||
|
C00006741
|
Malvidin 3-rhamnoside-5-glucoside
|
No. 1 | No. 15 |

|
||||
|
C00006728
|
Petunin
/ Petunidin 3,5-di-O-beta-D-glucoside |
No. 1 | No. 15 |
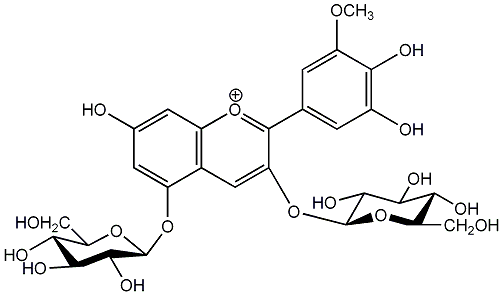
|
||||
|
C00006729
|
Petunidin 3-rhamnoside-5-glucoside
|
No. 1 | No. 15 |

|
||||
|
C00006710
|
Delphinidin 3-rhamnoside-5-glucoside
|
No. 1 | No. 15 |

|
||||
|
C00002374
|
Chrysanthemin
/ Cyanidin 3-O-glucoside |
CHEMBL257839
CHEMBL1197952 |
C114438
|
1 / 0 / 1 | No. 2 | No. 15 |
|
|
|
C00006653
|
Cyanidin 3-rhamnoside
|
No. 2 | No. 15 |

|
||||
|
C00006736
|
Malvidin 3-rhamnoside
|
No. 2 | No. 15 |

|
||||
|
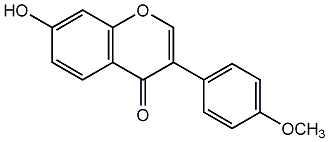
C00002525
|
Formononetin
/ 7-Hydroxy-4'-methoxyisoflavone |
CHEMBL242341
|
C007768
|
24 / 36 / 58 | 13 / 0 | No. 3 | No. 15 |
|
|
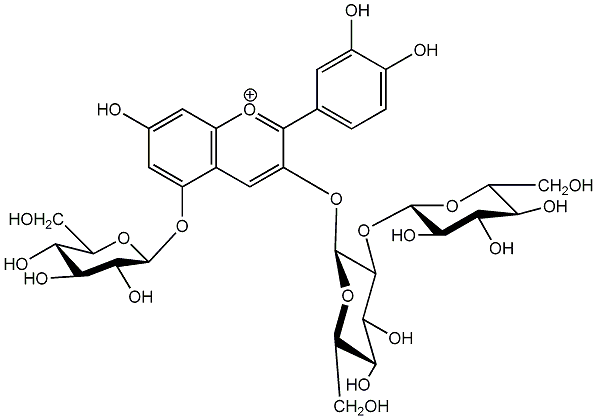
C00006674
|
Cyanidin 3-sophoroside-5-glucoside
|
No. 5 | No. 15 |
|
||||
|
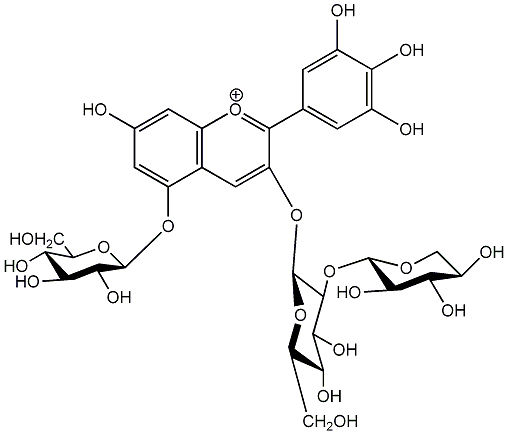
C00006715
|
Delphinidin 3-sambubioside-5-glucoside
|
No. 5 | No. 15 |
|
||||
|
C00006716
|
Delphinidin 3-sophoroside-5-glucoside
|
No. 5 | No. 15 |

|
||||
|
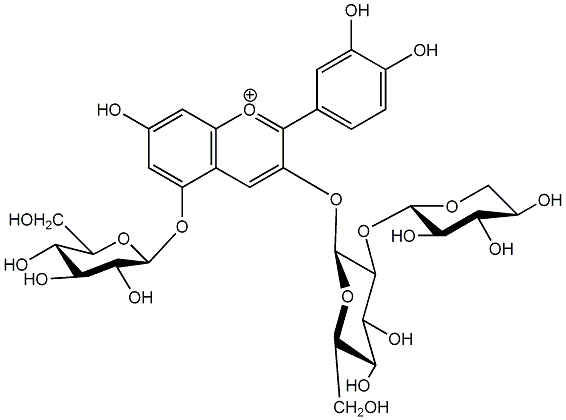
C00006673
|
Cyanidin 3-sambubioside-5-glucoside
|
No. 5 | No. 15 |
|
||||
|
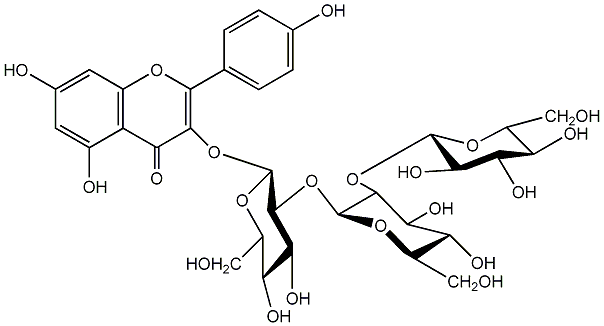
C00005208
|
Kaempferol 3-sophorotrioside
|
No. 5 | No. 15 |
|
||||
|
C00005988
|
Quercetin 3-(6''''-sinapylsophorotrioside)
|
No. 7 | No. 15 |

|
||||
|
C00005987
|
Quercetin 3-(6''''-ferulylsophorotrioside)
|
No. 7 | No. 15 |

|
||||
|
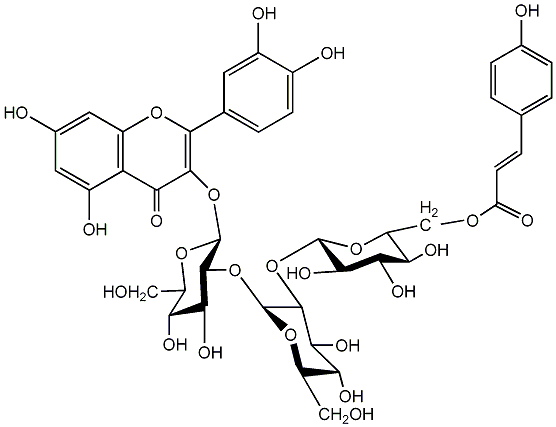
C00005986
|
Quercetin 3-(6''''-caffeylsophorotrioside)
|
No. 7 | No. 15 |
|
||||
|
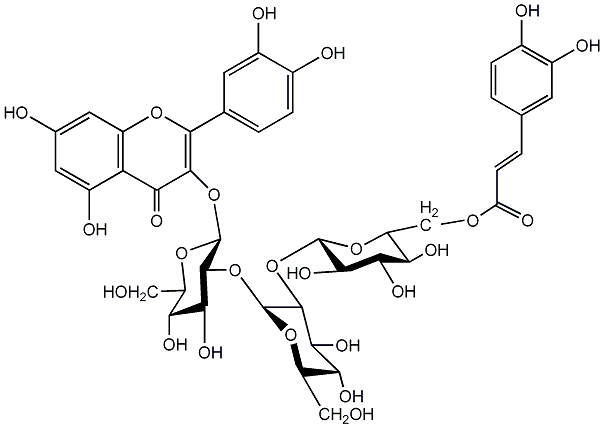
C00005985
|
Quercetin 3-(6''''-p-coumarylsophorotrioside)
|
No. 7 | No. 15 |
|
||||
|
C00001055
|
Isoorientin
/ Homoorientin / Lespecapitioside / Luteolin 6-C-beta-D-glucopyranoside / 2-(3,4-Dihydroxyphenyl)-6-beta-D-glucopyranosyl-5,7-dihydroxy-4H-1-benzopyran-4-one |
CHEMBL239559
CHEMBL1302308 |
C057912
|
23 / 14 / 17 | 0 / 1 | No. 22 | No. 15 |
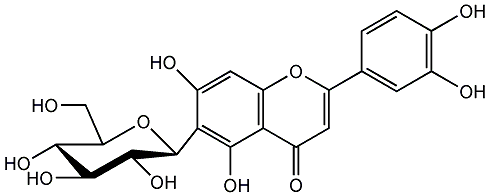
|
|
C00001059
|
Isovitexin
/ Saponaretin / Homovitexin / 6-C-beta-D-Glucopyranosylapigenin / 6-beta-D-Glucopyranosyl-4',5,7-trihydroxyflavone / 6-beta-D-Glucopyranosyl-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
CHEMBL465360
CHEMBL1601394 |
C049772
|
28 / 20 / 19 | No. 22 | No. 15 |

|
|
|
C00001110
|
Vitexin
/ Apigenin 8-C-glucoside / 8-D-Glucosyl-4',5,7-trihydroxyflavone / 8-beta-D-Glucopyranosyl-5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
CHEMBL487417
CHEMBL1332209 CHEMBL1357921 |
16 / 4 / 8 | No. 22 | No. 15 |
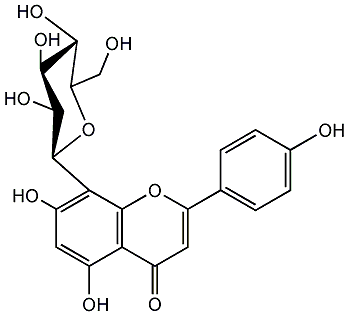
|
||
|
C00000977
|
Liquiritigenin
|
CHEMBL252642
CHEMBL271939 |
C083152
|
5 / 4 / 4 | 1 / 1 | No. 25 | No. 14 |
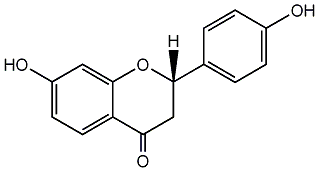
|
|
C00000649
|
2-Hydroxypseudobaptigenin
/ 2',7-Dihydroxy-4',5'-methylenedioxyisoflavone |
No. 27 | No. 15 |
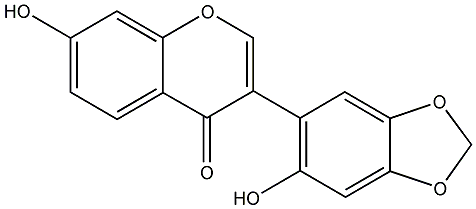
|
||||
|
C00009695
|
Anhydropisatin
/ Flemichapparin B / 6a,11a-Dehydropterocarpin |
No. 27 | No. 15 |
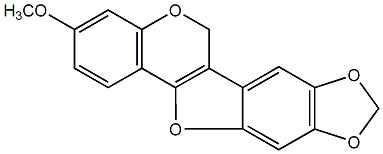
|
||||
|
C00002507
|
Castanin
/ Afromosin / Afrormosin / 7-Hydroxy-6,4'-dimethoxyisoflavone |
CHEMBL464404
|
C080240
|
7 / 14 / 39 | 1 / 0 | No. 35 | No. 15 |
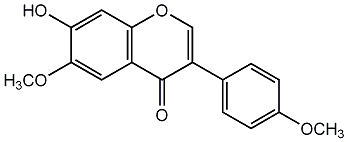
|
|
C00002571
|
Sayanedine
|
No. 35 | No. 15 |
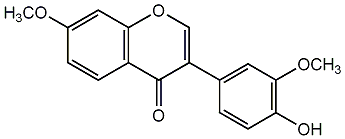
|
||||
|
C00000007
|
GA7
/ Gibberellin A7 |
No. 40 | No. 41 |
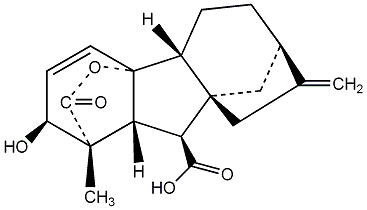
|
||||
|
C00000008
|
GA8
/ Gibberellin A8 |
No. 40 | No. 41 |
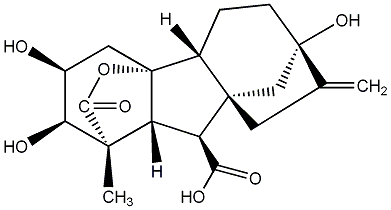
|
||||
|
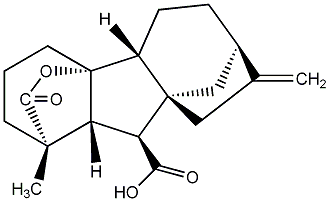
C00000009
|
GA9
/ Gibberellin A9 |
No. 40 | No. 41 |
|
||||
|
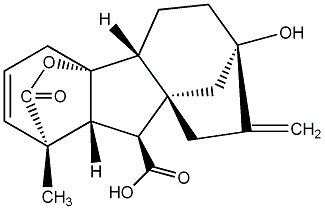
C00000005
|
GA5
/ Gibberellin A5 |
No. 40 | No. 41 |
|
||||
|
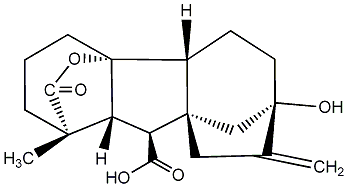
C00000020
|
GA20
/ Gibberellin A20 |
No. 40 | No. 41 |
|
||||
|
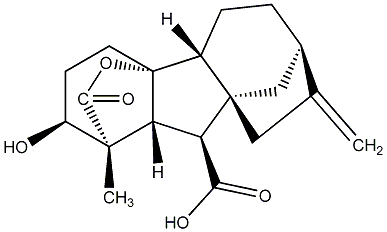
C00000004
|
GA4
/ Gibberellin A4 |
No. 40 | No. 41 |
|
||||
|
C00000081
|
GA81
/ Gibberellin A81 |
No. 40 | No. 41 |

|
||||
|
C00000216
|
GA91
/ Gibberellin A91 |
No. 40 | No. 41 |

|
||||
|
C00000051
|
GA51
/ Gibberellin A51 |
No. 40 | No. 41 |
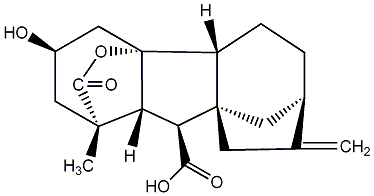
|
||||
|
C00000044
|
GA44
/ Gibberellin A44 |
No. 40 | No. 41 |
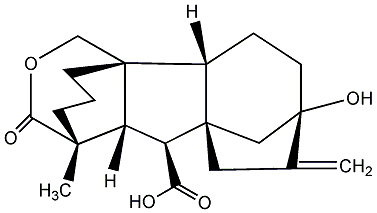
|
||||
|
C00000038
|
GA38
/ Gibberellin A38 |
No. 40 | No. 41 |
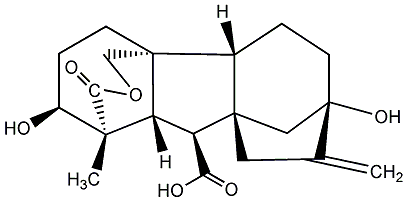
|
||||
|
C00000003
|
GA3
/ Gibberellin A3 |
CHEMBL513241
CHEMBL566653 CHEMBL1232952 CHEMBL1476967 CHEMBL1487394 |
C007842
|
10 / 18 / 17 | No. 40 | No. 41 |
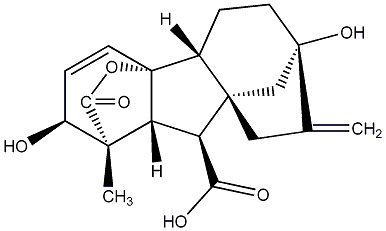
|
|
|
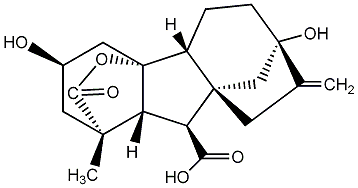
C00000029
|
GA29
/ Gibberellin A29 |
No. 40 | No. 41 |
|
||||
|
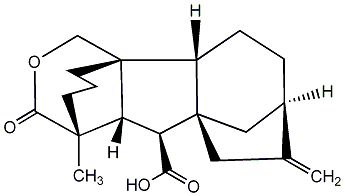
C00000015
|
GA15
/ Gibberellin A15 |
No. 40 | No. 41 |
|
||||
|
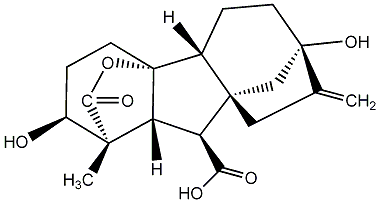
C00000001
|
GA1
/ Gibberellin A1 |
C422660
|
No. 40 | No. 41 |
|
|||
|
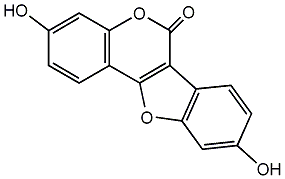
C00002514
|
Cumestrol
/ Coumestrol / 3,9-Dihydroxycoumestan |
CHEMBL30707
|
D003375
|
41 / 40 / 40 | 1795 / 2 | No. 54 | No. 17 |
|
|
C00009618
|
2-Methoxymedicarpin
|
No. 66 | No. 15 |

|
||||
|
C00009633
|
4-Hydroxy-2,3,9-trimethoxypterocarpan
|
No. 66 | No. 15 |

|
||||
|
C00006927
|
Isoliquiritigenin 2'-methy ether
/ 4,4'-Dihydroxy-2'-methoxychalcone |
CHEMBL253777
|
7 / 20 / 19 | No. 92 | No. 13 |
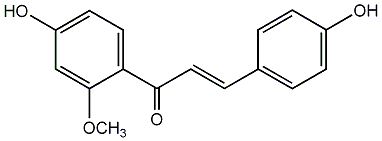
|
||
|
C00000650
|
(-)-Sophorol
/ (3R)-Sophorol |
No. 106 | No. 14 |

|
||||
|
C00035086
|
16-Dotriacontanol
/ Dotriacontan-16-ol |
No. 115 |

|
|||||
|
C00035139
|
13-Nonacosanol
/ Nonacosan-13-ol |
No. 115 |

|
|||||
|
C00035140
|
4-Nonacosanol
/ Nonacosan-14-ol |
No. 115 |

|
|||||
|
C00035084
|
14-Dotriacontanol
/ Dotriacontan-14-ol |
No. 115 |

|
|||||
|
C00035085
|
15-Dotriacontanol
/ Dotriacontan-15-ol |
No. 115 |

|
|||||
|
C00035141
|
Nonacosan-15-ol
|
No. 115 |

|
|||||
|
C00000298
|
GA97
/ Gibberellin A97 |
No. 187 | No. 41 |
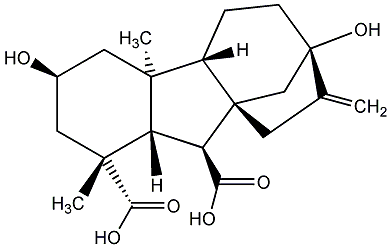
|
||||
|
C00000043
|
GA43
/ Gibberellin A43 |
No. 187 | No. 41 |
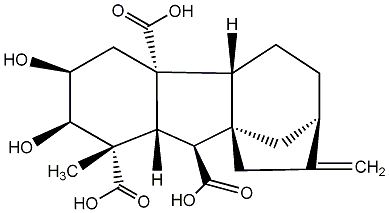
|
||||
|
C00000012
|
GA12
/ Gibberellin A12 |
C048462
|
No. 187 | No. 41 |

|
|||
|
C00000017
|
GA17
/ Gibberellin A17 |
No. 187 | No. 41 |
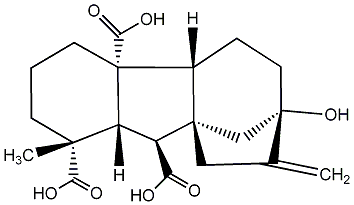
|
||||
|
C00000025
|
GA25
/ Gibberellin A25 |
No. 187 | No. 41 |
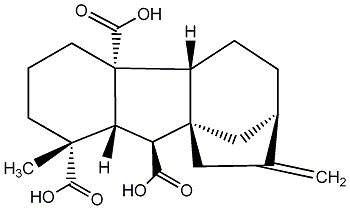
|
||||
|
C00000053
|
GA53
/ Gibberellin A53 |
No. 187 | No. 41 |
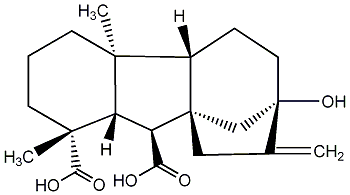
|
||||
|
C00000019
|
GA19
/ Gibberellin A19 |
C120175
|
No. 187 | No. 41 |
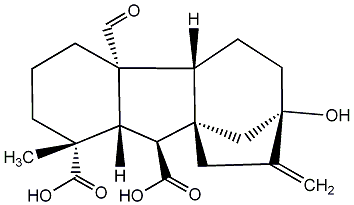
|
|||
|
C00000024
|
GA24
/ Gibberellin A24 |
C120176
|
No. 187 | No. 41 |
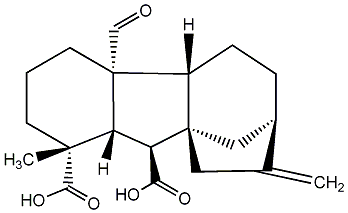
|
|||
|
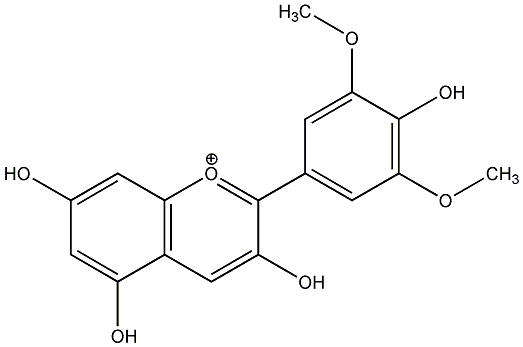
C00020647
|
Malvidin
|
CHEMBL255753
|
1 / 0 / 1 | No. 210 | No. 15 |
|
||
|
C00002390
|
Xenognosin A
|
CHEMBL1087026
|
No. 379 |

|
||||
|
C00019383
|
(+)-Hydroxypisatin
/ (6aR,11aR)2,6a-Dihydroxy-3-methoxy-8,9-methylenedioxypterocarpan |
No. 396 | No. 15 |

|
||||
|
C00000651
|
(+)-Pisatin
/ 6a-Hydroxy-3-methoxy-8,9-methylenedioxypterocarpan |
CHEMBL1784262
|
1 / 1 / 1 | No. 396 | No. 15 |

|
||
|
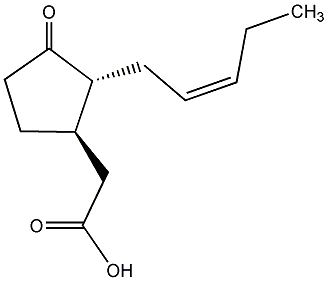
C00000218
|
Jasmonic acid
/ trans-Jasmonic acid / 3-Oxo-2-(2Z-pentenyl)cyclotentylacetic acid |
CHEMBL445499
CHEMBL449572 |
C011006
|
4 / 1 / 0 | No. 707 | No. 70 |
|
|
|
C00000108
|
IAM
/ Indole-3-acetamide / 1H-Indole-3-acetamide |
C015950
|
No. 710 | No. 4 |

|
|||
|
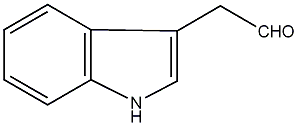
C00000109
|
Indole-3-acetaldehyde
|
C001655
|
No. 710 | No. 4 |
|
|||
|
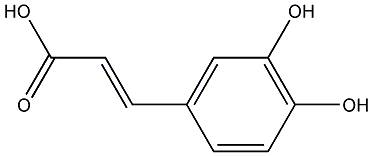
C00000615
|
Caffeic acid
|
CHEMBL145
CHEMBL1320034 |
68 / 64 / 63 | No. 904 | No. 6 |
|
||
|
C00000170
|
trans-Cinnamate
/ trans-Cinnamic acid |
CHEMBL27246
|
5 / 2 / 2 | No. 904 | No. 6 |
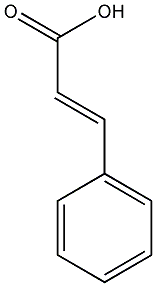
|
||
|
C00000580
|
trans-p-Coumaric acid
/ trans-p-Hydroxycinnamic acid |
CHEMBL66879
CHEMBL2336752 |
23 / 13 / 18 | No. 904 | No. 6 |
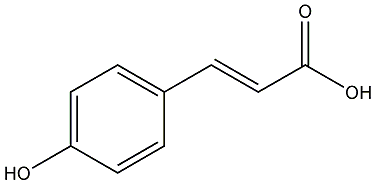
|
||
|
C00001435
|
Tyramine
|
CHEMBL11608
|
D014439
|
23 / 11 / 6 | 6 / 9 | No. 936 | No. 6 |
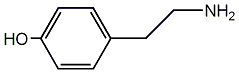
|
|
C00000099
|
2-Methylthioribosylzeatin
/ trans-Methylthioribosylzeatin / 2-Methylthioribosyl-trans-zeatin |
No. 989 |
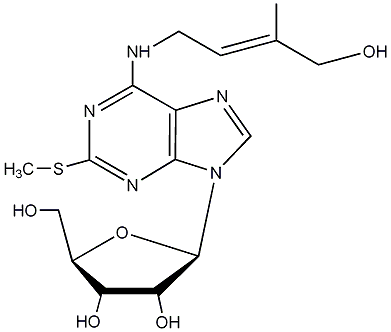
|
|||||
|
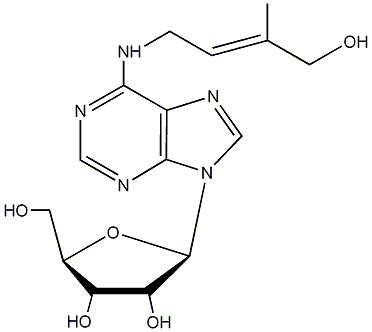
C00000095
|
9-Ribosyl-cis-zeatin
/ 6-(4-Hydroxy-3-methyl-cis-2-butenylamino)-9-beta-D-ribofuranosylpurine |
No. 989 |
|
|||||
|
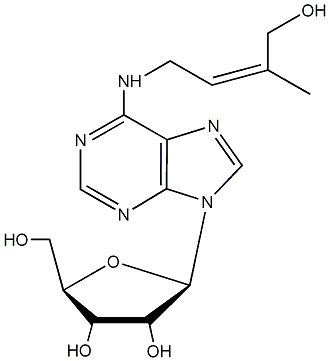
C00000096
|
9-Ribosyl-trans-zeatin
/ Zeatin-9-beta-D-ribofuranoside |
No. 989 |
|
|||||
|
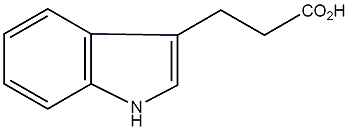
C00000115
|
Indol-3-propionic acid
/ 1H-Indole-3-propanoic acid |
CHEMBL207225
|
C015292
|
1 / 0 / 0 | No. 1432 | No. 4 |
|
|
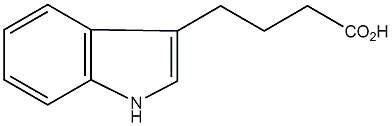
|
C00000116
|
IBA
/ Indole-3-butyric acid / 1H-Indole-3-butanoic acid |
CHEMBL582878
|
C014612
|
6 / 9 / 8 | No. 1432 | No. 4 |
|
|
|
C00000100
|
IAA
/ Indole-3-acetic acid |
CHEMBL82411
|
C030737
|
3 / 0 / 0 | 2 / 1 | No. 1432 | No. 4 |
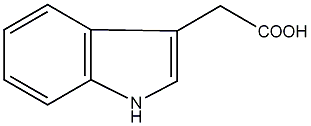
|
|
C00000134
|
(s)-(+)-Abscisic acid
/ 2,4-Pentadienoic acid |
CHEMBL288040
CHEMBL379808 CHEMBL1318134 CHEMBL1469719 CHEMBL1741460 CHEMBL1965138 |
D000040
|
10 / 9 / 10 | No. 1435 | No. 38 |
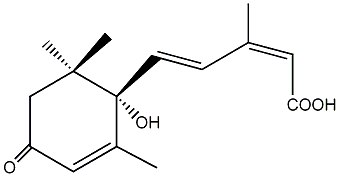
|
|
|
C00001369
|
(+)-gamma-Hydroxy-L-homoarginine
|
No. 1782 |
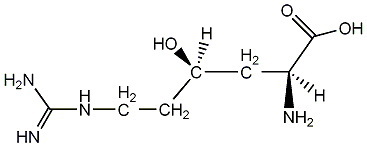
|
|||||
|
C00001429
|
Serotonin
/ Enteramin / Enteramine / DS substance / Thrombotonin / Thrombocytin / 5-Hydroxytryptamine |
CHEMBL39
CHEMBL1628597 |
D012701
|
78 / 35 / 22 | 32 / 40 | No. 1910 | No. 4 |
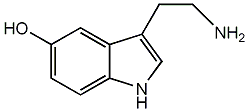
|
|
C00034649
|
Progesterone
|
CHEMBL103
CHEMBL125608 CHEMBL1335634 CHEMBL1489845 |
D011374
|
144 / 94 / 73 | 1905 / 95 | No. 2299 | No. 54 |
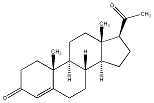
|
|
C00000750
|
Phenylacetic acid
|
CHEMBL1044
|
C025136
|
3 / 1 / 1 | 3 / 0 | No. 2481 |
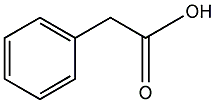
|
|
|
C00001358
|
L-Glutamic acid
|
CHEMBL76232
CHEMBL575060 |
37 / 11 / 11 | No. 2641 |
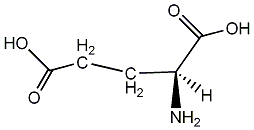
|
|||
|
C00001200
|
Pyruvic acid
|
CHEMBL1162144
|
D019289
|
5 / 2 / 2 | 10 / 4 | No. 2782 |
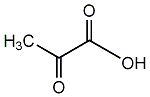
|
|
|
C00000094
|
2iP
/ N6-(delta2-Isopentenyl)adenine |
CHEMBL476189
|
C001478
|
5 / 2 / 3 | 18 / 0 | No. 2879 |
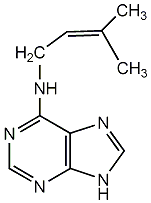
|
|
|
C00000091
|
trans-Zeatin
|
CHEMBL525239
|
D015026
|
2 / 3 / 2 | 0 / 1 | No. 2879 |
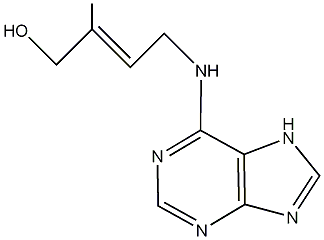
|
|
|
C00001424
|
Norepinephrine
/ L-Noradrenaline / (-)-Norepinephrine |
CHEMBL432
CHEMBL18824 CHEMBL1437 CHEMBL2311152 |
D009638
|
50 / 33 / 35 | 36 / 90 | No. 3197 |
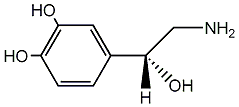
|
|
|
C00001366
|
L-Homoserine
|
CHEMBL11722
|
No. 3420 |
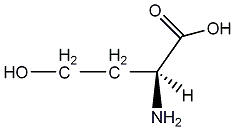
|
||||
|
C00000113
|
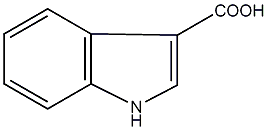
Indole-3-carboxylic acid
|
CHEMBL387527
|
C012382
|
1 / 3 / 4 | No. 3632 |
|
||
|
C00001183
|
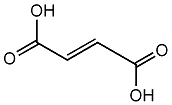
Fumaric acid
|
CHEMBL503160
CHEMBL539648 |
C032005
|
17 / 14 / 46 | 0 / 1 | No. 3766 |
|
|
|
C00001403
|
Cadaverine
|
CHEMBL119296
|
D002103
|
12 / 3 / 5 | No. 3767 |
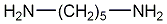
|
||
|
C00000103
|
4-Chloroindole-3-acetic acid methyl ester
|
C028267
|
No. 4225 |

|
||||
|
C00000101
|
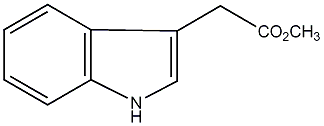
IAA methyl ester
/ Methyl indole-3-acetate |
No. 4225 |
|
|||||
|
C00000102
|
4-Chloro-IAA
/ 4-Chloroindole-3-acetic acid |
CHEMBL309993
|
C096198
|
1 / 0 / 0 | No. 4225 |
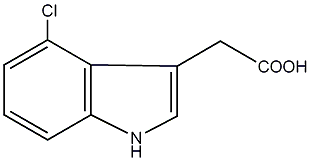
|
||
|
C00000112
|
Indole-3-carboxaldehyde
/ 1H-Indole-3-carboxaldehyde |
CHEMBL147741
|
C012381
|
2 / 1 / 1 | No. 4314 |
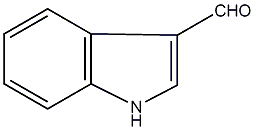
|
||
|
C00001120
|
D-Galacturonic acid
|
CHEMBL496672
CHEMBL1159524 CHEMBL2068684 |
13 / 14 / 14 | No. 4351 |
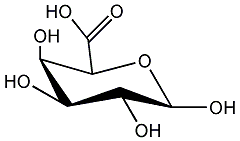
|
|||
|
C00000120
|
alpha-N-Carbomethoxyacetyl-D-4-chlorotryptophan
|
No. 4856 |

|
|||||
|
C00000121
|
alpha-N-Carboethoxyacetyl-D-4-chlorotryptophan
|
No. 4856 |

|
|||||
|
C00001375
|
Isowillardiine
|
No. 5581 |
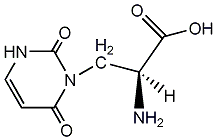
|
|||||
|
C00001267
|
Propane-1-thiol
|
C030762
|
1 / 0 | No. 7369 |
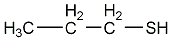
|
|||
|
C00001319
|
Pinolidoxin
|
CHEMBL504236
CHEMBL463173 |
No. 8564 |
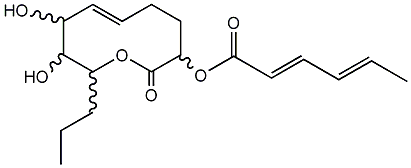
|
Human Protein / Gene in interactions
312 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000003 C00000094 C00000134 C00000615 C00001110 C00001120 C00001358 C00001424 C00001429 C00001435 C00002525 C00034649 | 0 / 1 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00000003 C00000094 C00000134 C00000615 C00001110 C00001120 C00001358 C00001429 C00001435 C00002525 C00034649 | 0 / 1 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000003 C00000094 C00000134 C00000615 C00001110 C00001120 C00001358 C00001429 C00001435 C00002525 C00034649 | 1 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00000003 C00000094 C00000134 C00000615 C00001110 C00001120 C00001358 C00001429 C00001435 C00002525 C00034649 | 1 / 1 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00000003 C00000094 C00000134 C00000615 C00001110 C00001120 C00001358 C00001429 C00001435 C00034649 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00000615 C00001055 C00001059 C00001110 C00001120 C00001183 C00001358 C00001424 C00002514 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00000615 C00001055 C00001059 C00001110 C00001358 C00001424 C00001435 C00002514 C00002525 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00000116 C00000134 C00000170 C00000580 C00000615 C00000651 C00002514 C00034649 | 1 / 1 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00000116 C00000134 C00001059 C00001183 C00001424 C00002514 C00002525 C00034649 | 0 / 0 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00000615 C00001055 C00001059 C00001424 C00001435 C00002514 C00002525 C00034649 | 0 / 0 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00000615 C00001055 C00001059 C00001424 C00002514 C00002525 C00034649 | 1 / 2 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00001055 C00001059 C00001183 C00001424 C00002507 C00002514 C00034649 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00000003 C00001110 C00001120 C00001183 C00001435 C00002507 C00002514 | 0 / 0 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00000615 C00001055 C00001059 C00001120 C00001424 C00002514 C00034649 | 1 / 1 |
| O00255 | Menin | Unclassified protein | C00000615 C00001055 C00001059 C00001424 C00002514 C00002525 C00034649 | 2 / 5 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00000615 C00001055 C00001059 C00001110 C00001403 C00001424 C00002514 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00000615 C00001059 C00001435 C00002514 C00002525 C00034649 | 0 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00000580 C00000615 C00001183 C00001403 C00001429 C00034649 | 1 / 2 |
| P02545 | Prelamin-A/C | Unclassified protein | C00000003 C00000615 C00001120 C00001424 C00002514 C00034649 | 11 / 10 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00001055 C00001059 C00001110 C00001358 C00001424 C00002514 | 0 / 0 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00000615 C00001358 C00001424 C00001429 C00001435 C00034649 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00000615 C00001055 C00001424 C00002514 C00002525 C00034649 | 4 / 3 |
| P10275 | Androgen receptor | NR3C4 | C00000003 C00000113 C00000116 C00000134 C00002514 C00034649 | 3 / 4 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00000091 C00000134 C00000615 C00001183 C00001429 C00034649 | 3 / 2 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00000580 C00000615 C00001183 C00001403 C00001429 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00001055 C00001059 C00001110 C00001120 C00002514 | 0 / 0 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00000580 C00000615 C00001183 C00001403 C00001429 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00000580 C00000615 C00001183 C00001403 C00001429 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00000615 C00001055 C00001059 C00001183 C00001424 | 0 / 0 |
| P15121 | Aldose reductase | Enzyme | C00000170 C00000580 C00000615 C00000750 C00001110 | 0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00000091 C00001059 C00001424 C00001429 C00002514 | 0 / 0 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00000580 C00000615 C00001403 C00001429 | 1 / 1 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00000615 C00001424 C00002514 C00002525 | 2 / 2 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00000580 C00000615 C00001403 C00001429 | 0 / 0 |
| P02768 | Serum albumin | Secreted protein | C00000100 C00000102 C00001120 C00034649 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00000615 C00001055 C00001059 C00002514 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00000580 C00000615 C00001429 C00034649 | 0 / 3 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00001055 C00001059 C00001110 C00002514 | 1 / 1 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00000615 C00001055 C00001059 C00002514 | 1 / 1 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00001424 C00001429 C00001435 C00034649 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00000615 C00001120 C00002514 C00034649 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00000580 C00000615 C00001403 C00001429 | 0 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00000580 C00000615 C00001403 C00001429 | 0 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00000580 C00000615 C00001403 C00001429 | 0 / 0 |
| Q9Y468 | Lethal(3)malignant brain tumor-like protein 1 | Unclassified protein | C00001055 C00001059 C00001110 C00001435 | 0 / 0 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00000615 C00001424 C00002514 C00034649 | 3 / 1 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00000580 C00000615 C00006927 C00034649 | 1 / 8 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00000580 C00000615 C00001403 C00001429 | 0 / 1 |
| O75751 | Solute carrier family 22 member 3 | Unclassified protein | C00001424 C00001429 C00001435 C00034649 | 0 / 0 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00000580 C00000615 C00001403 C00001429 | 1 / 2 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00000116 C00002514 C00002525 C00034649 | 0 / 0 |
| Q8TDS4 | Hydroxycarboxylic acid receptor 2 | Hydroxycarboxylic acid receptor | C00000170 C00000580 C00000615 C00001183 | 0 / 0 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00001055 C00001059 C00001110 C00001424 | 1 / 4 |
| P39748 | Flap endonuclease 1 | Enzyme | C00001055 C00001059 C00001424 C00002514 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00001055 C00001059 C00001110 C00001424 | 0 / 1 |
| P14679 | Tyrosinase | Oxidoreductase | C00000580 C00000615 C00001429 C00001435 | 4 / 2 |
| P14416 | D(2) dopamine receptor | Dopamine receptor | C00001424 C00001429 C00001435 C00034649 | 2 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00001424 C00002514 C00034649 | 2 / 3 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00000615 C00001429 C00034649 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00000170 C00000615 C00000977 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00000615 C00000977 C00034649 | 2 / 2 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00001358 C00001424 C00034649 | 0 / 0 |
| P52895 | Aldo-keto reductase family 1 member C2 | Enzyme | C00000218 C00000580 C00000615 | 1 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00000003 C00001183 C00001424 | 0 / 0 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00001358 C00001429 C00034649 | 0 / 0 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00001183 C00002507 C00002525 | 7 / 37 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00001358 C00001424 C00034649 | 0 / 1 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00001059 C00002507 C00034649 | 1 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00001358 C00001429 C00001435 | 2 / 0 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00001055 C00001059 C00001424 | 1 / 0 |
| P17405 | Sphingomyelin phosphodiesterase | Enzyme | C00001059 C00002507 C00002514 | 2 / 2 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00000116 C00002525 C00034649 | 5 / 3 |
| O15244 | Solute carrier family 22 member 2 | Drug uniporter | C00001424 C00001429 C00034649 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00000615 C00001424 C00034649 | 0 / 0 |
| Q14191 | Werner syndrome ATP-dependent helicase | Enzyme | C00001055 C00001059 C00002514 | 2 / 1 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00000615 C00001055 C00001059 | 0 / 0 |
| Q04828 | Aldo-keto reductase family 1 member C1 | Enzyme | C00000218 C00000580 C00000615 | 0 / 0 |
| P17516 | Aldo-keto reductase family 1 member C4 | Enzyme | C00000218 C00000580 C00000615 | 1 / 0 |
| Q9C0B1 | Alpha-ketoglutarate-dependent dioxygenase FTO | Unclassified protein | C00001183 C00001200 C00001358 | 1 / 2 |
| P42330 | Aldo-keto reductase family 1 member C3 | Enzyme | C00000218 C00000580 C00000615 | 0 / 0 |
| P19793 | Retinoic acid receptor RXR-alpha | NR2B1 | C00000116 C00002514 C00034649 | 0 / 0 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00000615 C00034649 | 5 / 2 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00001424 C00034649 | 1 / 0 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00000615 C00002525 | 3 / 3 |
| P22303 | Acetylcholinesterase | Hydrolase | C00001429 C00034649 | 1 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00001424 C00034649 | 0 / 0 |
| P18825 | Alpha-2C adrenergic receptor | Adrenergic receptor | C00001424 C00034649 | 0 / 0 |
| P06280 | Alpha-galactosidase A | Enzyme | C00000615 C00002514 | 1 / 1 |
| P28223 | 5-hydroxytryptamine receptor 2A | Serotonin receptor | C00001429 C00034649 | 0 / 0 |
| P50406 | 5-hydroxytryptamine receptor 6 | Serotonin receptor | C00001429 C00034649 | 0 / 0 |
| P28335 | 5-hydroxytryptamine receptor 2C | Serotonin receptor | C00001429 C00034649 | 0 / 0 |
| P31645 | Sodium-dependent serotonin transporter | Serotonin | C00001429 C00034649 | 2 / 0 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00001424 C00034649 | 0 / 0 |
| Q9GZT9 | Egl nine homolog 1 | Enzyme | C00000615 C00001183 | 1 / 1 |
| P54132 | Bloom syndrome protein | Enzyme | C00001424 C00002514 | 1 / 2 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00000615 C00002514 | 0 / 0 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00001435 C00034649 | 1 / 1 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00000170 C00000615 | 1 / 1 |
| O15245 | Solute carrier family 22 member 1 | Drug uniporter | C00001435 C00034649 | 0 / 0 |
| P28907 | ADP-ribosyl cyclase 1 | Enzyme | C00002374 C00020647 | 0 / 1 |
| Q16773 | Kynurenine--oxoglutarate transaminase 1 | Enzyme | C00000100 C00000115 | 0 / 0 |
| Q9NPD5 | Solute carrier organic anion transporter family member 1B3 | Electrochemical transporter | C00002514 C00034649 | 1 / 0 |
| P23975 | Sodium-dependent noradrenaline transporter | Norepinephrine | C00001424 C00034649 | 1 / 1 |
| P25100 | Alpha-1D adrenergic receptor | Adrenergic receptor | C00001424 C00034649 | 0 / 0 |
| P18089 | Alpha-2B adrenergic receptor | Adrenergic receptor | C00001424 C00034649 | 0 / 0 |
| P06239 | Tyrosine-protein kinase Lck | Src | C00000750 C00034649 | 0 / 1 |
| P03956 | Interstitial collagenase | M10A | C00000615 C00034649 | 0 / 1 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00001424 C00034649 | 0 / 0 |
| Q13093 | Platelet-activating factor acetylhydrolase | Enzyme | C00000580 C00000615 | 3 / 0 |
| P10145 | Interleukin-8 | Secreted protein | C00000134 C00034649 | 0 / 0 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00000615 C00034649 | 2 / 2 |
| P41595 | 5-hydroxytryptamine receptor 2B | Serotonin receptor | C00001429 C00034649 | 0 / 0 |
| P56937 | 3-keto-steroid reductase | Enzyme | C00002514 C00034649 | 0 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | C00000580 C00000615 | 0 / 0 |
| Q92731 | Estrogen receptor beta | NR3A2 | C00002514 C00034649 | 0 / 1 |
| O94956 | Solute carrier organic anion transporter family member 2B1 | Unclassified protein | C00002514 C00034649 | 0 / 0 |
| Q9Y6L6 | Solute carrier organic anion transporter family member 1B1 | Electrochemical transporter | C00002514 C00034649 | 1 / 0 |
| P46098 | 5-hydroxytryptamine receptor 3A | NS | C00001429 C00001435 | 0 / 0 |
| P35462 | D(3) dopamine receptor | Dopamine receptor | C00001429 C00034649 | 1 / 0 |
| P21917 | D(4) dopamine receptor | Dopamine receptor | C00001435 C00034649 | 0 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00002525 C00034649 | 4 / 1 |
| O75795 | UDP-glucuronosyltransferase 2B17 | Enzyme | C00001429 C00034649 | 1 / 0 |
| Q05BR4 | SLC16A10 protein | Unclassified protein | C00001200 C00001358 | 0 / 0 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00001055 C00002514 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00001424 C00001429 | 0 / 0 |
| P08913 | Alpha-2A adrenergic receptor | Adrenergic receptor | C00001424 C00034649 | 0 / 0 |
| P33527 | Multidrug resistance-associated protein 1 | drugs | C00002525 C00034649 | 0 / 0 |
| P51151 | Ras-related protein Rab-9A | Unclassified protein | C00002507 C00034649 | 0 / 0 |
| P38398 | Breast cancer type 1 susceptibility protein | Enzyme | C00002507 C00002525 | 4 / 2 |
| P63092 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | Other membrane protein | C00001059 | 7 / 3 |
| P54855 | UDP-glucuronosyltransferase 2B15 | Enzyme | C00001429 | 0 / 0 |
| P04035 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Oxidoreductase | C00034649 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00000003 | 2 / 0 |
| P11509 | Cytochrome P450 2A6 | Cytochrome P450 2A6 | C00034649 | 0 / 0 |
| P30988 | Calcitonin receptor | Calcitonin receptor | C00034649 | 0 / 0 |
| P35348 | Alpha-1A adrenergic receptor | Adrenergic receptor | C00001424 | 0 / 0 |
| P04278 | Sex hormone-binding globulin | Secreted protein | C00034649 | 0 / 0 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00034649 | 0 / 0 |
| P39086 | Glutamate receptor ionotropic, kainate 1 | NS | C00001358 | 0 / 0 |
| P43005 | Excitatory amino acid transporter 3 | Aspartate, glutamate and cysteine Na-symporter | C00001358 | 1 / 1 |
| P41968 | Melanocortin receptor 3 | Melanocortin receptor | C00034649 | 1 / 0 |
| P17948 | Vascular endothelial growth factor receptor 1 | Vegfr | C00034649 | 0 / 0 |
| P25101 | Endothelin-1 receptor | Endothelin receptor | C00034649 | 0 / 0 |
| P30411 | B2 bradykinin receptor | Bradykinin receptor | C00034649 | 0 / 0 |
| O15296 | Arachidonate 15-lipoxygenase B | Enzyme | C00001424 | 0 / 0 |
| P32245 | Melanocortin receptor 4 | Melanocortin receptor | C00034649 | 1 / 0 |
| P12104 | Fatty acid-binding protein, intestinal | Other cytosolic protein | C00034649 | 0 / 0 |
| P50052 | Type-2 angiotensin II receptor | Angiotensin receptor | C00034649 | 1 / 1 |
| P32238 | Cholecystokinin receptor type A | Cholecystokinin receptor | C00034649 | 0 / 0 |
| P08311 | Cathepsin G | S1A | C00034649 | 0 / 0 |
| Q99720 | Sigma non-opioid intracellular receptor 1 | Membrane receptor | C00034649 | 1 / 0 |
| P28566 | 5-hydroxytryptamine receptor 1E | Serotonin receptor | C00001429 | 0 / 0 |
| Q14416 | Metabotropic glutamate receptor 2 | Metabotropic glutamate receptor | C00001358 | 0 / 0 |
| P05164 | Myeloperoxidase | Enzyme | C00001429 | 1 / 2 |
| P32241 | Vasoactive intestinal polypeptide receptor 1 | Vasoactive intestinal peptide receptor | C00034649 | 0 / 0 |
| Q9GZT4 | Serine racemase | Enzyme | C00001424 | 0 / 0 |
| Q8TCC7 | Solute carrier family 22 member 8 | Antiporter | C00001120 | 0 / 0 |
| Q04206 | Transcription factor p65 | Transcription Factor | C00000977 | 0 / 0 |
| P25929 | Neuropeptide Y receptor type 1 | Neuropeptide Y receptor | C00034649 | 0 / 0 |
| P29274 | Adenosine receptor A2a | Adenosine receptor | C00034649 | 0 / 0 |
| P11308 | Transcriptional regulator ERG | Unclassified protein | C00001183 | 1 / 2 |
| Q9Y271 | Cysteinyl leukotriene receptor 1 | Leukotriene receptor | C00034649 | 0 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00034649 | 0 / 0 |
| P50224 | Sulfotransferase 1A3/1A4 | Enzyme | C00001424 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00002525 | 0 / 0 |
| P37288 | Vasopressin V1a receptor | Vasopressin and oxytocin receptor | C00034649 | 0 / 0 |
| P04150 | Glucocorticoid receptor | NR3C1 | C00034649 | 0 / 1 |
| P06401 | Progesterone receptor | NR3C3 | C00034649 | 0 / 1 |
| P08172 | Muscarinic acetylcholine receptor M2 | Acetylcholine receptor | C00034649 | 2 / 0 |
| P11229 | Muscarinic acetylcholine receptor M1 | Acetylcholine receptor | C00034649 | 0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00000977 | 2 / 2 |
| P14920 | D-amino-acid oxidase | Enzyme | C00001424 | 0 / 0 |
| P21554 | Cannabinoid receptor 1 | Cannabinoid receptor | C00034649 | 0 / 0 |
| Q14973 | Sodium/bile acid cotransporter | Bile acid | C00034649 | 0 / 0 |
| P16662 | UDP-glucuronosyltransferase 2B7 | Enzyme | C00001429 | 0 / 0 |
| P04626 | Receptor tyrosine-protein kinase erbB-2 | TK tyrosine-protein kinase EGFR subfamily | C00034649 | 5 / 9 |
| P20309 | Muscarinic acetylcholine receptor M3 | Acetylcholine receptor | C00034649 | 1 / 0 |
| Q9HBH1 | Peptide deformylase, mitochondrial | Enzyme | C00000100 | 0 / 0 |
| P27169 | Serum paraoxonase/arylesterase 1 | Enzyme | C00000750 | 1 / 0 |
| P21452 | Substance-K receptor | Neurokinin receptor | C00034649 | 0 / 0 |
| P42263 | Glutamate receptor 3 | NS | C00001358 | 1 / 1 |
| P12931 | Proto-oncogene tyrosine-protein kinase Src | Src | C00006927 | 0 / 0 |
| P51679 | C-C chemokine receptor type 4 | CC chemokine receptor | C00034649 | 0 / 0 |
| P51681 | C-C chemokine receptor type 5 | CC chemokine receptor | C00034649 | 3 / 0 |
| Q14831 | Metabotropic glutamate receptor 7 | Metabotropic glutamate receptor | C00001358 | 0 / 0 |
| P36544 | Neuronal acetylcholine receptor subunit alpha-7 | CHRN alpha | C00001429 | 0 / 0 |
| Q08209 | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Ser_Thr | C00034649 | 0 / 0 |
| P35968 | Vascular endothelial growth factor receptor 2 | Vegfr | C00006927 | 1 / 0 |
| O15303 | Metabotropic glutamate receptor 6 | Metabotropic glutamate receptor | C00001358 | 1 / 1 |
| P48039 | Melatonin receptor type 1A | Melatonin receptor | C00001429 | 0 / 0 |
| Q96RI1 | Bile acid receptor | NR1H4 | C00034649 | 0 / 0 |
| P08581 | Hepatocyte growth factor receptor | TK tyrosine-protein kinase MET subfamily | C00006927 | 2 / 3 |
| P35236 | Tyrosine-protein phosphatase non-receptor type 7 | Tyr | C00000615 | 0 / 0 |
| P41597 | C-C chemokine receptor type 2 | CC chemokine receptor | C00034649 | 1 / 0 |
| Q13002 | Glutamate receptor ionotropic, kainate 2 | NS | C00001358 | 1 / 1 |
| P08575 | Receptor-type tyrosine-protein phosphatase C | Enzyme | C00034649 | 2 / 1 |
| P56817 | Beta-secretase 1 | A1A | C00001435 | 0 / 0 |
| Q16478 | Glutamate receptor ionotropic, kainate 5 | NS | C00001358 | 0 / 0 |
| P00390 | Glutathione reductase, mitochondrial | Oxidoreductase | C00001358 | 0 / 0 |
| P06241 | Tyrosine-protein kinase Fyn | Src | C00034649 | 0 / 0 |
| P08253 | 72 kDa type IV collagenase | M10A | C00000615 | 1 / 3 |
| P25024 | C-X-C chemokine receptor type 1 | CXC chemokine receptor | C00034649 | 0 / 0 |
| Q9UNQ0 | ATP-binding cassette sub-family G member 2 | ATP binding cassette | C00034649 | 2 / 0 |
| Q92887 | Canalicular multispecific organic anion transporter 1 | Unclassified protein | C00034649 | 1 / 1 |
| P08183 | Multidrug resistance protein 1 | drug | C00034649 | 1 / 0 |
| Q16850 | Lanosterol 14-alpha demethylase | Cytochrome P450 51A1 | C00034649 | 0 / 0 |
| P51452 | Dual specificity protein phosphatase 3 | Ser_Thr_Tyr | C00000615 | 0 / 0 |
| P51449 | Nuclear receptor ROR-gamma | Nuclear hormone receptor subfamily 1 group F member 3 | C00034649 | 0 / 0 |
| P42261 | Glutamate receptor 1 | NS | C00001358 | 0 / 0 |
| P06133 | UDP-glucuronosyltransferase 2B4 | Enzyme | C00001429 | 0 / 0 |
| O76074 | cGMP-specific 3',5'-cyclic phosphodiesterase | PDE_5A | C00034649 | 0 / 0 |
| P00374 | Dihydrofolate reductase | Oxidoreductase | C00000112 | 1 / 1 |
| P04798 | Cytochrome P450 1A1 | Cytochrome P450 1A1 | C00002525 | 0 / 0 |
| P47898 | 5-hydroxytryptamine receptor 5A | Serotonin receptor | C00001429 | 0 / 0 |
| P34969 | 5-hydroxytryptamine receptor 7 | Serotonin receptor | C00001429 | 0 / 0 |
| P08912 | Muscarinic acetylcholine receptor M5 | Acetylcholine receptor | C00034649 | 0 / 0 |
| Q01959 | Sodium-dependent dopamine transporter | Dopamine | C00034649 | 1 / 0 |
| P35367 | Histamine H1 receptor | Histamine receptor | C00034649 | 0 / 0 |
| Q04609 | Glutamate carboxypeptidase 2 | M28B | C00001358 | 0 / 0 |
| P08173 | Muscarinic acetylcholine receptor M4 | Acetylcholine receptor | C00034649 | 0 / 0 |
| P25103 | Substance-P receptor | Neurokinin receptor | C00034649 | 0 / 0 |
| P25105 | Platelet-activating factor receptor | PAF receptor | C00034649 | 0 / 0 |
| P11362 | Fibroblast growth factor receptor 1 | Fgfr | C00006927 | 4 / 5 |
| P08185 | Corticosteroid-binding globulin | Secreted protein | C00034649 | 1 / 1 |
| P33032 | Melanocortin receptor 5 | Melanocortin receptor | C00034649 | 0 / 0 |
| Q14833 | Metabotropic glutamate receptor 4 | Metabotropic glutamate receptor | C00001358 | 0 / 0 |
| Q13255 | Metabotropic glutamate receptor 1 | Metabotropic glutamate receptor | C00001358 | 1 / 0 |
| P28222 | 5-hydroxytryptamine receptor 1B | Serotonin receptor | C00001429 | 0 / 0 |
| P30989 | Neurotensin receptor type 1 | Neurotensin receptor | C00001059 | 0 / 0 |
| P25021 | Histamine H2 receptor | Histamine receptor | C00034649 | 0 / 0 |
| P12268 | Inosine-5'-monophosphate dehydrogenase 2 | Oxidoreductase | C00000112 | 0 / 0 |
| P05181 | Cytochrome P450 2E1 | Cytochrome P450 2E1 | C00034649 | 0 / 0 |
| P23415 | Glycine receptor subunit alpha-1 | GLR alpha | C00001429 | 1 / 1 |
| Q6W5P4 | Neuropeptide S receptor | Neuropeptide receptor | C00034649 | 1 / 0 |
| Q99489 | D-aspartate oxidase | Enzyme | C00001424 | 0 / 0 |
| Q96RJ0 | Trace amine-associated receptor 1 | Trace amine receptor | C00001435 | 0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | C00034649 | 0 / 0 |
| P43004 | Excitatory amino acid transporter 2 | Aspartate and glutamate Na-symporter | C00001358 | 0 / 0 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | C00001429 | 5 / 1 |
| P27361 | Mitogen-activated protein kinase 3 | Erk | C00034649 | 0 / 0 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00001429 | 0 / 0 |
| P17252 | Protein kinase C alpha type | Alpha | C00034649 | 0 / 0 |
| P08235 | Mineralocorticoid receptor | NR3C2 | C00034649 | 2 / 2 |
| P08908 | 5-hydroxytryptamine receptor 1A | Serotonin receptor | C00001429 | 1 / 0 |
| P29466 | Caspase-1 | C14 | C00034649 | 0 / 0 |
| Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | Tyr | C00000615 | 4 / 2 |
| O00222 | Metabotropic glutamate receptor 8 | Metabotropic glutamate receptor | C00001358 | 0 / 0 |
| P28221 | 5-hydroxytryptamine receptor 1D | Serotonin receptor | C00001429 | 0 / 0 |
| P30542 | Adenosine receptor A1 | Adenosine receptor | C00034649 | 0 / 0 |
| P35368 | Alpha-1B adrenergic receptor | Adrenergic receptor | C00001424 | 0 / 0 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | C00002525 | 0 / 0 |
| Q13003 | Glutamate receptor ionotropic, kainate 3 | NS | C00001358 | 0 / 0 |
| P10721 | Mast/stem cell growth factor receptor Kit | Pdgfr | C00006927 | 4 / 3 |
| P24557 | Thromboxane-A synthase | Cytochrome P450 5A1 | C00034649 | 1 / 1 |
| P30939 | 5-hydroxytryptamine receptor 1F | Serotonin receptor | C00001429 | 0 / 0 |
| P05412 | Transcription factor AP-1 | Transcription Factor | C00000977 | 0 / 0 |
| P49286 | Melatonin receptor type 1B | Melatonin receptor | C00001429 | 0 / 1 |
| P49146 | Neuropeptide Y receptor type 2 | Neuropeptide Y receptor | C00034649 | 0 / 0 |
| Q14832 | Metabotropic glutamate receptor 3 | Metabotropic glutamate receptor | C00001358 | 0 / 0 |
| P35869 | Aryl hydrocarbon receptor | Transcription Factor | C00002514 | 0 / 0 |
| P25025 | C-X-C chemokine receptor type 2 | CXC chemokine receptor | C00034649 | 0 / 0 |
| P42262 | Glutamate receptor 2 | NS | C00001358 | 0 / 0 |
| P41594 | Metabotropic glutamate receptor 5 | Metabotropic glutamate receptor | C00001358 | 0 / 0 |
| P43003 | Excitatory amino acid transporter 1 | Aspartate and glutamate Na-symporter | C00001358 | 1 / 1 |
| P21802 | Fibroblast growth factor receptor 2 | TK tyrosine-protein kinase TLK subfamily | C00006927 | 9 / 3 |
| Q12794 | Hyaluronidase-1 | Enzyme | C00000615 | 1 / 2 |
| Q16539 | Mitogen-activated protein kinase 14 | p38 | C00034649 | 0 / 0 |
| P24298 | Alanine aminotransferase 1 | Enzyme | C00000615 | 0 / 0 |
| P08246 | Neutrophil elastase | S1A | C00034649 | 2 / 1 |
| Q13639 | 5-hydroxytryptamine receptor 4 | Serotonin receptor | C00001429 | 0 / 0 |
| P40225 | Thrombopoietin | Unclassified protein | C00034649 | 1 / 1 |
| P27338 | Amine oxidase [flavin-containing] B | Oxidoreductase | C00001435 | 0 / 0 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00000615 | 0 / 0 |
| Q7Z2W7 | Transient receptor potential cation channel subfamily M member 8 | Unclassified protein | C00001429 | 0 / 0 |
| P14618 | Pyruvate kinase PKM | Enzyme | C00001358 | 0 / 0 |
| Q16798 | NADP-dependent malic enzyme, mitochondrial | Enzyme | C00001183 | 0 / 0 |
| O95342 | Bile salt export pump | drug | C00034649 | 2 / 1 |
| Q99700 | Ataxin-2 | Unclassified protein | C00034649 | 1 / 1 |
| P31639 | Sodium/glucose cotransporter 2 | Glucose | C00002525 | 1 / 1 |
| Q9UN88 | Gamma-aminobutyric acid receptor subunit theta | GABA-A theta | C00001429 | 0 / 0 |
| P28472 | Gamma-aminobutyric acid receptor subunit beta-3 | GABA-A beta | C00001429 | 1 / 0 |
| P14867 | Gamma-aminobutyric acid receptor subunit alpha-1 | GABA-A alpha | C00001429 | 1 / 1 |
| P48169 | Gamma-aminobutyric acid receptor subunit alpha-4 | GABA-A alpha | C00001429 | 0 / 0 |
| O14764 | Gamma-aminobutyric acid receptor subunit delta | GABA-A delta | C00001429 | 1 / 1 |
| P31644 | Gamma-aminobutyric acid receptor subunit alpha-5 | GABA-A alpha | C00001429 | 0 / 0 |
| Q99928 | Gamma-aminobutyric acid receptor subunit gamma-3 | GABA-A gamma | C00001429 | 0 / 0 |
| P78334 | Gamma-aminobutyric acid receptor subunit epsilon | GABA-A epsilon | C00001429 | 0 / 0 |
| Q8N1C3 | Gamma-aminobutyric acid receptor subunit gamma-1 | GABA-A gamma | C00001429 | 0 / 0 |
| P34903 | Gamma-aminobutyric acid receptor subunit alpha-3 | GABA-A alpha | C00001429 | 0 / 0 |
| O00591 | Gamma-aminobutyric acid receptor subunit pi | GABA-A pi | C00001429 | 0 / 0 |
| P47870 | Gamma-aminobutyric acid receptor subunit beta-2 | GABA-A beta | C00001429 | 0 / 0 |
| P18507 | Gamma-aminobutyric acid receptor subunit gamma-2 | GABA-A gamma | C00001429 | 4 / 2 |
| P18505 | Gamma-aminobutyric acid receptor subunit beta-1 | GABA-A beta | C00001429 | 0 / 0 |
| Q16445 | Gamma-aminobutyric acid receptor subunit alpha-6 | GABA-A alpha | C00001429 | 0 / 0 |
| P47869 | Gamma-aminobutyric acid receptor subunit alpha-2 | GABA-A alpha | C00001429 | 1 / 0 |
| O15427 | Monocarboxylate transporter 4 | Unclassified protein | C00001200 | 0 / 0 |
| P13866 | Sodium/glucose cotransporter 1 | Glucose | C00002525 | 1 / 1 |
| Q92959 | Solute carrier organic anion transporter family member 2A1 | Unclassified protein | C00001200 | 1 / 0 |
| O60669 | Monocarboxylate transporter 2 | Unclassified protein | C00001200 | 0 / 0 |
| P35610 | Sterol O-acyltransferase 1 | Enzyme | C00034649 | 0 / 0 |
| Q8WXA8 | 5-hydroxytryptamine receptor 3C | NS | C00001429 | 0 / 0 |
| Q70Z44 | 5-hydroxytryptamine receptor 3D | NS | C00001429 | 0 / 0 |
| O95264 | 5-hydroxytryptamine receptor 3B | NS | C00001429 | 0 / 0 |
| A5X5Y0 | 5-hydroxytryptamine receptor 3E | NS | C00001429 | 0 / 0 |
| P07900 | Heat shock protein HSP 90-alpha | Other cytosolic protein | C00000615 | 0 / 0 |
| P08238 | Heat shock protein HSP 90-beta | Other cytosolic protein | C00000615 | 0 / 0 |
| P63165 | Small ubiquitin-related modifier 1 | Unclassified protein | C00002514 | 1 / 1 |
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | C00001429 | 3 / 0 |
| P48058 | Glutamate receptor 4 | NS | C00001358 | 0 / 0 |
| Q9Y2I1 | Nischarin | Other cytosolic protein | C00001424 | 0 / 0 |
| Q15125 | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | Enzyme | C00034649 | 1 / 1 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00034649 | 1 / 1 |
3398 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 4128 | MAOA, MAO-A | monoamine oxidase A (EC:1.4.3.4) |
C00001424
C00001429
C00001435
C00034649
|
| 1831 | TSC22D3, DIP, DSIPI, GILZ, TSC-22R, hDIP | TSC22 domain family, member 3 |
C00000977
C00002514
C00034649
|
| 1312 | COMT | catechol-O-methyltransferase (EC:2.1.1.6) |
C00001424
C00002514
C00034649
|
| 4790 | NFKB1, EBP-1, KBF1, NF-kB1, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
C00001424
C00002514
C00034649
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00001424
C00001429
C00034649
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00001424
C00001429
C00034649
|
| 5366 | PMAIP1, APR, NOXA | phorbol-12-myristate-13-acetate-induced protein 1 |
C00000094
C00002514
C00034649
|
| 6530 | SLC6A2, NAT1, NET, NET1, SLC6A5 | solute carrier family 6 (neurotransmitter transporter), member 2 |
C00001424
C00001429
C00001435
|
| 1487 | CTBP1, BARS | C-terminal binding protein 1 |
C00001200
C00002514
C00034649
|
| 6531 | SLC6A3, DAT, DAT1, PKDYS | solute carrier family 6 (neurotransmitter transporter), member 3 |
C00001424
C00001429
C00001435
|
| 6532 | SLC6A4, 5-HTT, 5-HTTLPR, 5HTT, HTT, OCD1, SERT, SERT1, hSERT | solute carrier family 6 (neurotransmitter transporter), member 4 |
C00001424
C00001429
C00001435
|
| 6818 | SULT1A3, HAST, HAST3, M-PST, ST1A3/ST1A4, ST1A5, STM, TL-PST | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (EC:2.8.2.1) |
C00001424
C00001429
C00001435
|
| 134864 | TAAR1, RP11-295F4.9, TA1, TAR1, TRAR1 | trace amine associated receptor 1 |
C00001424
C00001429
C00001435
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00002514
C00002525
C00034649
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00002514
C00002525
C00034649
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00002514
C00002525
C00034649
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00002514
C00002525
C00034649
|
| 11259 | FILIP1L, DOC-1, DOC1, GIP130, GIP90 | filamin A interacting protein 1-like |
C00002514
C00034649
|
| 5243 | ABCB1, ABC20, CD243, CLCS, GP170, MDR1, P-GP, PGY1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 (EC:3.6.3.44) |
C00002525
C00034649
|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00002507
C00002525
|
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002525
C00034649
|
| 2932 | GSK3B | glycogen synthase kinase 3 beta (EC:2.7.11.1 2.7.11.26) |
C00001429
C00034649
|
| 1586 | CYP17A1, CPT7, CYP17, P450C17, S17AH | cytochrome P450, family 17, subfamily A, polypeptide 1 (EC:1.14.99.9 4.1.2.30) |
C00001200
C00034649
|
| 3356 | HTR2A, 5-HT2A, HTR2 | 5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
C00001429
C00034649
|
| 3593 | IL12B, CLMF, CLMF2, IL-12B, NKSF, NKSF2 | interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
C00001429
C00034649
|
| 1080 | CFTR, ABC35, ABCC7, CF, CFTR/MRP, MRP7, TNR-CFTR, dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) (EC:3.6.3.49) |
C00001429
C00034649
|
| 6286 | S100P, MIG9 | S100 calcium binding protein P |
C00000094
C00034649
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00002514
C00002525
|
| 54577 | UGT1A7, UDPGT, UDPGT_1-7, UGT-1G, UGT1-07, UGT1.7, UGT1G | UDP glucuronosyltransferase 1 family, polypeptide A7 (EC:2.4.1.17) |
C00002514
C00002525
|
| 54576 | UGT1A8, UDPGT, UDPGT_1-8, UGT-1H, UGT1-08, UGT1.8, UGT1A8S, UGT1H | UDP glucuronosyltransferase 1 family, polypeptide A8 (EC:2.4.1.17) |
C00002514
C00002525
|
| 54600 | UGT1A9, HLUGP4, LUGP4, UDPGT, UDPGT_1-9, UGT-1I, UGT1-09, UGT1-9, UGT1.9, UGT1AI, UGT1I | UDP glucuronosyltransferase 1 family, polypeptide A9 (EC:2.4.1.17) |
C00002514
C00002525
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00001429
C00034649
|
| 3690 | ITGB3, BDPLT16, BDPLT2, CD61, GP3A, GPIIIa, GT | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
C00001429
C00034649
|
| 4353 | MPO | myeloperoxidase (EC:1.11.2.2) |
C00001429
C00034649
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00000100
C00034649
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00000100
C00002514
|
| 5782 | PTPN12, PTP-PEST, PTPG1 | protein tyrosine phosphatase, non-receptor type 12 (EC:3.1.3.48) |
C00000094
C00002514
|
| 2516 | NR5A1, AD4BP, ELP, FTZ1, FTZF1, POF7, SF-1, SF1, SPGF8, SRXY3 | nuclear receptor subfamily 5, group A, member 1 |
C00001200
C00034649
|
| 440 | ASNS, TS11 | asparagine synthetase (glutamine-hydrolyzing) (EC:6.3.5.4) |
C00000094
C00034649
|
| 6571 | SLC18A2, SVAT, SVMT, VAT2, VMAT2 | solute carrier family 18 (vesicular monoamine transporter), member 2 |
C00001424
C00001429
|
| 18 | ABAT, GABA-AT, GABAT, NPD009 | 4-aminobutyrate aminotransferase (EC:2.6.1.19 2.6.1.22) |
C00002514
C00034649
|
| 6570 | SLC18A1, CGAT, VAT1, VMAT1 | solute carrier family 18 (vesicular monoamine transporter), member 1 |
C00001424
C00001429
|
| 5825 | ABCD3, ABC43, PMP70, PXMP1, ZWS2 | ATP-binding cassette, sub-family D (ALD), member 3 |
C00002514
C00034649
|
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00001424
C00034649
|
| 4852 | NPY, PYY4 | neuropeptide Y |
C00001424
C00034649
|
| 9619 | ABCG1, ABC8, WHITE1 | ATP-binding cassette, sub-family G (WHITE), member 1 |
C00002514
C00034649
|
| 83451 | ABHD11, WBSCR21 | abhydrolase domain containing 11 |
C00002514
C00034649
|
| 26090 | ABHD12, ABHD12A, BEM46L2, C20orf22, PHARC, dJ965G21.2 | abhydrolase domain containing 12 (EC:3.1.1.23) |
C00002514
C00034649
|
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00001424
C00034649
|
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00001424
C00034649
|
| 39 | ACAT2 | acetyl-CoA acetyltransferase 2 (EC:2.3.1.9) |
C00002514
C00034649
|
| 10891 | PPARGC1A, LEM6, PGC-1(alpha), PGC-1v, PGC1, PGC1A, PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
C00001200
C00034649
|
| 5142 | PDE4B, DPDE4, PDE4B5, PDEIVB | phosphodiesterase 4B, cAMP-specific (EC:3.1.4.17) |
C00000094
C00034649
|
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00001429
C00034649
|
| 2778 | GNAS, AHO, C20orf45, GNAS1, GPSA, GSA, GSP, NESP, PHP1A, PHP1B, PHP1C, POH | GNAS complex locus |
C00001424
C00034649
|
| 2335 | FN1, CIG, ED-B, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | fibronectin 1 |
C00001424
C00034649
|
| 6352 | CCL5, D17S136E, RANTES, SCYA5, SIS-delta, SISd, TCP228, eoCP | chemokine (C-C motif) ligand 5 |
C00001424
C00034649
|
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00001424
C00034649
|
| 90 | ACVR1, ACTRI, ACVR1A, ACVRLK2, ALK2, FOP, SKR1, TSRI | activin A receptor, type I (EC:2.7.11.30) |
C00002514
C00034649
|
| 152 | ADRA2C, ADRA2L2, ADRA2RL2, ADRARL2, ALPHA2CAR | adrenoceptor alpha 2C |
C00001424
C00034649
|
| 195828 | ZNF367, AFF29, CDC14B, ZFF29 | zinc finger protein 367 |
C00002514
C00034649
|
| 54819 | ZCCHC10 | zinc finger, CCHC domain containing 10 |
C00002514
C00034649
|
| 8839 | WISP2, CCN5, CT58, CTGF-L | WNT1 inducible signaling pathway protein 2 |
C00002514
C00034649
|
| 150 | ADRA2A, ADRA2, ADRA2R, ADRAR, ALPHA2AAR, ZNF32 | adrenoceptor alpha 2A |
C00001424
C00002514
|
| 10785 | WDR4, TRM82, TRMT82 | WD repeat domain 4 |
C00002514
C00034649
|
| 11169 | WDHD1, AND-1, CHTF4, CTF4 | WD repeat and HMG-box DNA binding protein 1 |
C00002514
C00034649
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00002514
C00034649
|
| 10090 | UST, 2OST | uronyl-2-sulfotransferase |
C00002514
C00034649
|
| 7374 | UNG, DGU, HIGM4, HIGM5, UDG, UNG1, UNG15, UNG2 | uracil-DNA glycosylase (EC:3.2.2.27) |
C00002514
C00034649
|
| 7372 | UMPS, OPRT | uridine monophosphate synthetase (EC:2.4.2.10 4.1.1.23) |
C00002514
C00034649
|
| 7328 | UBE2H, E2-20K, GID3, UBC8, UBCH, UBCH2 | ubiquitin-conjugating enzyme E2H (EC:6.3.2.19) |
C00002514
C00034649
|
| 7298 | TYMS, HST422, TMS, TS | thymidylate synthetase (EC:2.1.1.45) |
C00002514
C00034649
|
| 7291 | TWIST1, ACS3, BPES2, BPES3, CRS1, SCS, TWIST, bHLHa38 | twist family bHLH transcription factor 1 |
C00002514
C00034649
|
| 7280 | TUBB2A, TUBB, TUBB2, dJ40E16.7 | tubulin, beta 2A class IIa |
C00002514
C00034649
|
| 203068 | TUBB, M40, OK/SW-cl.56, TUBB1, TUBB5 | tubulin, beta class I |
C00002514
C00034649
|
| 8458 | TTF2, HuF2 | transcription termination factor, RNA polymerase II |
C00002514
C00034649
|
| 11214 | AKAP13, AKAP-13, AKAP-Lbc, ARHGEF13, BRX, HA-3, Ht31, LBC, PRKA13, PROTO-LB, PROTO-LBC, c-lbc, p47 | A kinase (PRKA) anchor protein 13 |
C00002514
C00034649
|
| 214 | ALCAM, CD166, MEMD | activated leukocyte cell adhesion molecule |
C00002514
C00034649
|
| 55809 | TRERF1, BCAR2, HSA277276, RAPA, RP1-139D8.5, TREP132, TReP-132, dJ139D8.5 | transcriptional regulating factor 1 |
C00002514
C00034649
|
| 222 | ALDH3B2, ALDH8 | aldehyde dehydrogenase 3 family, member B2 (EC:1.2.1.5) |
C00002514
C00034649
|
| 22974 | TPX2, C20orf1, C20orf2, DIL-2, DIL2, FLS353, GD:C20orf1, HCA519, HCTP4, REPP86, p100 | TPX2, microtubule-associated |
C00002514
C00034649
|
| 7168 | TPM1, C15orf13, CMD1Y, CMH3, HTM-alpha, LVNC9, TMSA | tropomyosin 1 (alpha) |
C00002514
C00034649
|
| 7164 | TPD52L1, D53, hD53 | tumor protein D52-like 1 |
C00002514
C00034649
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00002514
C00034649
|
| 249 | ALPL, AP-TNAP, APTNAP, HOPS, TNAP, TNSALP | alkaline phosphatase, liver/bone/kidney (EC:3.1.3.1) |
C00002514
C00034649
|
| 10766 | TOB2, TOB4, TOBL, TROB2 | transducer of ERBB2, 2 |
C00002514
C00034649
|
| 27242 | TNFRSF21, BM-018, CD358, DR6 | tumor necrosis factor receptor superfamily, member 21 |
C00002514
C00034649
|
| 735524 |
C00002514
C00034649
|
||
| 64699 | TMPRSS3, DFNB10, DFNB8, ECHOS1, TADG12 | transmembrane protease, serine 3 (EC:3.4.21.-) |
C00002514
C00034649
|
| 27346 | TMEM97, MAC30 | transmembrane protein 97 |
C00002514
C00034649
|
| 9949 | AMMECR1, AMMERC1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
C00002514
C00034649
|
| 10329 | TMEM5, HP10481, MDDGA10 | transmembrane protein 5 |
C00002514
C00034649
|
| 84187 | TMEM164, bB360B22.3 | transmembrane protein 164 |
C00002514
C00034649
|
| 283578 | TMED8, FAM15B | transmembrane emp24 protein transport domain containing 8 |
C00002514
C00034649
|
| 53346 | TM6SF1 | transmembrane 6 superfamily member 1 |
C00002514
C00034649
|
| 7083 | TK1, TK2 | thymidine kinase 1, soluble (EC:2.7.1.21) |
C00002514
C00034649
|
| 54443 | ANLN, Scraps, scra | anillin, actin binding protein |
C00002514
C00034649
|
| 7078 | TIMP3, HSMRK222, K222, K222TA2, SFD | TIMP metallopeptidase inhibitor 3 |
C00002514
C00034649
|
| 79875 | THSD4, ADAMTSL-6, ADAMTSL6, FVSY9334, PRO34005 | thrombospondin, type I, domain containing 4 |
C00002514
C00034649
|
| 7057 | THBS1, THBS, THBS-1, TSP, TSP-1, TSP1 | thrombospondin 1 |
C00002514
C00034649
|
| 7050 | TGIF1, HPE4, TGIF | TGFB-induced factor homeobox 1 |
C00002514
C00034649
|
| 7042 | TGFB2, LDS4, TGF-beta2 | transforming growth factor, beta 2 |
C00002514
C00034649
|
| 306 | ANXA3, ANX3 | annexin A3 (EC:3.1.4.43) |
C00002514
C00034649
|
| 7035 | TFPI, EPI, LACI, TFI, TFPI1 | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
C00002514
C00034649
|
| 7033 | TFF3, ITF, P1B, TFI | trefoil factor 3 (intestinal) |
C00002514
C00034649
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00002514
C00034649
|
| 123036 | TC2N, C14orf47, C2CD1, MTAC2D1, Tac2-N | tandem C2 domains, nuclear |
C00002514
C00034649
|
| 4070 | TACSTD2, EGP-1, EGP1, GA733-1, GA7331, GP50, M1S1, TROP2 | tumor-associated calcium signal transducer 2 |
C00002514
C00034649
|
| 94122 | SYTL5, slp5 | synaptotagmin-like 5 |
C00002514
C00034649
|
| 9582 | APOBEC3B, A3B, APOBEC1L, ARCD3, ARP4, DJ742C19.2, PHRBNL, bK150C2.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
C00002514
C00034649
|
| 347 | APOD | apolipoprotein D |
C00002514
C00034649
|
| 94121 | SYTL4, SLP4 | synaptotagmin-like 4 |
C00002514
C00034649
|
| 360 | AQP3, AQP-3, GIL | aquaporin 3 (Gill blood group) |
C00002514
C00034649
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00002514
C00034649
|
| 55638 | SYBU, GOLSYN, OCSYN, SNPHL | syntabulin (syntaxin-interacting) |
C00002514
C00034649
|
| 55959 | SULF2, HSULF-2 | sulfatase 2 |
C00002514
C00034649
|
| 6809 | STX3, STX3A | syntaxin 3 |
C00002514
C00034649
|
| 396 | ARHGDIA, GDIA1, NPHS8, RHOGDI, RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha |
C00002514
C00034649
|
| 8614 | STC2, STC-2, STCRP | stanniocalcin 2 |
C00002514
C00034649
|
| 6781 | STC1, STC | stanniocalcin 1 |
C00002514
C00034649
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00002514
C00034649
|
| 6482 | ST3GAL1, Gal-NAc6S, SIAT4A, SIATFL, ST3GalA, ST3GalA.1, ST3GalIA, ST3GalIA,1, ST3O | ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (EC:2.4.99.4) |
C00002514
C00034649
|
| 8878 | SQSTM1, A170, OSIL, PDB3, ZIP3, p60, p62, p62B | sequestosome 1 |
C00002514
C00034649
|
| 58472 | SQRDL, PRO1975 | sulfide quinone reductase-like (yeast) |
C00002514
C00034649
|
| 57405 | SPC25, SPBC25, hSpc25 | SPC25, NDC80 kinetochore complex component |
C00002514
C00034649
|
| 6662 | SOX9, CMD1, CMPD1, SRA1 | SRY (sex determining region Y)-box 9 |
C00002514
C00034649
|
| 6659 | SOX4, EVI16 | SRY (sex determining region Y)-box 4 |
C00002514
C00034649
|
| 8835 | SOCS2, CIS2, Cish2, SOCS-2, SSI-2, SSI2, STATI2 | suppressor of cytokine signaling 2 |
C00002514
C00034649
|
| 259266 | ASPM, ASP, Calmbp1, MCPH5 | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
C00002514
C00034649
|
| 29028 | ATAD2, ANCCA, PRO2000 | ATPase family, AAA domain containing 2 (EC:3.6.1.3) |
C00002514
C00034649
|
| 29887 | SNX10, OPTB8 | sorting nexin 10 |
C00002514
C00034649
|
| 467 | ATF3 | activating transcription factor 3 |
C00002514
C00034649
|
| 10051 | SMC4, CAP-C, CAPC, SMC-4, SMC4L1, hCAP-C | structural maintenance of chromosomes 4 |
C00002514
C00034649
|
| 10592 | SMC2, CAP-E, CAPE, SMC-2, SMC2L1 | structural maintenance of chromosomes 2 |
C00002514
C00034649
|
| 481 | ATP1B1, ATP1B | ATPase, Na+/K+ transporting, beta 1 polypeptide |
C00002514
C00034649
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00002514
C00034649
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002514
C00034649
|
| 9368 | SLC9A3R1, EBP50, NHERF, NHERF-1, NHERF1, NPHLOP2 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
C00002514
C00034649
|
| 8140 | SLC7A5, 4F2LC, CD98, D16S469E, E16, LAT1, MPE16, hLAT1 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
C00002514
C00034649
|
| 27109 | ATP5S, ATPW, FB, HSU79253 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) (EC:3.6.1.14) |
C00002514
C00034649
|
| 23657 | SLC7A11, CCBR1, xCT | solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
C00002514
C00034649
|
| 5205 | ATP8B1, ATPIC, BRIC, FIC1, ICP1, PFIC, PFIC1 | ATPase, aminophospholipid transporter, class I, type 8B, member 1 (EC:3.6.3.1) |
C00002514
C00034649
|
| 6526 | SLC5A3, SMIT, SMIT1, SMIT2 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
C00002514
C00034649
|
| 64116 | SLC39A8, BIGM103, LZT-Hs6, ZIP8 | solute carrier family 39 (zinc transporter), member 8 |
C00002514
C00034649
|
| 2542 | SLC37A4, G6PT1, G6PT2, G6PT3, GSD1b, GSD1c, GSD1d, TRG-19, TRG19 | solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
C00002514
C00034649
|
| 6790 | AURKA, AIK, ARK1, AURA, AURORA2, BTAK, PPP1R47, STK15, STK6, STK7 | aurora kinase A (EC:2.7.11.1) |
C00002514
C00034649
|
| 1836 | SLC26A2, D5S1708, DTD, DTDST, EDM4, MST153, MSTP157 | solute carrier family 26 (anion exchanger), member 2 |
C00002514
C00034649
|
| 123096 | SLC25A29, C14orf69, CACL, ORNT3 | solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
C00002514
C00034649
|
| 6509 | SLC1A4, ASCT1, SATT | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
C00002514
C00034649
|
| 6502 | SKP2, FBL1, FBXL1, FLB1, p45 | S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
C00002514
C00034649
|
| 23411 | SIRT1, SIR2L1 | sirtuin 1 |
C00001200
C00002514
|
| 84002 | B3GNT5, B3GN-T5, beta3Gn-T5 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 (EC:2.4.1.206) |
C00002514
C00034649
|
| 6470 | SHMT1, CSHMT, SHMT | serine hydroxymethyltransferase 1 (soluble) (EC:2.1.2.1) |
C00002514
C00034649
|
| 79801 | SHCBP1, PAL | SHC SH2-domain binding protein 1 |
C00002514
C00034649
|
| 23677 | SH3BP4, BOG25, TTP | SH3-domain binding protein 4 |
C00002514
C00034649
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00002514
C00034649
|
| 9529 | BAG5, BAG-5 | BCL2-associated athanogene 5 |
C00002514
C00034649
|
| 151246 | SGOL2, SGO2, TRIPIN | shugoshin-like 2 (S. pombe) |
C00002514
C00034649
|
| 23678 | SGK3, CISK, SGK2, SGKL | serum/glucocorticoid regulated kinase family, member 3 (EC:2.7.11.1) |
C00002514
C00034649
|
| 83667 | SESN2, HI95, SES2, SEST2 | sestrin 2 |
C00002514
C00034649
|
| 10512 | SEMA3C, SEMAE, SemE | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
C00002514
C00034649
|
| 6319 | SCD, FADS5, MSTP008, SCD1, SCDOS | stearoyl-CoA desaturase (delta-9-desaturase) (EC:1.14.19.1) |
C00002514
C00034649
|
| 23328 | SASH1, SH3D6A, dJ323M4, dJ323M4.1 | SAM and SH3 domain containing 1 |
C00002514
C00034649
|
| 8412 | BCAR3, NSP2, SH2D3B | breast cancer anti-estrogen resistance 3 |
C00002514
C00034649
|
| 6281 | S100A10, 42C, ANX2L, ANX2LG, CAL1L, CLP11, Ca[1], GP11, P11, p10 | S100 calcium binding protein A10 |
C00002514
C00034649
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00002514
C00034649
|
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00002514
C00034649
|
| 255488 | RNF144B, IBRDC2, PIR2, bA528A10.3, p53RFP | ring finger protein 144B |
C00002514
C00034649
|
| 390 | RND3, ARHE, Rho8, RhoE, memB | Rho family GTPase 3 |
C00002514
C00034649
|
| 253260 | RICTOR, AVO3, PIA, hAVO3 | RPTOR independent companion of MTOR, complex 2 |
C00002514
C00034649
|
| 5997 | RGS2, G0S8 | regulator of G-protein signaling 2, 24kDa |
C00002514
C00034649
|
| 5932 | RBBP8, COM1, CTIP, JWDS, RIM, SAE2, SCKL2 | retinoblastoma binding protein 8 |
C00002514
C00034649
|
| 9821 | RB1CC1, CC1, FIP200 | RB1-inducible coiled-coil 1 |
C00002514
C00034649
|
| 10125 | RASGRP1, CALDAG-GEFI, CALDAG-GEFII, RASGRP, V, hRasGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
C00002514
C00034649
|
| 51655 | RASD1, AGS1, DEXRAS1, MGC:26290 | RAS, dexamethasone-induced 1 |
C00002514
C00034649
|
| 10743 | RAI1, SMCR, SMS | retinoic acid induced 1 |
C00002514
C00034649
|
| 29127 | RACGAP1, CYK4, HsCYK-4, ID-GAP, MgcRacGAP | Rac GTPase activating protein 1 |
C00002514
C00034649
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00002514
C00034649
|
| 641 | BLM, BS, RECQ2, RECQL2, RECQL3 | Bloom syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00002514
C00034649
|
| 11031 | RAB31, Rab22B | RAB31, member RAS oncogene family |
C00002514
C00034649
|
| 9536 | PTGES, MGST-IV, MGST1-L1, MGST1L1, MPGES, PGES, PIG12, PP102, PP1294, TP53I12, mPGES-1 | prostaglandin E synthase (EC:5.3.99.3) |
C00002514
C00034649
|
| 23362 | PSD3, EFA6R, HCA67 | pleckstrin and Sec7 domain containing 3 |
C00002514
C00034649
|
| 11098 | PRSS23, SIG13, SPUVE, ZSIG13 | protease, serine, 23 |
C00002514
C00034649
|
| 7916 | PRRC2A, BAT2, D6S51, D6S51E, G2 | proline-rich coiled-coil 2A |
C00002514
C00034649
|
| 222171 | PRR15 | proline rich 15 |
C00002514
C00034649
|
| 655 | BMP7, OP-1 | bone morphogenetic protein 7 |
C00002514
C00034649
|
| 23645 | PPP1R15A, GADD34 | protein phosphatase 1, regulatory subunit 15A |
C00002514
C00034649
|
| 5493 | PPL | periplakin |
C00002514
C00034649
|
| 10105 | PPIF, CYP3, CyP-M, Cyp-D, CypD | peptidylprolyl isomerase F (EC:5.2.1.8) |
C00002514
C00034649
|
| 84172 | POLR1B, RPA135, RPA2, Rpo1-2 | polymerase (RNA) I polypeptide B, 128kDa (EC:2.7.7.6) |
C00002514
C00034649
|
| 56937 | PMEPA1, STAG1, TMEPAI | prostate transmembrane protein, androgen induced 1 |
C00002514
C00034649
|
| 51090 | PLLP, PMLP, TM4SF11 | plasmolipin |
C00002514
C00034649
|
| 7262 | PHLDA2, BRW1C, BWR1C, HLDA2, IPL, TSSC3 | pleckstrin homology-like domain, family A, member 2 |
C00002514
C00034649
|
| 5241 | PGR, NR3C3, PR | progesterone receptor |
C00002514
C00034649
|
| 5238 | PGM3, AGM1, PAGM, PGM_3 | phosphoglucomutase 3 (EC:5.4.2.3) |
C00002514
C00034649
|
| 283209 | PGM2L1, BM32A | phosphoglucomutase 2-like 1 (EC:2.7.1.106) |
C00002514
C00034649
|
| 54704 | PDP1, PDH, PDP, PDPC, PPM2C | pyruvate dehyrogenase phosphatase catalytic subunit 1 (EC:3.1.3.43) |
C00002514
C00034649
|
| 9124 | PDLIM1, CLIM1, CLP-36, CLP36, hCLIM1 | PDZ and LIM domain 1 |
C00002514
C00034649
|
| 27250 | PDCD4, H731 | programmed cell death 4 (neoplastic transformation inhibitor) |
C00002514
C00034649
|
| 5046 | PCSK6, PACE4, SPC4 | proprotein convertase subtilisin/kexin type 6 |
C00002514
C00034649
|
| 5106 | PCK2, PEPCK, PEPCK-M, PEPCK2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) (EC:4.1.1.32) |
C00002514
C00034649
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00002514
C00034649
|
| 5098 | PCDHGC3, PC43, PCDH-GAMMA-C3, PCDH2 | protocadherin gamma subfamily C, 3 |
C00002514
C00034649
|
| 5087 | PBX1 | pre-B-cell leukemia homeobox 1 |
C00002514
C00034649
|
| 55872 | PBK, CT84, Nori-3, SPK, TOPK | PDZ binding kinase (EC:2.7.12.2) |
C00002514
C00034649
|
| 57103 | C12orf5, FR2BP, TIGAR | chromosome 12 open reading frame 5 (EC:3.1.3.46) |
C00002514
C00034649
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00001429
C00002514
|
| 89927 | C16orf45 | chromosome 16 open reading frame 45 |
C00002514
C00034649
|
| 9060 | PAPSS2, ATPSK2, BCYM4, SK2 | 3'-phosphoadenosine 5'-phosphosulfate synthase 2 (EC:2.7.1.25 2.7.7.4) |
C00002514
C00034649
|
| 5021 | OXTR, OT-R | oxytocin receptor |
C00002514
C00034649
|
| 10439 | OLFM1, AMY, NOE1, NOELIN1, OlfA | olfactomedin 1 |
C00002514
C00034649
|
| 55239 | OGFOD1, TPA1 | 2-oxoglutarate and iron-dependent oxygenase domain containing 1 (EC:1.14.11.-) |
C00002514
C00034649
|
| 51203 | NUSAP1, ANKT, BM037, LNP, NUSAP, PRO0310p1, Q0310, SAPL | nucleolar and spindle associated protein 1 |
C00002514
C00034649
|
| 4907 | NT5E, CALJA, CD73, E5NT, NT, NT5, NTE, eN, eNT | 5'-nucleotidase, ecto (CD73) (EC:3.1.3.5) |
C00002514
C00034649
|
| 2908 | NR3C1, GCCR, GCR, GR, GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
C00002514
C00034649
|
| 4886 | NPY1R, NPY1-R, NPYR | neuropeptide Y receptor Y1 |
C00002514
C00034649
|
| 4836 | NMT1, NMT | N-myristoyltransferase 1 (EC:2.3.1.97) |
C00002514
C00034649
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00002514
C00034649
|
| 57415 | C3orf14 | chromosome 3 open reading frame 14 |
C00002514
C00034649
|
| 182 | JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1 | jagged 1 |
C00000094
C00002514
|
| 4783 | NFIL3, E4BP4, IL3BP1, NF-IL3A, NFIL3A | nuclear factor, interleukin 3 regulated |
C00002514
C00034649
|
| 4753 | NELL2, NRP2 | NEL-like 2 (chicken) |
C00002514
C00034649
|
| 4751 | NEK2, HsPK21, NEK2A, NLK1 | NIMA-related kinase 2 (EC:2.7.11.1) |
C00002514
C00034649
|
| 10529 | NEBL, LASP2, LNEBL | nebulette |
C00002514
C00034649
|
| 8648 | NCOA1, F-SRC-1, KAT13A, RIP160, SRC1, bHLHe42, bHLHe74 | nuclear receptor coactivator 1 (EC:2.3.1.48) |
C00002514
C00034649
|
| 64151 | NCAPG, CAPG, CHCG, NY-MEL-3 | non-SMC condensin I complex, subunit G |
C00002514
C00034649
|
| 340719 | NANOS1, NOS1, SPGF12 | nanos homolog 1 (Drosophila) |
C00002514
C00034649
|
| 26509 | MYOF, FER1L3 | myoferlin |
C00002514
C00034649
|
| 4430 | MYO1B, myr1 | myosin IB |
C00002514
C00034649
|
| 29116 | MYLIP, IDOL, MIR | myosin regulatory light chain interacting protein (EC:6.3.2.-) |
C00002514
C00034649
|
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00002514
C00034649
|
| 771 | CA12, CAXII, HsT18816 | carbonic anhydrase XII (EC:4.2.1.1) |
C00002514
C00034649
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00002514
C00034649
|
| 4602 | MYB, Cmyb, c-myb, c-myb_CDS, efg | v-myb avian myeloblastosis viral oncogene homolog |
C00002514
C00034649
|
| 64968 | MRPS6, C21orf101, MRP-S6, RPMS6, S6mt | mitochondrial ribosomal protein S6 |
C00002514
C00034649
|
| 4325 | MMP16, C8orf57, MMP-X2, MT-MMP2, MT-MMP3, MT3-MMP | matrix metallopeptidase 16 (membrane-inserted) (EC:3.4.24.-) |
C00002514
C00034649
|
| 79682 | CENPU, CENP50, CENPU50, KLIP1, MLF1IP, PBIP1 | centromere protein U |
C00002514
C00034649
|
| 2872 | MKNK2, GPRK7, MNK2 | MAP kinase interacting serine/threonine kinase 2 (EC:2.7.11.1) |
C00002514
C00034649
|
| 817 | CAMK2D, CAMKD | calcium/calmodulin-dependent protein kinase II delta (EC:2.7.11.17) |
C00002514
C00034649
|
| 55450 | CAMK2N1, PRO1489, RP11-401M16.1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
C00002514
C00034649
|
| 4288 | MKI67, KIA, MIB-1 | marker of proliferation Ki-67 |
C00002514
C00034649
|
| 4256 | MGP, MGLAP, NTI | matrix Gla protein |
C00002514
C00034649
|
| 25840 | METTL7A, AAM-B | methyltransferase like 7A |
C00002514
C00034649
|
| 57082 | CASC5, AF15Q14, CT29, D40, KNL1, PPP1R55, Spc7, hKNL-1, hSpc105 | cancer susceptibility candidate 5 |
C00002514
C00034649
|
| 10988 | METAP2, MAP2, MNPEP, p67, p67eIF2 | methionyl aminopeptidase 2 (EC:3.4.11.18) |
C00002514
C00034649
|
| 4174 | MCM5, CDC46, P1-CDC46 | minichromosome maintenance complex component 5 (EC:3.6.4.12) |
C00002514
C00034649
|
| 857 | CAV1, BSCL3, CGL3, MSTP085, PPH3, VIP21, CAV | caveolin 1, caveolae protein, 22kDa |
C00002514
C00034649
|
| 4082 | MARCKS, 80K-L, MACS, PKCSL, PRKCSL | myristoylated alanine-rich protein kinase C substrate |
C00002514
C00034649
|
| 9448 | MAP4K4, FLH21957, HGK, MEKKK4, NIK | mitogen-activated protein kinase kinase kinase kinase 4 (EC:2.7.11.1) |
C00002514
C00034649
|
| 7804 | LRP8, APOER2, HSZ75190, LRP-8, MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
C00002514
C00034649
|
| 26018 | LRIG1, LIG-1, LIG1 | leucine-rich repeats and immunoglobulin-like domains 1 |
C00002514
C00034649
|
| 4001 | LMNB1, ADLD, LMN, LMN2, LMNB | lamin B1 |
C00002514
C00034649
|
| 29995 | LMCD1 | LIM and cysteine-rich domains 1 |
C00002514
C00034649
|
| 51474 | LIMA1, EPLIN, SREBP3 | LIM domain and actin binding 1 |
C00002514
C00034649
|
| 3964 | LGALS8, Gal-8, PCTA-1, PCTA1, Po66-CBP | lectin, galactoside-binding, soluble, 8 |
C00002514
C00034649
|
| 3953 | LEPR, CD295, LEP-R, LEPRD, OB-R, OBR | leptin receptor |
C00002514
C00034649
|
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00002514
C00034649
|
| 3897 | L1CAM, CAML1, CD171, HSAS, HSAS1, MASA, MIC5, N-CAM-L1, N-CAML1, NCAM-L1, S10, SPG1 | L1 cell adhesion molecule |
C00002514
C00034649
|
| 54800 | KLHL24, DRE1, KRIP6 | kelch-like family member 24 |
C00002514
C00034649
|
| 1316 | KLF6, BCD1, CBA1, COPEB, CPBP, GBF, PAC1, ST12, ZF9 | Kruppel-like factor 6 |
C00002514
C00034649
|
| 9314 | KLF4, EZF, GKLF | Kruppel-like factor 4 (gut) |
C00002514
C00034649
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00002514
C00034649
|
| 9928 | KIF14 | kinesin family member 14 |
C00002514
C00034649
|
| 9768 | NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | KIAA0101 |
C00002514
C00034649
|
| 3728 | JUP, ARVD12, CTNNG, DP3, DPIII, PDGB, PKGB | junction plakoglobin |
C00002514
C00034649
|
| 133746 | JMY, WHDC1L3 | junction mediating and regulatory protein, p53 cofactor |
C00002514
C00034649
|
| 891 | CCNB1, CCNB | cyclin B1 |
C00002514
C00034649
|
| 9133 | CCNB2, HsT17299 | cyclin B2 |
C00002514
C00034649
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00002514
C00034649
|
| 10625 | IVNS1ABP, FLARA3, HSPC068, KLHL39, ND1, NS-1, NS1-BP, NS1BP | influenza virus NS1A binding protein |
C00002514
C00034649
|
| 3708 | ITPR1, INSP3R1, IP3R, IP3R1, SCA15, SCA16, SCA29 | inositol 1,4,5-trisphosphate receptor, type 1 |
C00002514
C00034649
|
| 3673 | ITGA2, BR, CD49B, GPIa, HPA-5, VLA-2, VLAA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
C00002514
C00034649
|
| 8660 | IRS2, IRS-2 | insulin receptor substrate 2 |
C00002514
C00034649
|
| 3667 | IRS1, HIRS-1 | insulin receptor substrate 1 |
C00002514
C00034649
|
| 100133941 | CD24, CD24A | CD24 molecule |
C00002514
C00034649
|
| 3664 | IRF6, LPS, OFC6, PIT, PPS, PPS1, VWS, VWS1 | interferon regulatory factor 6 |
C00002514
C00034649
|
| 960 | CD44, CDW44, CSPG8, ECMR-III, HCELL, HUTCH-I, IN, LHR, MC56, MDU2, MDU3, MIC4, Pgp1 | CD44 molecule (Indian blood group) |
C00002514
C00034649
|
| 128239 | IQGAP3 | IQ motif containing GTPase activating protein 3 |
C00002514
C00034649
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00002514
C00034649
|
| 8555 | CDC14B, CDC14B3, Cdc14B1, Cdc14B2, hCDC14B | cell division cycle 14B (EC:3.1.3.16 3.1.3.48) |
C00002514
C00034649
|
| 8821 | INPP4B | inositol polyphosphate-4-phosphatase, type II, 105kDa (EC:3.1.3.66) |
C00002514
C00034649
|
| 3625 | INHBB | inhibin, beta B |
C00002514
C00034649
|
| 3613 | IMPA2 | inositol(myo)-1(or 4)-monophosphatase 2 (EC:3.1.3.25) |
C00002514
C00034649
|
| 3572 | IL6ST, CD130, CDW130, GP130, IL-6RB | interleukin 6 signal transducer (gp130, oncostatin M receptor) |
C00002514
C00034649
|
| 3558 | IL2, IL-2, TCGF, lymphokine | interleukin 2 |
C00002514
C00034649
|
| 3554 | IL1R1, CD121A, D2S1473, IL-1R-alpha, IL1R, IL1RA, P80 | interleukin 1 receptor, type I |
C00002514
C00034649
|
| 990 | CDC6, CDC18L, HsCDC18, HsCDC6 | cell division cycle 6 |
C00002514
C00034649
|
| 55540 | IL17RB, CRL4, EVI27, IL17BR, IL17RH1 | interleukin 17 receptor B |
C00002514
C00034649
|
| 3488 | IGFBP5, IBP5 | insulin-like growth factor binding protein 5 |
C00002514
C00034649
|
| 83461 | CDCA3, GRCC8, TOME-1 | cell division cycle associated 3 |
C00002514
C00034649
|
| 3487 | IGFBP4, BP-4, HT29-IGFBP, IBP4, IGFBP-4 | insulin-like growth factor binding protein 4 |
C00002514
C00034649
|
| 3485 | IGFBP2, IBP2, IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa |
C00002514
C00034649
|
| 10964 | IFI44L, C1orf29 | interferon-induced protein 44-like |
C00002514
C00034649
|
| 389792 | IER5L, bA247A12.2 | immediate early response 5-like |
C00002514
C00034649
|
| 8870 | IER3, DIF-2, DIF2, GLY96, IEX-1, IEX-1L, IEX1, PRG1 | immediate early response 3 |
C00002514
C00034649
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00002514
C00034649
|
| 3422 | IDI1, IPP1, IPPI1 | isopentenyl-diphosphate delta isomerase 1 (EC:5.3.3.2) |
C00002514
C00034649
|
| 3399 | ID3, HEIR-1, bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
C00002514
C00034649
|
| 55755 | CDK5RAP2, C48, Cep215, MCPH3 | CDK5 regulatory subunit associated protein 2 |
C00002514
C00034649
|
| 90161 | HS6ST2 | heparan sulfate 6-O-sulfotransferase 2 (EC:2.8.2.-) |
C00002514
C00034649
|
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00002514
C00034649
|
| 1028 | CDKN1C, BWCR, BWS, KIP2, WBS, p57, p57Kip2 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
C00002514
C00034649
|
| 55602 | CDKN2AIP, CARF | CDKN2A interacting protein |
C00002514
C00034649
|
| 1030 | CDKN2B, CDK4I, INK4B, MTS2, P15, TP15, p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
C00002514
C00034649
|
| 1031 | CDKN2C, INK4C, p18, p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
C00002514
C00034649
|
| 1033 | CDKN3, CDI1, CIP2, KAP, KAP1 | cyclin-dependent kinase inhibitor 3 (EC:3.1.3.16 3.1.3.48) |
C00002514
C00034649
|
| 3161 | HMMR, CD168, IHABP, RHAMM | hyaluronan-mediated motility receptor (RHAMM) |
C00002514
C00034649
|
| 9324 | HMGN3, PNAS-25, TRIP7 | high mobility group nucleosomal binding domain 3 |
C00002514
C00034649
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00002514
C00034649
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00002514
C00034649
|
| 1052 | CEBPD, C/EBP-delta, CELF, CRP3, NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta |
C00002514
C00034649
|
| 3159 | HMGA1, HMG-R, HMGA1A, HMGIY | high mobility group AT-hook 1 |
C00002514
C00034649
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00002514
C00034649
|
| 55355 | HJURP, FAKTS, URLC9, hFLEG1 | Holliday junction recognition protein |
C00002514
C00034649
|
| 1063 | CENPF, CENF, PRO1779, hcp-1 | centromere protein F, 350/400kDa |
C00002514
C00034649
|
| 8346 | HIST1H2BI, H2B/k, H2BFK | histone cluster 1, H2bi |
C00002514
C00034649
|
| 8345 | HIST1H2BH, H2B/j, H2BFJ | histone cluster 1, H2bh |
C00002514
C00034649
|
| 3017 | HIST1H2BD, H2B.1B, H2B/b, H2BFB, HIRIP2, dJ221C16.6 | histone cluster 1, H2bd |
C00002514
C00034649
|
| 3006 | HIST1H1C, H1.2, H1C, H1F2, H1s-1 | histone cluster 1, H1c |
C00002514
C00034649
|
| 28996 | HIPK2, PRO0593 | homeodomain interacting protein kinase 2 (EC:2.7.11.1) |
C00002514
C00034649
|
| 55839 | CENPN, BM039, C16orf60, CENP-N, ICEN32 | centromere protein N |
C00002514
C00034649
|
| 3280 | HES1, HES-1, HHL, HRY, bHLHb39 | hes family bHLH transcription factor 1 |
C00002514
C00034649
|
| 55166 | CENPQ, C6orf139, CENP-Q | centromere protein Q |
C00002514
C00034649
|
| 3070 | HELLS, LSH, PASG, SMARCA6 | helicase, lymphoid-specific |
C00002514
C00034649
|
| 26959 | HBP1 | HMG-box transcription factor 1 |
C00002514
C00034649
|
| 3033 | HADH, HAD, HADH1, HADHSC, HCDH, HHF4, MSCHAD, SCHAD | hydroxyacyl-CoA dehydrogenase (EC:1.1.1.35) |
C00002514
C00034649
|
| 94239 | H2AFV, H2A.Z-2, H2AV | H2A histone family, member V |
C00002514
C00034649
|
| 51512 | GTSE1, B99 | G-2 and S-phase expressed 1 |
C00002514
C00034649
|
| 55165 | CEP55, C10orf3, CT111, URCC6 | centrosomal protein 55kDa |
C00002514
C00034649
|
| 9702 | CEP57, MVA2, PIG8, TSP57 | centrosomal protein 57kDa |
C00002514
C00034649
|
| 9569 | GTF2IRD1, BEN, CREAM1, GTF3, MUSTRD1, RBAP2, WBS, WBSCR11, WBSCR12, hMusTRD1alpha1 | GTF2I repeat domain containing 1 |
C00002514
C00034649
|
| 80273 | GRPEL1, HMGE | GrpE-like 1, mitochondrial (E. coli) |
C00002514
C00034649
|
| 9687 | GREB1 | growth regulation by estrogen in breast cancer 1 |
C00002514
C00034649
|
| 2887 | GRB10, GRB-IR, Grb-10, IRBP, MEG1, RSS | growth factor receptor-bound protein 10 |
C00002514
C00034649
|
| 9052 | GPRC5A, GPCR5A, RAI3, RAIG1 | G protein-coupled receptor, family C, group 5, member A |
C00002514
C00034649
|
| 2805 | GOT1, ASTQTL1, GIG18, cAspAT, cCAT | glutamic-oxaloacetic transaminase 1, soluble (EC:2.6.1.1 2.6.1.3) |
C00002514
C00034649
|
| 51053 | GMNN, Gem | geminin, DNA replication inhibitor |
C00002514
C00034649
|
| 79833 | GEMIN6 | gem (nuclear organelle) associated protein 6 |
C00002514
C00034649
|
| 2669 | GEM, KIR | GTP binding protein overexpressed in skeletal muscle |
C00002514
C00034649
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00002514
C00034649
|
| 2634 | GBP2 | guanylate binding protein 2, interferon-inducible |
C00002514
C00034649
|
| 283431 | GAS2L3, G2L3 | growth arrest-specific 2 like 3 |
C00002514
C00034649
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00002514
C00034649
|
| 1123 | CHN1, ARHGAP2, CHN, DURS2, NC, RHOGAP2 | chimerin 1 |
C00002514
C00034649
|
| 56994 | CHPT1, CPT, CPT1 | choline phosphotransferase 1 (EC:2.7.8.2) |
C00002514
C00034649
|
| 1138 | CHRNA5, LNCR2 | cholinergic receptor, nicotinic, alpha 5 (neuronal) |
C00002514
C00034649
|
| 6624 | FSCN1, FAN1, HSN, SNL, p55 | fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
C00002514
C00034649
|
| 122786 | FRMD6, C14orf31, EX1, Willin, c14_5320 | FERM domain containing 6 |
C00002514
C00034649
|
| 10370 | CITED2, ASD8, MRG-1, MRG1, P35SRJ, VSD2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
C00002514
C00034649
|
| 26586 | CKAP2, LB1, TMAP, se20-10 | cytoskeleton associated protein 2 |
C00002514
C00034649
|
| 257019 | FRMD3, 4.1O, EPB41L4O, EPB41LO, P410 | FERM domain containing 3 |
C00002514
C00034649
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00002514
C00034649
|
| 2289 | FKBP5, AIG6, FKBP51, FKBP54, P54, PPIase, Ptg-10 | FK506 binding protein 5 (EC:5.2.1.8) |
C00002514
C00034649
|
| 7431 | VIM, CTRCT30 | vimentin |
C00000094
C00034649
|
| 1163 | CKS1B, CKS1, PNAS-16, PNAS-18, ckshs1 | CDC28 protein kinase regulatory subunit 1B |
C00002514
C00034649
|
| 2274 | FHL2, AAG11, DRAL, FHL-2, SLIM-3, SLIM3 | four and a half LIM domains 2 |
C00002514
C00034649
|
| 2271 | FH, HLRCC, LRCC, MCL, MCUL1 | fumarate hydratase (EC:4.2.1.2) |
C00002514
C00034649
|
| 9076 | CLDN1, CLD1, ILVASC, SEMP1 | claudin 1 |
C00002514
C00034649
|
| 1364 | CLDN4, CPE-R, CPER, CPETR, CPETR1, WBSCR8, hCPE-R | claudin 4 |
C00002514
C00034649
|
| 2224 | FDPS, FPPS, FPS | farnesyl diphosphate synthase (EC:2.5.1.1 2.5.1.10) |
C00002514
C00034649
|
| 2222 | FDFT1, DGPT, ERG9, SQS, SS | farnesyl-diphosphate farnesyltransferase 1 (EC:2.5.1.21) |
C00002514
C00034649
|
| 55215 | FANCI, KIAA1794 | Fanconi anemia, complementation group I |
C00002514
C00034649
|
| 131583 | FAM43A | family with sequence similarity 43, member A |
C00002514
C00034649
|
| 165215 | FAM171B, KIAA1946 | family with sequence similarity 171, member B |
C00002514
C00034649
|
| 642273 | FAM110C | family with sequence similarity 110, member C |
C00002514
C00034649
|
| 3992 | FADS1, D5D, FADS6, FADSD5, LLCDL1, TU12 | fatty acid desaturase 1 (EC:1.14.19.-) |
C00002514
C00034649
|
| 2150 | F2RL1, GPR11, PAR2 | coagulation factor II (thrombin) receptor-like 1 |
C00002514
C00034649
|
| 2149 | F2R, CF2R, HTR, PAR-1, PAR1, TR | coagulation factor II (thrombin) receptor |
C00002514
C00034649
|
| 2146 | EZH2, ENX-1, ENX1, EZH1, EZH2b, KMT6, KMT6A, WVS, WVS2 | enhancer of zeste homolog 2 (Drosophila) (EC:2.1.1.43) |
C00002514
C00034649
|
| 54206 | ERRFI1, GENE-33, MIG-6, MIG6, RALT | ERBB receptor feedback inhibitor 1 |
C00002514
C00034649
|
| 8507 | ENC1, CCL28, ENC-1, KLHL35, KLHL37, NRPB, PIG10, TP53I10 | ectodermal-neural cortex 1 (with BTB domain) |
C00002514
C00034649
|
| 79071 | ELOVL6, FACE, FAE, LCE | ELOVL fatty acid elongase 6 (EC:2.3.1.199) |
C00002514
C00034649
|
| 1960 | EGR3, EGR-3, PILOT | early growth response 3 |
C00002514
C00034649
|
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00002514
C00034649
|
| 7498 | XDH, XO, XOR | xanthine dehydrogenase (EC:1.17.1.4 1.17.3.2) |
C00001429
C00034649
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00002514
C00034649
|
| 1942 | EFNA1, B61, ECKLG, EFL1, EPLG1, LERK-1, LERK1, TNFAIP4 | ephrin-A1 |
C00002514
C00034649
|
| 80303 | EFHD1, MST133, MSTP133, SWS2 | EF-hand domain family, member D1 |
C00002514
C00034649
|
| 2202 | EFEMP1, DHRD, DRAD, FBLN3, FBNL, FIBL-3, MLVT, MTLV, S1-5 | EGF containing fibulin-like extracellular matrix protein 1 |
C00002514
C00034649
|
| 1906 | EDN1, ET1, HDLCQ7, PPET1 | endothelin 1 |
C00002514
C00034649
|
| 10682 | EBP, CDPX2, CHO2, CPX, CPXD | emopamil binding protein (sterol isomerase) (EC:5.3.3.5) |
C00002514
C00034649
|
| 79733 | E2F8, E2F-8 | E2F transcription factor 8 |
C00002514
C00034649
|
| 56940 | DUSP22, JKAP, JSP-1, JSP1, LMW-DSP2, LMWDSP2, MKP-x, MKPX, VHX | dual specificity phosphatase 22 (EC:3.1.3.16 3.1.3.48) |
C00002514
C00034649
|
| 132864 | CPEB2, CPE-BP2, CPEB-2, hCPEB-2 | cytoplasmic polyadenylation element binding protein 2 |
C00002514
C00034649
|
| 1843 | DUSP1, CL100, HVH1, MKP-1, MKP1, PTPN10 | dual specificity phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00002514
C00034649
|
| 23234 | DNAJC9, HDJC9, JDD1, SB73 | DnaJ (Hsp40) homolog, subfamily C, member 9 |
C00002514
C00034649
|
| 4189 | DNAJB9, ERdj4, MDG-1, MDG1, MST049, MSTP049 | DnaJ (Hsp40) homolog, subfamily B, member 9 |
C00002514
C00034649
|
| 1746 | DLX2, TES-1, TES1 | distal-less homeobox 2 |
C00002514
C00034649
|
| 9787 | DLGAP5, DLG7, HURP | discs, large (Drosophila) homolog-associated protein 5 |
C00002514
C00034649
|
| 81624 | DIAPH3, AN, AUNA1, DIA2, DRF3, NSDAN, diap3, mDia2 | diaphanous-related formin 3 |
C00002514
C00034649
|
| 1717 | DHCR7, SLOS | 7-dehydrocholesterol reductase (EC:1.3.1.21) |
C00002514
C00034649
|
| 55789 | DEPDC1B, BRCC3, XTP1 | DEP domain containing 1B |
C00002514
C00034649
|
| 55635 | DEPDC1, DEP.8, DEPDC1-V2, DEPDC1A, SDP35 | DEP domain containing 1 |
C00002514
C00034649
|
| 10675 | CSPG5, NGC | chondroitin sulfate proteoglycan 5 (neuroglycan C) |
C00002514
C00034649
|
| 81566 | CSRNP2, C12orf2, C12orf22, FAM130A1, PPP1R72, TAIP-12 | cysteine-serine-rich nuclear protein 2 |
C00002514
C00034649
|
| 1475 | CSTA, AREI, STF1, STFA | cystatin A (stefin A) |
C00002514
C00034649
|
| 7913 | DEK, D6S231E | DEK oncogene |
C00002514
C00034649
|
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00002514
C00034649
|
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative |
C00000094
C00002514
|
| 23564 | DDAH2, DDAH, DDAHII, G6a, NG30 | dimethylarginine dimethylaminohydrolase 2 (EC:3.5.3.18) |
C00002514
C00034649
|
| 1627 | DBN1, D0S117E | drebrin 1 |
C00002514
C00034649
|
| 1595 | CYP51A1, CP51, CYP51, CYPL1, LDM, P450-14DM, P450L1 | cytochrome P450, family 51, subfamily A, polypeptide 1 (EC:1.14.13.70) |
C00002514
C00034649
|
| 26999 | CYFIP2, PIR121 | cytoplasmic FMR1 interacting protein 2 |
C00002514
C00034649
|
| 80777 | CYB5B, CYB5-M, CYPB5M, OMB5 | cytochrome b5 type B (outer mitochondrial membrane) |
C00002514
C00034649
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00002514
C00034649
|
| 1515 | CTSV, CATL2, CTSL2, CTSU | cathepsin V (EC:3.4.22.43) |
C00002514
C00034649
|
| 1512 | CTSH, ACC-4, ACC-5, CPSB, minichain | cathepsin H (EC:3.4.22.16) |
C00002514
C00034649
|
| 5443 | POMC, ACTH, CLIP, LPH, MSH, NPP, POC | proopiomelanocortin |
C00034649
|
| 1508 | CTSB, APPS, CPSB | cathepsin B (EC:3.4.22.1) |
C00002514
|
| 2017 | CTTN, EMS1 | cortactin |
C00002514
|
| 55917 | CTTNBP2NL | CTTNBP2 N-terminal like |
C00002514
|
| 9646 | CTR9, SH2BP1, TSBP, p150, p150TSP | CTR9, Paf1/RNA polymerase II complex component |
C00002514
|
| 256643 | CXorf23 | chromosome X open reading frame 23 |
C00002514
|
| 51523 | CXXC5, CF5, RINF, WID | CXXC finger protein 5 |
C00002514
|
| 51797 |
C00002514
|
||
| 51706 | CYB5R1, B5R.1, B5R1, B5R2, NQO3A2, humb5R2 | cytochrome b5 reductase 1 (EC:1.6.2.2) |
C00002514
|
| 1501 | CTNND2, GT24, NPRAP | catenin (cadherin-associated protein), delta 2 |
C00002514
|
| 120227 | CYP2R1 | cytochrome P450, family 2, subfamily R, polypeptide 1 (EC:1.14.13.159) |
C00002514
|
| 1500 | CTNND1, CAS, CTNND, P120CAS, P120CTN, p120, p120(CAS), p120(CTN) | catenin (cadherin-associated protein), delta 1 |
C00002514
|
| 3491 | CYR61, CCN1, GIG1, IGFBP10 | cysteine-rich, angiogenic inducer, 61 |
C00002514
|
| 9266 | CYTH2, ARNO, CTS18, CTS18.1, PSCD2, PSCD2L, SEC7L, Sec7p-L, Sec7p-like | cytohesin 2 |
C00002514
|
| 23002 | DAAM1 | dishevelled associated activator of morphogenesis 1 |
C00002514
|
| 26007 | DAK, NET45 | dihydroxyacetone kinase 2 homolog (S. cerevisiae) (EC:2.7.1.29 4.6.1.15 2.7.1.28) |
C00002514
|
| 57291 | DANCR, AGU2, ANCR, KIAA0114, SNHG13 | differentiation antagonizing non-protein coding RNA |
C00002514
|
| 55157 | DARS2, ASPRS, LBSL, MT-ASPRS, RP3-383J4.2 | aspartyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.12) |
C00002514
|
| 10926 | DBF4, ASK, CHIF, DBF4A, ZDBF1 | DBF4 homolog (S. cerevisiae) |
C00002514
|
| 8727 | CTNNAL1, CLLP, alpha-CATU | catenin (cadherin-associated protein), alpha-like 1 |
C00002514
|
| 90379 | DCAF15, C19orf72 | DDB1 and CUL4 associated factor 15 |
C00002514
|
| 26094 | DCAF4, WDR21, WDR21A | DDB1 and CUL4 associated factor 4 |
C00002514
|
| 8816 | DCAF5, BCRG2, BCRP2, D14S1461E, WDR22 | DDB1 and CUL4 associated factor 5 |
C00002514
|
| 100528464 |
C00002514
|
||
| 1633 | DCK | deoxycytidine kinase (EC:2.7.1.74) |
C00002514
|
| 9201 | DCLK1, CL1, CLICK1, DCAMKL1, DCDC3A, DCLK | doublecortin-like kinase 1 (EC:2.7.11.1) |
C00002514
|
| 9937 | DCLRE1A, PSO2, SNM1, SNM1A | DNA cross-link repair 1A |
C00002514
|
| 64858 | DCLRE1B, APOLLO, SNM1B, SNMIB | DNA cross-link repair 1B |
C00002514
|
| 28960 | DCPS, DCS1, HINT-5, HINT5, HSL1 | decapping enzyme, scavenger (EC:3.6.1.59) |
C00002514
|
| 79077 | DCTPP1, RS21C6, XTP3TPA | dCTP pyrophosphatase 1 (EC:3.6.1.12) |
C00002514
|
| 51496 | CTDSPL2, HSPC129 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 (EC:3.1.3.-) |
C00002514
|
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00002514
|
| 326709 |
C00002514
|
||
| 1478 | CSTF2, CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
C00002514
|
| 1663 | DDX11, CHL1, CHLR1, KRG2, WABS | DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 (EC:3.6.4.13) |
C00002514
|
| 68278 |
C00002514
|
||
| 54606 | DDX56, DDX21, DDX26, NOH61 | DEAD (Asp-Glu-Ala-Asp) box helicase 56 (EC:3.6.4.13) |
C00002514
|
| 1477 | CSTF1, CstF-50, CstFp50 | cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
C00002514
|
| 57706 | DENND1A, FAM31A, KIAA1608, RP11-230L22.3 | DENN/MADD domain containing 1A |
C00002514
|
| 163486 | DENND1B, C1ORF18, C1orf218, FAM31B | DENN/MADD domain containing 1B |
C00002514
|
| 79958 | DENND1C, FAM31C | DENN/MADD domain containing 1C |
C00002514
|
| 8562 | DENR, DRP, DRP1, SMAP-3 | density-regulated protein |
C00002514
|
| 1459 | CSNK2A2, CK2A2, CSNK2A1 | casein kinase 2, alpha prime polypeptide (EC:2.7.11.1) |
C00002514
|
| 1456 | CSNK1G3, CKI-gamma_3, CSNK1G3L | casein kinase 1, gamma 3 (EC:2.7.11.1) |
C00002514
|
| 420357 |
C00002514
|
||
| 51071 | DERA, DEOC | deoxyribose-phosphate aldolase (putative) (EC:4.1.2.4) |
C00002514
|
| 51029 | DESI2, C1orf121, DESI, DESI1, DeSI-2, FAM152A, PNAS-4, PPPDE1 | desumoylating isopeptidase 2 |
C00002514
|
| 53944 | CSNK1G1, CK1gamma1 | casein kinase 1, gamma 1 (EC:2.7.11.1) |
C00002514
|
| 1719 | DHFR, DHFRP1, DYR | dihydrofolate reductase (EC:1.5.1.3) |
C00002514
|
| 10202 | DHRS2, HEP27, SDR25C1 | dehydrogenase/reductase (SDR family) member 2 |
C00002514
|
| 51635 | DHRS7, SDR34C1, retDSR4, retSDR4 | dehydrogenase/reductase (SDR family) member 7 |
C00002514
|
| 55526 | DHTKD1, AMOXAD, CMT2Q | dehydrogenase E1 and transketolase domain containing 1 (EC:1.2.4.2) |
C00002514
|
| 1452 | CSNK1A1, CK1, CK1a, CKIa, HLCDGP1, PRO2975 | casein kinase 1, alpha 1 (EC:2.7.11.1) |
C00002514
|
| 23405 | DICER1, DCR1, Dicer, HERNA, MNG1, RMSE2 | dicer 1, ribonuclease type III (EC:3.1.26.-) |
C00002514
|
| 22982 | DIP2C, KIAA0934 | DIP2 disco-interacting protein 2 homolog C (Drosophila) |
C00002514
|
| 85458 | DIXDC1, CCD1 | DIX domain containing 1 |
C00002514
|
| 1736 | DKC1, CBF5, DKC, DKCX, NAP57, NOLA4, XAP101 | dyskeratosis congenita 1, dyskerin |
C00002514
|
| 1737 | DLAT, DLTA, PDC-E2, PDCE2 | dihydrolipoamide S-acetyltransferase (EC:2.3.1.12) |
C00002514
|
| 79469 | DLEU2L, BCMSUNL | deleted in lymphocytic leukemia 2-like |
C00002514
|
| 1434 | CSE1L, CAS, CSE1, XPO2 | CSE1 chromosome segregation 1-like (yeast) |
C00002514
|
| 1407 | CRY1, PHLL1 | cryptochrome 1 (photolyase-like) |
C00002514
|
| 1657 | DMXL1 | Dmx-like 1 |
C00002514
|
| 1763 | DNA2, DNA2L, PEOA6, hDNA2 | DNA replication helicase/nuclease 2 (EC:3.6.4.12) |
C00002514
|
| 10294 | DNAJA2, CPR3, DJ3, DJA2, DNAJ, DNJ3, HIRIP4, PRO3015, RDJ2 | DnaJ (Hsp40) homolog, subfamily A, member 2 |
C00002514
|
| 1397 | CRIP2, CRIP, CRP2, ESP1 | cysteine-rich protein 2 |
C00002514
|
| 64215 | DNAJC1, DNAJL1, ERdj1, HTJ1, MTJ1 | DnaJ (Hsp40) homolog, subfamily C, member 1 |
C00002514
|
| 23341 | DNAJC16 | DnaJ (Hsp40) homolog, subfamily C, member 16 |
C00002514
|
| 148327 | CREB3L4, AIBZIP, ATCE1, CREB3, CREB4, JAL, hJAL | cAMP responsive element binding protein 3-like 4 |
C00002514
|
| 1777 | DNASE2, DNASE2A, DNL, DNL2 | deoxyribonuclease II, lysosomal (EC:3.1.22.1) |
C00002514
|
| 10059 | DNM1L, DLP1, DRP1, DVLP, DYMPLE, EMPF, HDYNIV | dynamin 1-like (EC:3.6.5.5) |
C00002514
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00002514
|
| 81704 | DOCK8, MRD2, ZIR8 | dedicator of cytokinesis 8 |
C00002514
|
| 29980 | DONSON, B17, C21orf60 | downstream neighbor of SON |
C00002514
|
| 9980 | DOPEY2, 21orf5, C21orf5 | dopey family member 2 |
C00002514
|
| 1798 | DPAGT1, ALG7, CDG-Ij, CDG1J, CMSTA2, D11S366, DGPT, DPAGT, DPAGT2, G1PT, GPT, UAGT, UGAT | dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) (EC:2.7.8.15) |
C00002514
|
| 8818 | DPM2, CDG1U | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (EC:2.4.1.83) |
C00002514
|
| 23333 | DPY19L1 | dpy-19-like 1 (C. elegans) |
C00002514
|
| 1824 | DSC2, ARVD11, CDHF2, DG2, DGII/III, DSC3 | desmocollin 2 |
C00002514
|
| 380353 |
C00002514
|
||
| 79980 | DSN1, C20orf172, KNL3, MIS13, dJ469A13.2, hKNL-3 | DSN1, MIS12 kinetochore complex component |
C00002514
|
| 1837 | DTNA, D18S892E, DRP3, DTN, DTN-A, LVNC1 | dystrobrevin, alpha |
C00002514
|
| 51692 | CPSF3, CPSF-73, CPSF73 | cleavage and polyadenylation specific factor 3, 73kDa |
C00002514
|
| 11221 | DUSP10, MKP-5, MKP5 | dual specificity phosphatase 10 (EC:3.1.3.16 3.1.3.48) |
C00002514
|
| 1363 | CPE | carboxypeptidase E (EC:3.4.17.10) |
C00002514
|
| 1854 | DUT, dUTPase | deoxyuridine triphosphatase (EC:3.6.1.23) |
C00002514
|
| 51143 | DYNC1LI1, DNCLI1, LIC1 | dynein, cytoplasmic 1, light intermediate chain 1 |
C00002514
|
| 6990 | DYNLT3, RP3, TCTE1L, TCTEX1L | dynein, light chain, Tctex-type 3 |
C00002514
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002514
|
| 1870 | E2F2, E2F-2 | E2F transcription factor 2 |
C00002514
|
| 1875 | E2F5, E2F-5 | E2F transcription factor 5, p130-binding |
C00002514
|
| 144455 | E2F7 | E2F transcription factor 7 |
C00002514
|
| 33136 |
C00002514
|
||
| 7464 | CORO2A, CLIPINB, IR10, WDR2 | coronin, actin binding protein, 2A |
C00002514
|
| 1894 | ECT2, ARHGEF31 | epithelial cell transforming sequence 2 oncogene |
C00002514
|
| 80267 | EDEM3, C1orf22 | ER degradation enhancer, mannosidase alpha-like 3 (EC:3.2.1.113) |
C00002514
|
| 10085 | EDIL3, DEL1 | EGF-like repeats and discoidin I-like domains 3 |
C00002514
|
| 23603 | CORO1C, HCRNN4 | coronin, actin binding protein, 1C |
C00002514
|
| 8726 | EED, HEED, WAIT1 | embryonic ectoderm development |
C00002514
|
| 1915 | EEF1A1, CCS-3, CCS3, EE1A1, EEF-1, EEF1A, EF-Tu, EF1A, GRAF-1EF, HNGC:16303, LENG7, PTI1, eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 |
C00002514
|
| 57175 | CORO1B, CORONIN-2 | coronin, actin binding protein, 1B |
C00002514
|
| 10229 | COQ7, CAT5, CLK-1, CLK1 | coenzyme Q7 homolog, ubiquinone (yeast) |
C00002514
|
| 27235 | COQ2, CL640, COQ10D1, MSA1 | coenzyme Q2 4-hydroxybenzoate polyprenyltransferase (EC:2.5.1.39) |
C00002514
|
| 9318 | COPS2, ALIEN, CSN2, SGN2, TRIP15 | COP9 signalosome subunit 2 |
C00002514
|
| 1949 | EFNB3, EFL6, EPLG8, LERK8 | ephrin-B3 |
C00002514
|
| 9343 | EFTUD2, MFDGA, MFDM, SNRNP116, Snrp116, Snu114, U5-116KD | elongation factor Tu GTP binding domain containing 2 |
C00002514
|
| 54939 | COMMD4 | COMM domain containing 4 |
C00002514
|
| 81578 | COL21A1, COLA1L, dJ682J15.1, dJ708F5.1 | collagen, type XXI, alpha 1 |
C00002514
|
| 79813 | EHMT1, EUHMTASE1, Eu-HMTase1, FP13812, GLP, GLP1, KMT1D, bA188C12.1 | euchromatic histone-lysine N-methyltransferase 1 (EC:2.1.1.43) |
C00002514
|
| 9451 | EIF2AK3, PEK, PERK, WRS | eukaryotic translation initiation factor 2-alpha kinase 3 (EC:2.7.11.1) |
C00002514
|
| 1965 | EIF2S1, EIF-2, EIF-2A, EIF-2alpha, EIF2, EIF2A | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
C00002514
|
| 1979 | EIF4EBP2, 4EBP2, PHASII | eukaryotic translation initiation factor 4E binding protein 2 |
C00002514
|
| 8672 | EIF4G3, eIF-4G_3, eIF4G_3, eIF4GII | eukaryotic translation initiation factor 4 gamma, 3 |
C00002514
|
| 2001 | ELF5, ESE2 | E74-like factor 5 (ets domain transcription factor) |
C00002514
|
| 2002 | ELK1 | ELK1, member of ETS oncogene family |
C00002514
|
| 2004 | ELK3, ERP, NET, SAP2 | ELK3, ETS-domain protein (SRF accessory protein 2) |
C00002514
|
| 63916 | ELMO2, CED-12, CED12, ELMO-2 | engulfment and cell motility 2 |
C00002514
|
| 54898 | ELOVL2, SSC2 | ELOVL fatty acid elongase 2 (EC:2.3.1.199) |
C00002514
|
| 60481 | ELOVL5, HELO1, dJ483K16.1 | ELOVL fatty acid elongase 5 (EC:2.3.1.199) |
C00002514
|
| 57511 | COG6, CDG2L, COD2, SHNS | component of oligomeric golgi complex 6 |
C00002514
|
| 79993 | ELOVL7 | ELOVL fatty acid elongase 7 (EC:2.3.1.199) |
C00002514
|
| 23587 | ELP5, C17orf81, DERP6, MST071, MSTP071 | elongator acetyltransferase complex subunit 5 |
C00002514
|
| 54859 | ELP6, C3orf75, TMEM103 | elongator acetyltransferase complex subunit 6 |
C00002514
|
| 51016 | EMC9, C14orf122, FAM158A | ER membrane protein complex subunit 9 |
C00002514
|
| 2010 | EMD, EDMD, LEMD5, STA | emerin |
C00002514
|
| 146956 | EME1, MMS4L, SLX2A | essential meiotic structure-specific endonuclease 1 |
C00002514
|
| 10466 | COG5, CDG2I, GOLTC1, GTC90 | component of oligomeric golgi complex 5 |
C00002514
|
| 23052 | ENDOD1 | endonuclease domain containing 1 |
C00002514
|
| 55556 | ENOSF1, HSRTSBETA, RTS, TYMSAS | enolase superfamily member 1 |
C00002514
|
| 954 | ENTPD2, CD39L1, NTPDase-2 | ectonucleoside triphosphate diphosphohydrolase 2 (EC:3.6.1.-) |
C00002514
|
| 9583 | ENTPD4, LALP70, LAP70, LYSAL1, NTPDase-4, UDPase | ectonucleoside triphosphate diphosphohydrolase 4 (EC:3.6.1.6) |
C00002514
|
| 2037 | EPB41L2, 4.1-G, 4.1G | erythrocyte membrane protein band 4.1-like 2 |
C00002514
|
| 57669 | EPB41L5, BE37, YMO1 | erythrocyte membrane protein band 4.1 like 5 |
C00002514
|
| 2043 | EPHA4, HEK8, SEK, TYRO1 | EPH receptor A4 (EC:2.7.10.1) |
C00002514
|
| 58513 | EPS15L1, EPS15R | epidermal growth factor receptor pathway substrate 15-like 1 |
C00002514
|
| 2064 | ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU, NGL, TKR1 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (EC:2.7.10.1) |
C00002514
|
| 2065 | ERBB3, ErbB-3, HER3, LCCS2, MDA-BF-1, c-erbB-3, c-erbB3, erbB3-S, p180-ErbB3, p45-sErbB3, p85-sErbB3 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 (EC:2.7.10.1) |
C00002514
|
| 2066 | ERBB4, ALS19, HER4, p180erbB4 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 (EC:2.7.10.1) |
C00002514
|
| 54821 | ERCC6L, PICH, RAD26L | excision repair cross-complementing rodent repair deficiency, complementation group 6-like (EC:3.6.4.12) |
C00002514
|
| 112479 | ERI2, EXOD1 | ERI1 exoribonuclease family member 2 (EC:3.1.-.-) |
C00002514
|
| 25839 | COG4, CDG2J, COD1 | component of oligomeric golgi complex 4 |
C00002514
|
| 157570 | ESCO2, 2410004I17Rik, EFO2, RBS | establishment of sister chromatid cohesion N-acetyltransferase 2 (EC:2.3.1.-) |
C00002514
|
| 9700 | ESPL1, ESP1, SEPA | extra spindle pole bodies homolog 1 (S. cerevisiae) (EC:3.4.22.49) |
C00002514
|
| 57488 | ESYT2, CHR2SYT, E-Syt2, FAM62B | extended synaptotagmin-like protein 2 |
C00002514
|
| 55500 | ETNK1, EKI, EKI_1, EKI1, Nbla10396 | ethanolamine kinase 1 (EC:2.7.1.82) |
C00002514
|
| 7813 | EVI5, NB4S | ecotropic viral integration site 5 |
C00002514
|
| 9156 | EXO1, HEX1, hExoI | exonuclease 1 |
C00002514
|
| 55770 | EXOC2, SEC5, SEC5L1, Sec5p | exocyst complex component 2 |
C00002514
|
| 23404 | EXOSC2, RRP4, Rrp4p, hRrp4p, p7 | exosome component 2 |
C00002514
|
| 51010 | EXOSC3, PCH1B, RP11-3J10.8, RRP40, Rrp40p, bA3J10.7, hRrp-40, p10 | exosome component 3 |
C00002514
|
| 11340 | EXOSC8, CIP3, EAP2, OIP2, RP11-421P11.3, RRP43, Rrp43p, bA421P11.3, p9 | exosome component 8 (EC:3.1.13.-) |
C00002514
|
| 5393 | EXOSC9, PM/Scl-75, PMSCL1, RRP45, Rrp45p, p5, p6 | exosome component 9 (EC:3.1.13.-) |
C00002514
|
| 2131 | EXT1, EXT, LGCR, LGS, TRPS2, TTV | exostosin glycosyltransferase 1 (EC:2.4.1.225 2.4.1.224) |
C00002514
|
| 1690 | COCH, COCH-5B2, COCH5B2, DFNA9, DFNA31 | cochlin |
C00002514
|
| 7430 | EZR, CVIL, CVL, VIL2 | ezrin |
C00002514
|
| 23242 | COBL | cordon-bleu WH2 repeat protein |
C00002514
|
| 55744 | COA1, C7orf44, MITRAC15 | cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
C00002514
|
| 2171 | FABP5, E-FABP, EFABP, KFABP, PA-FABP, PAFABP | fatty acid binding protein 5 (psoriasis-associated) |
C00002514
|
| 152189 | CMTM8, CKLFSF8, CKLFSF8-V2 | CKLF-like MARVEL transmembrane domain containing 8 |
C00002514
|
| 2184 | FAH | fumarylacetoacetate hydrolase (fumarylacetoacetase) (EC:3.7.1.2) |
C00002514
|
| 9214 | FAIM3, FCMR, TOSO | Fas apoptotic inhibitory molecule 3 |
C00002514
|
| 83641 | FAM107B, C10orf45 | family with sequence similarity 107, member B |
C00002514
|
| 90362 | FAM110B, C8orf72 | family with sequence similarity 110, member B |
C00002514
|
| 84319 | CMSS1, C3orf26 | cms1 ribosomal small subunit homolog (yeast) |
C00002514
|
| 63901 | FAM111A, GCLEB, KCS2 | family with sequence similarity 111, member A |
C00002514
|
| 374393 | FAM111B, CANP | family with sequence similarity 111, member B |
C00002514
|
| 159090 | FAM122B, SPACIA2 | family with sequence similarity 122B |
C00002514
|
| 285172 | FAM126B, HYCC2 | family with sequence similarity 126, member B |
C00002514
|
| 317662 | FAM149B1, KIAA0974 | family with sequence similarity 149, member B1 |
C00002514
|
| 145483 | FAM161B, C14orf44, c14_5547 | family with sequence similarity 161, member B |
C00002514
|
| 26049 | FAM169A, SLAP75 | family with sequence similarity 169, member A |
C00002514
|
| 56942 | CMC2, 2310061C15Rik, C16orf61 | C-x(9)-C motif containing 2 |
C00002514
|
| 400451 | FAM174B | family with sequence similarity 174, member B |
C00002514
|
| 55719 | FAM178A, C10orf6 | family with sequence similarity 178, member A |
C00002514
|
| 9413 | FAM189A2, C9orf61, RP11-548B3.1, X123 | family with sequence similarity 189, member A2 |
C00002514
|
| 116151 | FAM210B, 5A3, C20orf108, dJ1167H4.1 | family with sequence similarity 210, member B |
C00002514
|
| 56204 | FAM214A, KIAA1370 | family with sequence similarity 214, member A |
C00002514
|
| 29902 | FAM216A, C12orf24, HSU79274 | family with sequence similarity 216, member A |
C00002514
|
| 55731 | FAM222B, C17orf63 | family with sequence similarity 222, member B |
C00002514
|
| 1191 | CLU, APO-J, APOJ, CLI, CLU1, CLU2, KUB1, NA1/NA2, SGP-2, SGP2, SP-40, TRPM-2, TRPM2 | clusterin |
C00002514
|
| 54855 | FAM46C | family with sequence similarity 46, member C |
C00002514
|
| 51571 | FAM49B, L1 | family with sequence similarity 49, member B |
C00002514
|
| 79850 | FAM57A, CT120 | family with sequence similarity 57, member A |
C00002514
|
| 54478 | FAM64A, CATS, RCS1 | family with sequence similarity 64, member A |
C00002514
|
| 2175 | FANCA, FA, FA-H, FA1, FAA, FACA, FAH, FANCH | Fanconi anemia, complementation group A |
C00002514
|
| 2177 | FANCD2, FA-D2, FA4, FACD, FAD, FAD2, FANCD | Fanconi anemia, complementation group D2 |
C00002514
|
| 2189 | FANCG, FAG, XRCC9 | Fanconi anemia, complementation group G |
C00002514
|
| 2055 | CLN8, C8orf61, EPMR | ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) |
C00002514
|
| 55120 | FANCL, FAAP43, PHF9, POG | Fanconi anemia, complementation group L |
C00002514
|
| 10056 | FARSB, FARSLB, FRSB, PheHB, PheRS | phenylalanyl-tRNA synthetase, beta subunit (EC:6.1.1.20) |
C00002514
|
| 54751 | FBLIM1, CAL, FBLP-1, FBLP1 | filamin binding LIM protein 1 |
C00002514
|
| 26234 | FBXL5, FBL4, FBL5, FLR1 | F-box and leucine-rich repeat protein 5 |
C00002514
|
| 81545 | FBXO38, Fbx38, MOKA | F-box protein 38 |
C00002514
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00002514
|
| 51077 | FCF1, Bka, C14orf111, UTP24 | FCF1 rRNA-processing protein |
C00002514
|
| 54982 | CLN6, CLN4A, HsT18960, nclf | ceroid-lipofuscinosis, neuronal 6, late infantile, variant |
C00002514
|
| 6249 | CLIP1, CLIP, CLIP-170, CLIP170, CYLN1, RSN | CAP-GLY domain containing linker protein 1 |
C00002514
|
| 2237 | FEN1, FEN-1, MF1, RAD2 | flap structure-specific endonuclease 1 |
C00002514
|
| 2260 | FGFR1, BFGFR, CD331, CEK, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 | fibroblast growth factor receptor 1 (EC:2.7.10.1) |
C00002514
|
| 23155 | CLCC1, MCLC | chloride channel CLIC-like 1 |
C00002514
|
| 2273 | FHL1, FHL-1, FHL1A, FHL1B, FLH1A, KYOT, SLIM, SLIM-1, SLIM1, SLIMMER, XMPMA | four and a half LIM domains 1 |
C00002514
|
| 1164 | CKS2, CKSHS2 | CDC28 protein kinase regulatory subunit 2 |
C00002514
|
| 9158 | FIBP, FGFIBP, FIBP-1 | fibroblast growth factor (acidic) intracellular binding protein |
C00002514
|
| 63979 | FIGNL1 | fidgetin-like 1 |
C00002514
|
| 51192 | CKLF, C32, CKLF1, CKLF2, CKLF3, CKLF4, UCK-1 | chemokine-like factor |
C00002514
|
| 51303 | FKBP11, FKBP19 | FK506 binding protein 11, 19 kDa (EC:5.2.1.8) |
C00002514
|
| 2288 | FKBP4, FKBP51, FKBP52, FKBP59, HBI, Hsp56, PPIase, p52 | FK506 binding protein 4, 59kDa (EC:5.2.1.8) |
C00002514
|
| 9793 | CKAP5, CHTOG, MSPS, TOG, TOGp, ch-TOG | cytoskeleton associated protein 5 |
C00002514
|
| 63943 | FKBPL, DIR1, NG7, WISP39 | FK506 binding protein like |
C00002514
|
| 201163 | FLCN, BHD, FLCL | folliculin |
C00002514
|
| 114793 | FMNL2, FHOD2 | formin-like 2 |
C00002514
|
| 23048 | FNBP1, FBP17 | formin binding protein 1 |
C00002514
|
| 54874 | FNBP1L, C1orf39, TOCA1 | formin binding protein 1-like |
C00002514
|
| 64778 | FNDC3B, FAD104, PRO4979, YVTM2421 | fibronectin type III domain containing 3B |
C00002514
|
| 10970 | CKAP4, CLIMP-63, ERGIC-63, p63 | cytoskeleton-associated protein 4 |
C00002514
|
| 2297 | FOXD1, FKHL8, FREAC4 | forkhead box D1 |
C00002514
|
| 2305 | FOXM1, FKHL16, FOXM1B, HFH-11, HFH11, HNF-3, INS-1, MPHOSPH2, MPP-2, MPP2, PIG29, TGT3, TRIDENT | forkhead box M1 |
C00002514
|
| 494532 |
C00002514
|
||
| 93986 | FOXP2, CAGH44, SPCH1, TNRC10 | forkhead box P2 |
C00002514
|
| 80020 | FOXRED2, ERFAD | FAD-dependent oxidoreductase domain containing 2 |
C00002514
|
| 150468 | CKAP2L | cytoskeleton associated protein 2-like |
C00002514
|
| 11113 | CIT, CRIK, STK21 | citron (rho-interacting, serine/threonine kinase 21) (EC:2.7.11.1) |
C00002514
|
| 55847 | CISD1, C10orf70, ZCD1, mitoNEET | CDGSH iron sulfur domain 1 |
C00002514
|
| 24140 | FTSJ1, CDLIV, MRX44, MRX9, SPB1, TRMT7 | FtsJ RNA methyltransferase homolog 1 (E. coli) (EC:2.1.1.205) |
C00002514
|
| 29960 | FTSJ2, FJH1, MRM2 | FtsJ RNA methyltransferase homolog 2 (E. coli) (EC:2.1.1.-) |
C00002514
|
| 2517 | FUCA1, FUCA | fucosidase, alpha-L- 1, tissue (EC:3.2.1.51) |
C00002514
|
| 79443 | FYCO1, CATC2, CTRCT18, RUFY3, ZFYVE7 | FYVE and coiled-coil domain containing 1 |
C00002514
|
| 2535 | FZD2, Fz2, fz-2, fzE2, hFz2 | frizzled family receptor 2 |
C00002514
|
| 55632 | G2E3, KIAA1333, PHF7B | G2/M-phase specific E3 ubiquitin protein ligase |
C00002514
|
| 9908 | G3BP2 | GTPase activating protein (SH3 domain) binding protein 2 |
C00002514
|
| 126626 | GABPB2 | GA binding protein transcription factor, beta subunit 2 |
C00002514
|
| 555843 |
C00002514
|
||
| 90480 | GADD45GIP1, CKBBP2, CRIF1, MRP-L59, PLINP, PLINP-1, PRG6, Plinp1 | growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
C00002514
|
| 51083 | GAL, GAL-GMAP, GALN, GLNN, GMAP | galanin/GMAP prepropeptide |
C00002514
|
| 2588 | GALNS, GALNAC6S, GAS, GalN6S, MPS4A | galactosamine (N-acetyl)-6-sulfate sulfatase (EC:3.1.6.4) |
C00002514
|
| 2589 | GALNT1, GALNAC-T1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (EC:2.4.1.41) |
C00002514
|
| 55568 | GALNT10, GALNACT10, PPGALNACT10, PPGANTASE10 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) (EC:2.4.1.41) |
C00002514
|
| 51809 | GALNT7, GALNAC-T7, GalNAcT7 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (EC:2.4.1.41) |
C00002514
|
| 23193 | GANAB, G2AN, GLUII | glucosidase, alpha; neutral AB (EC:3.2.1.84) |
C00002514
|
| 2618 | GART, AIRS, GARS, GARTF, PAIS, PGFT, PRGS | phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase (EC:2.1.2.2 6.3.3.1 6.3.4.13) |
C00002514
|
| 11200 | CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53, hCds1 | checkpoint kinase 2 (EC:2.7.11.1) |
C00002514
|
| 2624 | GATA2, DCML, MONOMAC, NFE1B | GATA binding protein 2 |
C00002514
|
| 2625 | GATA3, HDR, HDRS | GATA binding protein 3 |
C00002514
|
| 57798 | GATAD1, CMD2B, ODAG, RG083M05.2 | GATA zinc finger domain containing 1 |
C00002514
|
| 1111 | CHEK1, CHK1 | checkpoint kinase 1 (EC:2.7.11.1) |
C00002514
|
| 2644 | GCHFR, GFRP, HsT16933, P35 | GTP cyclohydrolase I feedback regulator |
C00002514
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00002514
|
| 84181 | CHD6, CHD-6, CHD5, RIGB | chromodomain helicase DNA binding protein 6 (EC:3.6.4.12) |
C00002514
|
| 79153 | GDPD3 | glycerophosphodiester phosphodiesterase domain containing 3 |
C00002514
|
| 1106 | CHD2, EEOC | chromodomain helicase DNA binding protein 2 (EC:3.6.4.12) |
C00002514
|
| 8487 | GEMIN2, SIP1, SIP1-delta | gem (nuclear organelle) associated protein 2 |
C00002514
|
| 50628 | GEMIN4, HC56, HCAP1, HHRF-1, p97 | gem (nuclear organelle) associated protein 4 |
C00002514
|
| 8208 | CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSPH7, MPP7 | chromatin assembly factor 1, subunit B (p60) |
C00002514
|
| 2674 | GFRA1, GDNFR, GDNFRA, GFR-ALPHA-1, RET1L, RETL1, TRNR1 | GDNF family receptor alpha 1 |
C00002514
|
| 23062 | GGA2, VEAR | golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
C00002514
|
| 79017 | GGCT, C7orf24, CRF21, GCTG, GGC | gamma-glutamylcyclotransferase (EC:2.3.2.4) |
C00002514
|
| 8836 | GGH, GH | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (EC:3.4.19.9) |
C00002514
|
| 9837 | GINS1, PSF1 | GINS complex subunit 1 (Psf1 homolog) |
C00002514
|
| 51659 | GINS2, HSPC037, PSF2, Pfs2 | GINS complex subunit 2 (Psf2 homolog) |
C00002514
|
| 64785 | GINS3, PSF3 | GINS complex subunit 3 (Psf3 homolog) |
C00002514
|
| 84296 | GINS4, SLD5 | GINS complex subunit 4 (Sld5 homolog) |
C00002514
|
| 2717 | GLA, GALA | galactosidase, alpha (EC:3.2.1.22) |
C00002514
|
| 2733 | GLE1, GLE1L, LCCS, LCCS1, hGLE1 | GLE1 RNA export mediator |
C00002514
|
| 11146 | GLMN, FAP, FAP48, FAP68, FKBPAP, GLML, GVM, VMGLOM | glomulin, FKBP associated protein |
C00002514
|
| 2744 | GLS, AAD20, GAC, GAM, GLS1, KGA | glutaminase (EC:3.5.1.2) |
C00002514
|
| 10036 | CHAF1A, CAF-1, CAF1, CAF1B, CAF1P150, P150 | chromatin assembly factor 1, subunit A (p150) |
C00002514
|
| 8833 | GMPS | guanine monphosphate synthase (EC:6.3.5.2) |
C00002514
|
| 2770 | GNAI1, Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
C00002514
|
| 2776 | GNAQ, CMC1, G-ALPHA-q, GAQ, SWS | guanine nucleotide binding protein (G protein), q polypeptide |
C00002514
|
| 54552 | GNL3L | guanine nucleotide binding protein-like 3 (nucleolar)-like |
C00002514
|
| 2799 | GNS, G6S | glucosamine (N-acetyl)-6-sulfatase (EC:3.1.6.14) |
C00002514
|
| 2801 | GOLGA2, GM130 | golgin A2 |
C00002514
|
| 494143 | CHAC2 | ChaC, cation transport regulator homolog 2 (E. coli) |
C00002514
|
| 8733 | GPAA1, GAA1, hGAA1 | glycosylphosphatidylinositol anchor attachment 1 |
C00002514
|
| 253635 | GPATCH11, CCDC75, CENP-Y, CENPY | G patch domain containing 11 |
C00002514
|
| 55668 | GPATCH2L, C14orf118 | G patch domain containing 2-like |
C00002514
|
| 54865 | GPATCH4, GPATC4 | G patch domain containing 4 |
C00002514
|
| 2820 | GPD2, GDH2, GPDM, mGPDH | glycerol-3-phosphate dehydrogenase 2 (mitochondrial) (EC:1.1.5.3) |
C00002514
|
| 2852 | GPER1, CEPR, CMKRL2, DRY12, FEG-1, GPCR-Br, GPER, GPR30, LERGU, LERGU2, LyGPR | G protein-coupled estrogen receptor 1 |
C00002514
|
| 7107 | GPR137B, TM7SF1 | G protein-coupled receptor 137B |
C00002514
|
| 26996 | GPR160, GPCR1, GPCR150 | G protein-coupled receptor 160 |
C00002514
|
| 57530 | CGN | cingulin |
C00002514
|
| 55890 | GPRC5C, RAIG-3, RAIG3 | G protein-coupled receptor, family C, group 5, member C |
C00002514
|
| 29899 | GPSM2, CMCS, DFNB82, LGN, PINS | G-protein signaling modulator 2 |
C00002514
|
| 380153 |
C00002514
|
||
| 493869 | GPX8, EPLA847, GPx-8, GSHPx-8, UNQ847 | glutathione peroxidase 8 (putative) (EC:1.11.1.9) |
C00002514
|
| 57655 | GRAMD1A, KIAA1533 | GRAM domain containing 1A |
C00002514
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00002514
|
| 64793 | CEP85, CCDC21 | centrosomal protein 85kDa |
C00002514
|
| 57822 | GRHL3, SOM, TFCP2L4 | grainyhead-like 3 (Drosophila) |
C00002514
|
| 2870 | GRK6, GPRK6 | G protein-coupled receptor kinase 6 (EC:2.7.11.16) |
C00002514
|
| 2896 | GRN, CLN11, GEP, GP88, PCDGF, PEPI, PGRN | granulin |
C00002514
|
| 84131 | CEP78, C9orf81, IP63 | centrosomal protein 78kDa |
C00002514
|
| 83903 | GSG2, HASPIN | germ cell associated 2 (haspin) (EC:2.7.11.1) |
C00002514
|
| 2968 | GTF2H4, P52, TFB2, TFIIH | general transcription factor IIH, polypeptide 4, 52kDa |
C00002514
|
| 285753 | CEP57L1, C6orf182, bA487F23.2, cep57R | centrosomal protein 57kDa-like 1 |
C00002514
|
| 100196261 |
C00002514
|
||
| 51454 | GULP1, CED-6, CED6, GULP | GULP, engulfment adaptor PTB domain containing 1 |
C00002514
|
| 55125 | CEP192, PPP1R62 | centrosomal protein 192kDa |
C00002514
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00002514
|
| 3015 | H2AFZ, H2A.Z-1, H2A.z, H2A/z, H2AZ | H2A histone family, member Z |
C00002514
|
| 3021 | H3F3B, H3.3B | H3 histone, family 3B (H3.3B) |
C00002514
|
| 22995 | CEP152, MCPH4, MCPH9, SCKL5 | centrosomal protein 152kDa |
C00002514
|
| 8520 | HAT1, KAT1 | histone acetyltransferase 1 (EC:2.3.1.48) |
C00002514
|
| 54930 | HAUS4, C14orf94 | HAUS augmin-like complex, subunit 4 |
C00002514
|
| 55559 | HAUS7, UCHL5IP, UIP1 | HAUS augmin-like complex, subunit 7 |
C00002514
|
| 93323 | HAUS8, DGT4, HICE1, NY-SAR-48 | HAUS augmin-like complex, subunit 8 |
C00002514
|
| 145508 | CEP128, C14orf145, C14orf61, LEDP/132 | centrosomal protein 128kDa |
C00002514
|
| 50865 | HEBP1, HBP, HEBP | heme binding protein 1 |
C00002514
|
| 79654 | HECTD3, RP11-69J16.1 | HECT domain containing E3 ubiquitin protein ligase 3 |
C00002514
|
| 387103 | CENPW, C6orf173, CENP-W, CUG2 | centromere protein W |
C00002514
|
| 85441 | HELZ2, PDIP-1, PRIC285 | helicase with zinc finger 2, transcriptional coactivator |
C00002514
|
| 8925 | HERC1, p532, p619 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
C00002514
|
| 64224 | HERPUD2 | HERPUD family member 2 |
C00002514
|
| 79172 | CENPO, CENP-O, ICEN-36, MCM21R | centromere protein O |
C00002514
|
| 204851 | HIPK1, Myak, Nbak2 | homeodomain interacting protein kinase 1 (EC:2.7.11.1) |
C00002514
|
| 79019 | CENPM, C22orf18, CENP-M, PANE1, bK250D10.2 | centromere protein M |
C00002514
|
| 8479 | HIRIP3 | HIRA interacting protein 3 |
C00002514
|
| 91687 | CENPL, C1orf155, CENP-L, RP3-383J4.1, dJ383J4.3 | centromere protein L |
C00002514
|
| 8334 | HIST1H2AC, H2A/l, H2AFL, dJ221C16.4 | histone cluster 1, H2ac |
C00002514
|
| 3012 | HIST1H2AE, H2A.1, H2A.2, H2A/a, H2AFA | histone cluster 1, H2ae |
C00002514
|
| 8969 | HIST1H2AG, H2A.1b, H2A/p, H2AFP, H2AG, pH2A/f | histone cluster 1, H2ag |
C00002514
|
| 8329 | HIST1H2AI, H2A/c, H2AFC | histone cluster 1, H2ai |
C00002514
|
| 8330 | HIST1H2AK, H2A/d, H2AFD | histone cluster 1, H2ak |
C00002514
|
| 8347 | HIST1H2BC, H2B.1, H2B/l, H2BFL, dJ221C16.3 | histone cluster 1, H2bc |
C00002514
|
| 55835 | CENPJ, BM032, CENP-J, CPAP, LAP, LIP1, MCPH6, SASS4, SCKL4, Sas-4 | centromere protein J |
C00002514
|
| 8344 | HIST1H2BE, H2B.h, H2B/h, H2BFH, dJ221C16.8 | histone cluster 1, H2be |
C00002514
|
| 8343 | HIST1H2BF, H2B/g, H2BFG | histone cluster 1, H2bf |
C00002514
|
| 8339 | HIST1H2BG, H2B.1A, H2B/a, H2BFA, dJ221C16.8 | histone cluster 1, H2bg |
C00002514
|
| 2491 | CENPI, CENP-I, FSHPRH1, LRPR1, Mis6 | centromere protein I |
C00002514
|
| 64946 | CENPH | centromere protein H |
C00002514
|
| 85236 | HIST1H2BK, H2B/S, H2BFAiii, H2BFT, H2BK | histone cluster 1, H2bk |
C00002514
|
| 8351 | HIST1H3D, H3/b, H3FB | histone cluster 1, H3d |
C00002514
|
| 8357 | HIST1H3H, H3/k, H3F1K, H3FK | histone cluster 1, H3h |
C00002514
|
| 8365 | HIST1H4H, H4/h, H4FH | histone cluster 1, H4h |
C00002514
|
| 8349 | HIST2H2BE, GL105, H2B, H2B.1, H2BFQ, H2BGL105, H2BQ | histone cluster 2, H2be |
C00002514
|
| 92815 | HIST3H2A | histone cluster 3, H2a |
C00002514
|
| 3096 | HIVEP1, CIRIP, CRYBP1, MBP-1, PRDII-BF1, Schnurri-1, ZAS1, ZNF40, ZNF40A | human immunodeficiency virus type I enhancer binding protein 1 |
C00002514
|
| 1062 | CENPE, CENP-E, KIF10, PPP1R61 | centromere protein E, 312kDa |
C00002514
|
| 1952 | CELSR2, CDHF10, EGFL2, Flamingo1, MEGF3 | cadherin, EGF LAG seven-pass G-type receptor 2 |
C00002514
|
| 9425 | CDYL, CDYL1 | chromodomain protein, Y-like (EC:2.3.1.48) |
C00002514
|
| 55573 | CDV3, H41 | CDV3 homolog (mouse) |
C00002514
|
| 81620 | CDT1, DUP, RIS2 | chromatin licensing and DNA replication factor 1 |
C00002514
|
| 1040 | CDS1, CDS | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 (EC:2.7.7.41) |
C00002514
|
| 3187 | HNRNPH1, HNRPH, HNRPH1, hnRNPH | heterogeneous nuclear ribonucleoprotein H1 (H) |
C00002514
|
| 3189 | HNRNPH3, 2H9, HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) |
C00002514
|
| 3192 | HNRNPU, HNRPU, SAF-A, U21.1, hnRNP_U | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
C00002514
|
| 100195139 |
C00002514
|
||
| 3229 | HOXC13, ECTD9, HOX3, HOX3G | homeobox C13 |
C00002514
|
| 3223 | HOXC6, CP25, HHO.C8, HOX3, HOX3C | homeobox C6 |
C00002514
|
| 3251 | HPRT1, HGPRT, HPRT | hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8) |
C00002514
|
| 6792 | CDKL5, EIEE2, ISSX, STK9 | cyclin-dependent kinase-like 5 (EC:2.7.11.22) |
C00002514
|
| 8630 | HSD17B6, HSE, RODH, SDR9C6 | hydroxysteroid (17-beta) dehydrogenase 6 (EC:1.1.1.62 1.1.1.239 1.1.1.105) |
C00002514
|
| 7923 | HSD17B8, D6S2245E, FABG, FABGL, H2-KE6, HKE6, KE6, RING2, SDR30C1, dJ1033B10.9 | hydroxysteroid (17-beta) dehydrogenase 8 (EC:1.1.1.62 1.1.1.239) |
C00002514
|
| 7184 | HSP90B1, ECGP, GP96, GRP94, TRA1 | heat shock protein 90kDa beta (Grp94), member 1 |
C00002514
|
| 22824 | HSPA4L, APG-1, HSPH3, Osp94 | heat shock 70kDa protein 4-like |
C00002514
|
| 3313 | HSPA9, CSA, GRP-75, GRP75, HSPA9B, MOT, MOT2, MTHSP75, PBP74 | heat shock 70kDa protein 9 (mortalin) |
C00002514
|
| 219844 | HYLS1, HLS | hydrolethalus syndrome 1 |
C00002514
|
| 25764 | HYPK, C15orf63 | huntingtin interacting protein K |
C00002514
|
| 3382 | ICA1, ICA69, ICAp69 | islet cell autoantigen 1, 69kDa |
C00002514
|
| 23463 | ICMT, HSTE14, MST098, MSTP098, PCCMT, PCMT, PPMT | isoprenylcysteine carboxyl methyltransferase (EC:2.1.1.100) |
C00002514
|
| 10263 | CDK2AP2, DOC-1R, p14 | cyclin-dependent kinase 2 associated protein 2 |
C00002514
|
| 8621 | CDK13, CDC2L, CDC2L5, CHED, hCDK13 | cyclin-dependent kinase 13 (EC:2.7.11.23 2.7.11.22) |
C00002514
|
| 3423 | IDS, MPS2, SIDS | iduronate 2-sulfatase (EC:3.1.6.13) |
C00002514
|
| 9592 | IER2, ETR101 | immediate early response 2 |
C00002514
|
| 1010 | CDH12, CDHB | cadherin 12, type 2 (N-cadherin 2) |
C00002514
|
| 51278 | IER5, SBBI48 | immediate early response 5 |
C00002514
|
| 55143 | CDCA8, BOR, BOREALIN, DasraB, MESRGP | cell division cycle associated 8 |
C00002514
|
| 55536 | CDCA7L, JPO2, R1, RAM2 | cell division cycle associated 7-like |
C00002514
|
| 3433 | IFIT2, G10P2, GARG-39, IFI-54, IFI-54K, IFI54, IFIT-2, ISG-54_K, ISG-54K, ISG54, P54, cig42 | interferon-induced protein with tetratricopeptide repeats 2 |
C00002514
|
| 7866 | IFRD2, IFNRP, SKMc15, SM15 | interferon-related developmental regulator 2 |
C00002514
|
| 55764 | IFT122, CED, CED1, SPG, WDR10, WDR10p, WDR140 | intraflagellar transport 122 homolog (Chlamydomonas) |
C00002514
|
| 90410 | IFT20 | intraflagellar transport 20 homolog (Chlamydomonas) |
C00002514
|
| 112752 | IFT43, C14orf179, CED3 | intraflagellar transport 43 homolog (Chlamydomonas) |
C00002514
|
| 113130 | CDCA5, SORORIN | cell division cycle associated 5 |
C00002514
|
| 55038 | CDCA4, HEPP, SEI-3/HEPP | cell division cycle associated 4 |
C00002514
|
| 157313 | CDCA2, Repo-Man | cell division cycle associated 2 |
C00002514
|
| 79713 | IGFLR1, TMEM149, U2AF1L4 | IGF-like family receptor 1 |
C00002514
|
| 445438 |
C00002514
|
||
| 8317 | CDC7, CDC7L1, HsCDC7, Hsk1, huCDC7 | cell division cycle 7 (EC:2.7.11.1) |
C00002514
|
| 988 | CDC5L, CDC5, CDC5-LIKE, CEF1, PCDC5RP, dJ319D22.1 | cell division cycle 5-like |
C00002514
|
| 31052 |
C00002514
|
||
| 50604 | IL20, IL-20, IL10D, ZCYTO10 | interleukin 20 |
C00002514
|
| 9578 | CDC42BPB, MRCKB | CDC42 binding protein kinase beta (DMPK-like) (EC:2.7.11.1) |
C00002514
|
| 3608 | ILF2, NF45, PRO3063 | interleukin enhancer binding factor 2 |
C00002514
|
| 10994 | ILVBL, 209L8, AHAS, ILV2H | ilvB (bacterial acetolactate synthase)-like (EC:2.2.1.-) |
C00002514
|
| 196294 | IMMP1L, IMP1, IMP1-LIKE | IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
C00002514
|
| 92856 | IMP4, BXDC4 | IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
C00002514
|
| 996 | CDC27, ANAPC3, APC3, CDC27Hs, D0S1430E, D17S978E, HNUC, NUC2 | cell division cycle 27 |
C00002514
|
| 10207 | INADL, Cipp, PATJ, hINADL | InaD-like (Drosophila) |
C00002514
|
| 3619 | INCENP | inner centromere protein antigens 135/155kDa |
C00002514
|
| 995 | CDC25C, CDC25, PPP1R60 | cell division cycle 25C (EC:3.1.3.48) |
C00002514
|
| 283899 | INO80E, CCDC95 | INO80 complex subunit E |
C00002514
|
| 993 | CDC25A, CDC25A2 | cell division cycle 25A (EC:3.1.3.48) |
C00002514
|
| 966 | CD59, 16.3A5, 1F5, EJ16, EJ30, EL32, G344, HRF-20, HRF20, MAC-IP, MACIF, MEM43, MIC11, MIN1, MIN2, MIN3, MIRL, MSK21, p18-20 | CD59 molecule, complement regulatory protein |
C00002514
|
| 25896 | INTS7, C1orf73, INT7 | integrator complex subunit 7 |
C00002514
|
| 51447 | IP6K2, IHPK2, PIUS | inositol hexakisphosphate kinase 2 (EC:2.7.4.21) |
C00002514
|
| 51194 | IPO11, RanBP11 | importin 11 |
C00002514
|
| 55705 | IPO9, Imp9 | importin 9 |
C00002514
|
| 124152 | IQCK | IQ motif containing K |
C00002514
|
| 1604 | CD55, CR, CROM, DAF, TC | CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
C00002514
|
| 359948 | IRF2BP2 | interferon regulatory factor 2 binding protein 2 |
C00002514
|
| 51293 | CD320, 8D6, 8D6A, TCBLR | CD320 molecule |
C00002514
|
| 10379 | IRF9, IRF-9, ISGF3, ISGF3G, p48 | interferon regulatory factor 9 |
C00002514
|
| 9236 | CCPG1, CPR8 | cell cycle progression 1 |
C00002514
|
| 10983 | CCNI, CYC1, CYI | cyclin I |
C00002514
|
| 51477 | ISYNA1, INO1, INOS, IPS, IPS_1, IPS-1 | inositol-3-phosphate synthase 1 (EC:5.5.1.4) |
C00002514
|
| 901 | CCNG2 | cyclin G2 |
C00002514
|
| 3682 | ITGAE, CD103, HUMINAE | integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
C00002514
|
| 23421 | ITGB3BP, CENP-R, CENPR, HSU37139, NRIF3, TAP20 | integrin beta 3 binding protein (beta3-endonexin) |
C00002514
|
| 899 | CCNF, FBX1, FBXO1 | cyclin F |
C00002514
|
| 9134 | CCNE2, CYCE2 | cyclin E2 |
C00002514
|
| 890 | CCNA2, CCN1, CCNA | cyclin A2 |
C00002514
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00002514
|
| 3726 | JUNB, AP-1 | jun B proto-oncogene |
C00002514
|
| 461205 |
C00002514
|
||
| 7994 | KAT6A, MOZ, MYST3, RUNXBP2, ZC2HC6A, ZNF220 | K(lysine) acetyltransferase 6A (EC:2.3.1.48) |
C00002514
|
| 84056 | KATNAL1 | katanin p60 subunit A-like 1 (EC:3.6.4.3) |
C00002514
|
| 8645 | KCNK5, K2p5.1, TASK-2, TASK2 | potassium channel, subfamily K, member 5 |
C00002514
|
| 54793 | KCTD9, BTBD27 | potassium channel tetramerization domain containing 9 |
C00002514
|
| 23028 | KDM1A, AOF2, BHC110, KDM1, LSD1 | lysine (K)-specific demethylase 1A |
C00002514
|
| 221656 | KDM1B, AOF1, C6orf193, LSD2, bA204B7.3, dJ298J15.2 | lysine (K)-specific demethylase 1B |
C00002514
|
| 55818 | KDM3A, JHDM2A, JHMD2A, JMJD1, JMJD1A, TSGA | lysine (K)-specific demethylase 3A |
C00002514
|
| 5927 | KDM5A, RBBP-2, RBBP2, RBP2, JARID1A | lysine (K)-specific demethylase 5A |
C00002514
|
| 10765 | KDM5B, CT31, JARID1B, PLU-1, PLU1, PUT1, RBBP2H1A | lysine (K)-specific demethylase 5B |
C00002514
|
| 9703 | BCOX, BCOX1, CT101 | KIAA0100 |
C00002514
|
| 440193 | CCDC88C, DAPLE, HKRP2, KIAA1509 | coiled-coil domain containing 88C |
C00002514
|
| 9764 | KIAA0513 |
C00002514
|
|
| 23247 | KIAA0556 |
C00002514
|
|
| 9786 | Talpid3 | KIAA0586 |
C00002514
|
| 23379 | KIAA0947 |
C00002514
|
|
| 23325 | SWIP | KIAA1033 |
C00002514
|
| 57189 | LCHN, PRO2561 | KIAA1147 |
C00002514
|
| 57179 | p60MONOX | KIAA1191 |
C00002514
|
| 57482 | KIAA1211 |
C00002514
|
|
| 57221 | A7322, ARFGEF3, BIG3, C6orf92, PPP1R33, dJ171N11.1 | KIAA1244 |
C00002514
|
| 57535 | EIG121 | KIAA1324 |
C00002514
|
| 57650 | CIP2A, p90 | KIAA1524 |
C00002514
|
| 57498 | KIDINS220, ARMS | kinase D-interacting substrate, 220kDa |
C00002514
|
| 3832 | KIF11, EG5, HKSP, KNSL1, MCLMR, TRIP5 | kinesin family member 11 |
C00002514
|
| 131076 | CCDC58 | coiled-coil domain containing 58 |
C00002514
|
| 56992 | KIF15, HKLP2, KNSL7, NY-BR-62 | kinesin family member 15 |
C00002514
|
| 81930 | KIF18A, MS-KIF18A | kinesin family member 18A |
C00002514
|
| 146909 | KIF18B | kinesin family member 18B |
C00002514
|
| 79714 | CCDC51 | coiled-coil domain containing 51 |
C00002514
|
| 11004 | KIF2C, KNSL6, MCAK | kinesin family member 2C |
C00002514
|
| 3800 | KIF5C, CDCBM2, KINN, NKHC, NKHC-2, NKHC2 | kinesin family member 5C |
C00002514
|
| 3833 | KIFC1, HSET, KNSL2 | kinesin family member C1 |
C00002514
|
| 11278 | KLF12, AP-2rep, AP2REP | Kruppel-like factor 12 |
C00002514
|
| 100286684 |
C00002514
|
||
| 57003 | CCDC47, MSTP041 | coiled-coil domain containing 47 |
C00002514
|
| 687 | KLF9, BTEB, BTEB1 | Kruppel-like factor 9 |
C00002514
|
| 23008 | KLHDC10, slim | kelch domain containing 10 |
C00002514
|
| 116138 | KLHDC3, PEAS, RP1-20C7.3, dJ20C7.3 | kelch domain containing 3 |
C00002514
|
| 91057 | CCDC34, NY-REN-41, RAMA3 | coiled-coil domain containing 34 |
C00002514
|
| 51088 | KLHL5 | kelch-like family member 5 |
C00002514
|
| 346689 | KLRG2, CLEC15B | killer cell lectin-like receptor subfamily G, member 2 |
C00002514
|
| 8085 | KMT2D, AAD10, ALR, CAGL114, KABUK1, KMS, MLL2, MLL4, TNRC21 | lysine (K)-specific methyltransferase 2D (EC:2.1.1.43) |
C00002514
|
| 55904 | KMT2E, HDCMC04P, MLL5 | lysine (K)-specific methyltransferase 2E (EC:2.1.1.43) |
C00002514
|
| 90417 | KNSTRN, C15orf23, SKAP, TRAF4AF1 | kinetochore-localized astrin/SPAG5 binding protein |
C00002514
|
| 9735 | KNTC1, ROD | kinetochore associated 1 |
C00002514
|
| 779317 |
C00002514
|
||
| 3839 | KPNA3, IPOA4, SRP1, SRP1gamma, SRP4, hSRP1 | karyopherin alpha 3 (importin alpha 4) |
C00002514
|
| 3840 | KPNA4, IPOA3, QIP1, SRP3 | karyopherin alpha 4 (importin alpha 3) |
C00002514
|
| 3845 | KRAS, C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RASK2 | Kirsten rat sarcoma viral oncogene homolog |
C00002514
|
| 51315 | KRCC1, CHBP2 | lysine-rich coiled-coil 1 |
C00002514
|
| 144501 | KRT80, KB20 | keratin 80 |
C00002514
|
| 3895 | KTN1, CG1, KNT, MU-RMS-40.19 | kinectin 1 (kinesin receptor) |
C00002514
|
| 374969 | CCDC23 | coiled-coil domain containing 23 |
C00002514
|
| 79944 | L2HGDH, C14orf160 | L-2-hydroxyglutarate dehydrogenase (EC:1.1.99.2) |
C00002514
|
| 114294 | LACTB, G24, MRPL56 | lactamase, beta |
C00002514
|
| 8270 | LAGE3, CVG5, DXS9879E, DXS9951E, ESO3, ITBA2 | L antigen family, member 3 |
C00002514
|
| 3916 | LAMP1, CD107a, LAMPA, LGP120 | lysosomal-associated membrane protein 1 |
C00002514
|
| 55323 | LARP6, ACHN | La ribonucleoprotein domain family, member 6 |
C00002514
|
| 3930 | LBR, DHCR14B, LMN2R, PHA, TDRD18 | lamin B receptor |
C00002514
|
| 9836 | LCMT2, PPM2, TYW4 | leucine carboxyl methyltransferase 2 |
C00002514
|
| 154467 | CCDC167, C6orf129, HSPC265 | coiled-coil domain containing 167 |
C00002514
|
| 64770 | CCDC14 | coiled-coil domain containing 14 |
C00002514
|
| 3956 | LGALS1, GAL1, GBP | lectin, galactoside-binding, soluble, 1 |
C00002514
|
| 150275 | CCDC117, dJ366L4.1 | coiled-coil domain containing 117 |
C00002514
|
| 9355 | LHX2, LH2, hLhx2 | LIM homeobox 2 |
C00002514
|
| 3978 | LIG1 | ligase I, DNA, ATP-dependent (EC:6.5.1.1) |
C00002514
|
| 3981 | LIG4 | ligase IV, DNA, ATP-dependent (EC:6.5.1.1) |
C00002514
|
| 55013 | CCDC109B, MCUb | coiled-coil domain containing 109B |
C00002514
|
| 91750 | LIN52, C14orf46, c14_5549 | lin-52 homolog (C. elegans) |
C00002514
|
| 8825 | LIN7A, LIN-7A, LIN7, MALS-1, TIP-33, VELI1 | lin-7 homolog A (C. elegans) |
C00002514
|
| 55327 | LIN7C, LIN-7-C, LIN-7C, MALS-3, MALS3, VELI3 | lin-7 homolog C (C. elegans) |
C00002514
|
| 145978 | LINC00052, NCRNA00052, TMEM83 | long intergenic non-protein coding RNA 52 |
C00002514
|
| 112869 | CCDC101, SGF29, STAF36 | coiled-coil domain containing 101 |
C00002514
|
| 23468 | CBX5, HP1, HP1A | chromobox homolog 5 |
C00002514
|
| 84823 | LMNB2, LAMB2, LMN2 | lamin B2 |
C00002514
|
| 9361 | LONP1, LON, LONP, LonHS, PIM1, PRSS15, hLON | lon peptidase 1, mitochondrial |
C00002514
|
| 8535 | CBX4, NBP16, PC2 | chromobox homolog 4 |
C00002514
|
| 26020 | LRP10, LRP9, MST087 | low density lipoprotein receptor-related protein 10 |
C00002514
|
| 873 | CBR1, CBR, SDR21C1, hCBR1 | carbonyl reductase 1 (EC:1.1.1.189 1.1.1.197 1.1.1.184) |
C00002514
|
| 122769 | LRR1, 4-1BBLRR, LRR-1, PPIL5 | leucine rich repeat protein 1 |
C00002514
|
| 80131 | LRRC8E | leucine rich repeat containing 8 family, member E |
C00002514
|
| 57819 | LSM2, C6orf28, G7B, YBL026W, snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00002514
|
| 25804 | LSM4, GRP, YER112W | LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00002514
|
| 51690 | LSM7, YNL147W | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00002514
|
| 55692 | LUC7L, LUC7B1, Luc7, SR+89, hLuc7B1 | LUC7-like (S. cerevisiae) |
C00002514
|
| 55646 | LYAR, ZC2HC2, ZLYAR | Ly1 antibody reactive |
C00002514
|
| 4067 | LYN, JTK8, p53Lyn, p56Lyn | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (EC:2.7.10.2) |
C00002514
|
| 130574 | LYPD6 | LY6/PLAUR domain containing 6 |
C00002514
|
| 4085 | MAD2L1, HSMAD2, MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) |
C00002514
|
| 10459 | MAD2L2, MAD2B, POLZ2, REV7 | MAD2 mitotic arrest deficient-like 2 (yeast) |
C00002514
|
| 23764 | MAFF, U-MAF, hMafF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
C00002514
|
| 10916 | MAGED2, 11B6, BCG-1, BCG1, HCA10, MAGE-D2 | melanoma antigen family D, 2 |
C00002514
|
| 9223 | MAGI1, AIP-3, AIP3, BAIAP1, BAP-1, BAP1, MAGI-1, TNRC19, WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 |
C00002514
|
| 55110 | MAGOHB, MGN2, mago, magoh | mago-nashi homolog B (Drosophila) |
C00002514
|
| 378938 | MALAT1, HCN, LINC00047, MALAT-1, NCRNA00047, NEAT2, PRO2853 | metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
C00002514
|
| 4124 | MAN2A1, GOLIM7, MANA2, MANII | mannosidase, alpha, class 2A, member 1 (EC:3.2.1.114) |
C00002514
|
| 79694 | MANEA, ENDO, hEndo | mannosidase, endo-alpha (EC:3.2.1.130) |
C00002514
|
| 4131 | MAP1B, FUTSCH, MAP5 | microtubule-associated protein 1B |
C00002514
|
| 81631 | MAP1LC3B, ATG8F, LC3B, MAP1A/1BLC3, MAP1LC3B-a | microtubule-associated protein 1 light chain 3 beta |
C00002514
|
| 5606 | MAP2K3, MAPKK3, MEK3, MKK3, PRKMK3, SAPKK-2, SAPKK2 | mitogen-activated protein kinase kinase 3 (EC:2.7.12.2) |
C00002514
|
| 868 | CBLB, Cbl-b, RNF56 | Cbl proto-oncogene B, E3 ubiquitin protein ligase |
C00002514
|
| 64844 | MARCH7, AXO, AXOT, MARCH-VII, RNF177 | membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
C00002514
|
| 865 | CBFB, PEBP2B | core-binding factor, beta subunit |
C00002514
|
| 375449 | MAST4 | microtubule associated serine/threonine kinase family member 4 (EC:2.7.11.1) |
C00002514
|
| 84930 | MASTL, GREATWALL, GW, GWL, MAST-L, RP11-85G18.2, THC2, hGWL | microtubule associated serine/threonine kinase-like (EC:2.7.11.1) |
C00002514
|
| 4148 | MATN3, DIPOA, EDM5, HOA, OADIP, OS2 | matrilin 3 |
C00002514
|
| 4149 | MAX, bHLHd4 | MYC associated factor X |
C00002514
|
| 4150 | MAZ, PUR1, Pur-1, SAF-1, SAF-2, SAF-3, ZF87, ZNF801, Zif87 | MYC-associated zinc finger protein (purine-binding transcription factor) |
C00002514
|
| 151963 | MB21D2, C3orf59 | Mab-21 domain containing 2 |
C00002514
|
| 4162 | MCAM, CD146, MUC18 | melanoma cell adhesion molecule |
C00002514
|
| 55388 | MCM10, CNA43, DNA43 | minichromosome maintenance complex component 10 |
C00002514
|
| 4171 | MCM2, BM28, CCNL1, CDCL1, D3S3194, MITOTIN, cdc19 | minichromosome maintenance complex component 2 (EC:3.6.4.12) |
C00002514
|
| 4172 | MCM3, HCC5, P1-MCM3, P1.h, RLFB | minichromosome maintenance complex component 3 (EC:3.6.4.12) |
C00002514
|
| 4173 | MCM4, CDC21, CDC54, NKCD, NKGCD, P1-CDC21, hCdc21 | minichromosome maintenance complex component 4 (EC:3.6.4.12) |
C00002514
|
| 831 | CAST, BS-17 | calpastatin |
C00002514
|
| 4175 | MCM6, MCG40308, Mis5, P105MCM | minichromosome maintenance complex component 6 (EC:3.6.4.12) |
C00002514
|
| 4176 | MCM7, CDC47, MCM2, P1.1-MCM3, P1CDC47, P85MCM, PNAS146 | minichromosome maintenance complex component 7 (EC:3.6.4.12) |
C00002514
|
| 84515 | MCM8, C20orf154, dJ967N21.5 | minichromosome maintenance complex component 8 (EC:3.6.4.12) |
C00002514
|
| 9656 | MDC1, NFBD1 | mediator of DNA-damage checkpoint 1 |
C00002514
|
| 4201 | MEA1, HYS, MEA | male-enhanced antigen 1 |
C00002514
|
| 64769 | MEAF6, C1orf149, CENP-28, EAF6, NY-SAR-91 | MYST/Esa1-associated factor 6 |
C00002514
|
| 4205 | MEF2A, ADCAD1, RSRFC4, RSRFC9, mef2 | myocyte enhancer factor 2A |
C00002514
|
| 4213 | MEIS3P1, MEIS3, MEIS4, MRG2 | Meis homeobox 3 pseudogene 1 |
C00002514
|
| 9833 | MELK, HPK38 | maternal embryonic leucine zipper kinase (EC:2.7.11.1 2.7.10.2) |
C00002514
|
| 56257 | MEPCE, BCDIN3 | methylphosphate capping enzyme |
C00002514
|
| 59274 | MESDC1 | mesoderm development candidate 1 |
C00002514
|
| 9994 | CASP8AP2, CED-4, FLASH, RIP25 | caspase 8 associated protein 2 |
C00002514
|
| 4234 | METTL1, C12orf1, TRM8, TRMT8, YDL201w | methyltransferase like 1 (EC:2.1.1.33) |
C00002514
|
| 23589 | CARHSP1, CRHSP-24, CSDC1 | calcium regulated heat stable protein 1, 24kDa |
C00002514
|
| 4236 | MFAP1 | microfibrillar-associated protein 1 |
C00002514
|
| 23269 | MGA, MAD5, MXD5 | MGA, MAX dimerization protein |
C00002514
|
| 11282 | MGAT4B, GNT-IV, GNT-IVB | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (EC:2.4.1.145) |
C00002514
|
| 4249 | MGAT5, GNT-V, GNT-VA | mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase (EC:2.4.1.155) |
C00002514
|
| 10724 | MGEA5, MEA5, NCOAT, OGA | meningioma expressed antigen 5 (hyaluronidase) (EC:2.3.1.48 3.2.1.169) |
C00002514
|
| 11343 | MGLL, HU-K5, HUK5, MAGL, MGL | monoglyceride lipase (EC:3.1.1.23) |
C00002514
|
| 22900 | CARD8, CARDINAL, DACAR, DAKAR, NDPP, NDPP1, TUCAN | caspase recruitment domain family, member 8 |
C00002514
|
| 85377 | MICALL1, MICAL-L1, MIRAB13 | MICAL-like 1 |
C00002514
|
| 79778 | MICALL2, JRAB, MICAL-L2 | MICAL-like 2 |
C00002514
|
| 4277 | MICB, PERB11.2 | MHC class I polypeptide-related sequence B |
C00002514
|
| 84864 | MINA, MDIG, MINA53, NO52, ROX | MYC induced nuclear antigen |
C00002514
|
| 406991 | MIR21, MIRN21, hsa-mir-21, miR-21, miRNA21 | microRNA 21 |
C00002514
|
| 55320 | MIS18BP1, C14orf106, HSA242977, KNL2, M18BP1 | MIS18 binding protein 1 |
C00002514
|
| 124583 | CANT1, DBQD, SCAN-1, SCAN1, SHAPY | calcium activated nucleotidase 1 (EC:3.6.1.6) |
C00002514
|
| 57118 | CAMK1D, CKLiK, CaM-K1, CaMKID | calcium/calmodulin-dependent protein kinase ID (EC:2.7.11.17) |
C00002514
|
| 4291 | MLF1 | myeloid leukemia factor 1 |
C00002514
|
| 813 | CALU | calumenin |
C00002514
|
| 4292 | MLH1, COCA2, FCC2, HNPCC, HNPCC2, hMLH1 | mutL homolog 1 |
C00002514
|
| 197259 | MLKL | mixed lineage kinase domain-like |
C00002514
|
| 326625 | MMAB, ATR, cblB, cob | methylmalonic aciduria (cobalamin deficiency) cblB type (EC:2.5.1.17) |
C00002514
|
| 808 | CALM3, PHKD, PHKD3 | calmodulin 3 (phosphorylase kinase, delta) (EC:2.7.11.19) |
C00002514
|
| 55329 | MNS1, SPATA40 | meiosis-specific nuclear structural 1 |
C00002514
|
| 64112 | MOAP1, MAP-1, PNMA4 | modulator of apoptosis 1 |
C00002514
|
| 9643 | MORF4L2, MORFL2, MRGX | mortality factor 4 like 2 |
C00002514
|
| 56180 | MOSPD1, DJ473B4 | motile sperm domain containing 1 |
C00002514
|
| 10198 | MPHOSPH9, MPP-9, MPP9 | M-phase phosphoprotein 9 |
C00002514
|
| 744 | MPPED2, 239FB, C11orf8 | metallophosphoesterase domain containing 2 |
C00002514
|
| 4361 | MRE11A, ATLD, HNGS1, MRE11, MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
C00002514
|
| 78988 | MRP63, bMRP63 | mitochondrial ribosomal protein 63 |
C00002514
|
| 51253 | MRPL37, L37mt, MRP-L2, MRP-L37, MRPL2, RPML2 | mitochondrial ribosomal protein L37 |
C00002514
|
| 64949 | MRPS26, C20orf193, GI008, MRP-S13, MRP-S26, MRPS13, NY-BR-87, RPMS13, dJ534B8.3 | mitochondrial ribosomal protein S26 |
C00002514
|
| 23107 | MRPS27, MRP-S27, S27mt | mitochondrial ribosomal protein S27 |
C00002514
|
| 799 | CALCR, CRT, CT-R, CTR, CTR1 | calcitonin receptor |
C00002514
|
| 100009675 |
C00002514
|
||
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00002514
|
| 4437 | MSH3, DUP, MRP1 | mutS homolog 3 |
C00002514
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00002514
|
| 4488 | MSX2, CRS2, FPP, HOX8, MSH, PFM, PFM1 | msh homeobox 2 |
C00002514
|
| 4515 | MTCP1, P13MTCP1, p8MTCP1 | mature T-cell proliferation 1 |
C00002514
|
| 51537 | MTFP1, MTP18 | mitochondrial fission process 1 |
C00002514
|
| 113115 | MTFR2, DUFD1, FAM54A | mitochondrial fission regulator 2 |
C00002514
|
| 92170 | MTG1, GTP, GTPBP7, RP11-108K14.2 | mitochondrial ribosome-associated GTPase 1 |
C00002514
|
| 4522 | MTHFD1, MTHFC, MTHFD | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase (EC:1.5.1.5 6.3.4.3 3.5.4.9) |
C00002514
|
| 9110 | MTMR4, FYVE-DSP2, ZFYVE11 | myotubularin related protein 4 (EC:3.1.3.48) |
C00002514
|
| 57509 | MTUS1, ATBP, ATIP, ICIS, MP44, MTSG1 | microtubule associated tumor suppressor 1 |
C00002514
|
| 9961 | MVP, LRP, VAULT1 | major vault protein |
C00002514
|
| 10608 | MXD4, MAD4, MST149, MSTP149, bHLHc12 | MAX dimerization protein 4 |
C00002514
|
| 4601 | MXI1, MAD2, MXD2, MXI, bHLHc11 | MAX interactor 1, dimerization protein |
C00002514
|
| 439921 | MXRA7, PS1TP1, TMAP1 | matrix-remodelling associated 7 |
C00002514
|
| 27101 | CACYBP, GIG5, RP1-102G20.6, S100A6BP, SIP | calcyclin binding protein |
C00002514
|
| 4605 | MYBL2, B-MYB, BMYB | v-myb avian myeloblastosis viral oncogene homolog-like 2 |
C00002514
|
| 157983 | C9orf66, RP11-59O6.1 | chromosome 9 open reading frame 66 |
C00002514
|
| 26292 | MYCBP, AMY-1 | MYC binding protein |
C00002514
|
| 23077 | MYCBP2, PAM | MYC binding protein 2, E3 ubiquitin protein ligase |
C00002514
|
| 79784 | MYH14, DFNA4, DFNA4A, MHC16, MYH17, NMHC_II-C, NMHC-II-C, PNMHH, myosin | myosin, heavy chain 14, non-muscle (EC:3.6.4.1) |
C00002514
|
| 55071 | C9orf40 | chromosome 9 open reading frame 40 |
C00002514
|
| 80179 | MYO19, MYOHD1 | myosin XIX |
C00002514
|
| 401546 | C9orf152, bA470J20.2 | chromosome 9 open reading frame 152 |
C00002514
|
| 4644 | MYO5A, GS1, MYH12, MYO5, MYR12 | myosin VA (heavy chain 12, myoxin) |
C00002514
|
| 4645 | MYO5B | myosin VB |
C00002514
|
| 4646 | MYO6, DFNA22, DFNB37 | myosin VI |
C00002514
|
| 286257 | C9orf142 | chromosome 9 open reading frame 142 |
C00002514
|
| 440145 | MZT1, C13orf37, MOZART1 | mitotic spindle organizing protein 1 |
C00002514
|
| 9683 | N4BP1 | NEDD4 binding protein 1 |
C00002514
|
| 90634 | N4BP2L1, CG018 | NEDD4 binding protein 2-like 1 |
C00002514
|
| 80218 | NAA50, MAK3, NAT13, NAT13P, NAT5, NAT5P, SAN, hNAT5, hSAN | N(alpha)-acetyltransferase 50, NatE catalytic subunit (EC:2.3.1.-) |
C00002514
|
| 4664 | NAB1 | NGFI-A binding protein 1 (EGR1 binding protein 1) |
C00002514
|
| 133686 | NADK2, C5orf33, MNADK, NADKD1 | NAD kinase 2, mitochondrial (EC:2.7.1.23) |
C00002514
|
| 254778 | C8orf46 | chromosome 8 open reading frame 46 |
C00002514
|
| 140838 | NANP, C20orf147, HDHD4, dJ694B14.3 | N-acetylneuraminic acid phosphatase (EC:3.1.3.29) |
C00002514
|
| 4676 | NAP1L4, NAP1L4b, NAP2, NAP2L, hNAP2 | nucleosome assembly protein 1-like 4 |
C00002514
|
| 100195127 |
C00002514
|
||
| 4678 | NASP, FLB7527, PRO1999 | nuclear autoantigenic sperm protein (histone-binding) |
C00002514
|
| 26960 | NBEA, BCL8B, LYST2 | neurobeachin |
C00002514
|
| 65065 | NBEAL1, A530083I02Rik, ALS2CR16, ALS2CR17 | neurobeachin-like 1 |
C00002514
|
| 55672 | NBPF1, AB13, AB14, AB23, AD2, NBG, NBPF | neuroblastoma breakpoint family, member 1 |
C00002514
|
| 200030 | NBPF11 | neuroblastoma breakpoint family, member 11 |
C00002514
|
| 9918 | NCAPD2, CAP-D2, CNAP1, hCAP-D2 | non-SMC condensin I complex, subunit D2 |
C00002514
|
| 23310 | NCAPD3, CAP-D3, CAPD3, hCAP-D3, hHCP-6, hcp-6 | non-SMC condensin II complex, subunit D3 |
C00002514
|
| 78996 | C7orf49, MRI | chromosome 7 open reading frame 49 |
C00002514
|
| 54892 | NCAPG2, CAP-G2, CAPG2, LUZP5, MTB, hCAP-G2 | non-SMC condensin II complex, subunit G2 |
C00002514
|
| 23397 | NCAPH, BRRN1, CAP-H | non-SMC condensin I complex, subunit H |
C00002514
|
| 29781 | NCAPH2, CAPH2 | non-SMC condensin II complex, subunit H2 |
C00002514
|
| 4686 | NCBP1, CBP80, NCBP, Sto1 | nuclear cap binding protein subunit 1, 80kDa |
C00002514
|
| 63920 | C5orf54, Buster3, ZBED8 | chromosome 5 open reading frame 54 |
C00002514
|
| 10499 | NCOA2, GRIP1, KAT13C, NCoA-2, SRC2, TIF2, bHLHe75 | nuclear receptor coactivator 2 |
C00002514
|
| 8202 | NCOA3, ACTR, AIB-1, AIB1, CAGH16, CTG26, KAT13B, RAC3, SRC-3, SRC3, TNRC14, TNRC16, TRAM-1, bHLHe42, pCIP | nuclear receptor coactivator 3 (EC:2.3.1.48) |
C00002514
|
| 135112 | NCOA7, ERAP140, ESNA1, Nbla00052, Nbla10993, TLDC4, dJ187J11.3 | nuclear receptor coactivator 7 |
C00002514
|
| 55706 | NDC1, NET3, TMEM48 | NDC1 transmembrane nucleoporin |
C00002514
|
| 54820 | NDE1, HOM-TES-87, LIS4, NDE, NUDE, NUDE1 | nudE neurodevelopment protein 1 |
C00002514
|
| 80762 | NDFIP1, N4WBP5 | Nedd4 family interacting protein 1 |
C00002514
|
| 4728 | NDUFS8, CI-23k, CI23KD, TYKY | NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) (EC:1.6.5.3 1.6.99.3) |
C00002514
|
| 4723 | NDUFV1, CI-51K, CI51KD, UQOR1 | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa (EC:1.6.5.3 1.6.99.3) |
C00002514
|
| 283131 | NEAT1, LINC00084, NCRNA00084, TncRNA, VINC | nuclear paraspeckle assembly transcript 1 (non-protein coding) |
C00002514
|
| 375444 | C5orf34 | chromosome 5 open reading frame 34 |
C00002514
|
| 23327 | NEDD4L, NEDD4-2, NEDD4.2, RSP5, hNEDD4-2 | neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00002514
|
| 4739 | NEDD9, CAS-L, CAS2, CASL, CASS2, HEF1 | neural precursor cell expressed, developmentally down-regulated 9 |
C00002514
|
| 55247 | NEIL3, FGP2, FPG2, NEI3, hFPG2, hNEI3 | nei endonuclease VIII-like 3 (E. coli) (EC:4.2.99.18) |
C00002514
|
| 727 | C5, C5a, C5b, CPAMD4 | complement component 5 |
C00002514
|
| 201725 | C4orf46 | chromosome 4 open reading frame 46 |
C00002514
|
| 81831 | NETO2, BTCL2, NEOT2 | neuropilin (NRP) and tolloid (TLL)-like 2 |
C00002514
|
| 4758 | NEU1, NANH, NEU, SIAL1 | sialidase 1 (lysosomal sialidase) (EC:3.2.1.18) |
C00002514
|
| 54492 | NEURL1B, NEURL3, hNeur2, neur2 | neuralized E3 ubiquitin protein ligase 1B (EC:6.3.2.-) |
C00002514
|
| 84901 | NFATC2IP, ESC2, NIP45, RAD60 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein |
C00002514
|
| 401152 | C4orf3 | chromosome 4 open reading frame 3 |
C00002514
|
| 79669 | C3orf52, TTMP | chromosome 3 open reading frame 52 |
C00002514
|
| 205327 | C2orf69 | chromosome 2 open reading frame 69 |
C00002514
|
| 22981 | NINL, NLP, dJ691N24.1 | ninein-like |
C00002514
|
| 79570 | NKAIN1, FAM77C | Na+/K+ transporting ATPase interacting 1 |
C00002514
|
| 112770 | C1orf85, NCU-G1 | chromosome 1 open reading frame 85 |
C00002514
|
| 9397 | NMT2 | N-myristoyltransferase 2 (EC:2.3.1.97) |
C00002514
|
| 10874 | NMU | neuromedin U |
C00002514
|
| 23530 | NNT, GCCD4 | nicotinamide nucleotide transhydrogenase (EC:1.6.1.2) |
C00002514
|
| 64318 | NOC3L, AD24, C10orf117, FAD24 | nucleolar complex associated 3 homolog (S. cerevisiae) |
C00002514
|
| 100380365 |
C00002514
|
||
| 9221 | NOLC1, NOPP130, NOPP140, NS5ATP13, P130 | nucleolar and coiled-body phosphoprotein 1 |
C00002514
|
| 64434 | NOM1, C7orf3, SGD1 | nucleolar protein with MIF4G domain 1 |
C00002514
|
| 4854 | NOTCH3, CADASIL, CASIL, IMF2 | notch 3 |
C00002514
|
| 64067 | NPAS3, MOP6, PASD6, bHLHe12 | neuronal PAS domain protein 3 |
C00002514
|
| 9520 | NPEPPS, AAP-S, MP100, PSA | aminopeptidase puromycin sensitive (EC:3.4.11.14) |
C00002514
|
| 27031 | NPHP3, MKS7, NPH3, RHPD, RHPD1 | nephronophthisis 3 (adolescent) |
C00002514
|
| 10360 | NPM3, PORMIN, TMEM123 | nucleophosmin/nucleoplasmin 3 |
C00002514
|
| 255743 | NPNT, EGFL6L, POEM | nephronectin |
C00002514
|
| 149563 | C1orf64, ERRF, RP11-5P18.4 | chromosome 1 open reading frame 64 |
C00002514
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00002514
|
| 4835 | NQO2, DHQV, DIA6, NMOR2, QR2 | NAD(P)H dehydrogenase, quinone 2 (EC:1.10.99.2) |
C00002514
|
| 7181 | NR2C1, TR2 | nuclear receptor subfamily 2, group C, member 1 |
C00002514
|
| 126382 | NR2C2AP, TRA16 | nuclear receptor 2C2-associated protein |
C00002514
|
| 7026 | NR2F2, ARP1, COUPTFB, COUPTFII, NF-E3, NR2F1, SVP40, TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 |
C00002514
|
| 2063 | NR2F6, EAR-2, EAR2, ERBAL2 | nuclear receptor subfamily 2, group F, member 6 |
C00002514
|
| 199920 | C1orf168 | chromosome 1 open reading frame 168 |
C00002514
|
| 4929 | NR4A2, HZF-3, NOT, NURR1, RNR1, TINUR | nuclear receptor subfamily 4, group A, member 2 |
C00002514
|
| 4897 | NRCAM | neuronal cell adhesion molecule |
C00002514
|
| 11270 | NRM, NRM29 | nurim (nuclear envelope membrane protein) |
C00002514
|
| 79762 | C1orf115, RP11-322F10.4 | chromosome 1 open reading frame 115 |
C00002514
|
| 59277 | NTN4, PRO3091 | netrin 4 |
C00002514
|
| 9891 | NUAK1, ARK5 | NUAK family, SNF1-like kinase, 1 (EC:2.7.11.1) |
C00002514
|
| 4521 | NUDT1, MTH1 | nudix (nucleoside diphosphate linked moiety X)-type motif 1 (EC:3.6.1.55 3.6.1.56) |
C00002514
|
| 131870 | NUDT16 | nudix (nucleoside diphosphate linked moiety X)-type motif 16 (EC:3.6.1.62) |
C00002514
|
| 11051 | NUDT21, CFIM25, CPSF5 | nudix (nucleoside diphosphate linked moiety X)-type motif 21 |
C00002514
|
| 57122 | NUP107, NUP84 | nucleoporin 107kDa |
C00002514
|
| 9631 | NUP155, N155 | nucleoporin 155kDa |
C00002514
|
| 23165 | NUP205, C7orf14 | nucleoporin 205kDa |
C00002514
|
| 23225 | NUP210, GP210, POM210 | nucleoporin 210kDa |
C00002514
|
| 129401 | NUP35, MP44, NP44, NUP53 | nucleoporin 35kDa |
C00002514
|
| 79023 | NUP37, p37 | nucleoporin 37kDa |
C00002514
|
| 10762 | NUP50, NPAP60, NPAP60L | nucleoporin 50kDa |
C00002514
|
| 54830 | NUP62CL | nucleoporin 62kDa C-terminal like |
C00002514
|
| 79902 | NUP85, Nup75 | nucleoporin 85kDa |
C00002514
|
| 55732 | C1orf112, RP1-97P20.1 | chromosome 1 open reading frame 112 |
C00002514
|
| 91775 | NXPE3, FAM55C, MST115 | neurexophilin and PC-esterase domain family, member 3 |
C00002514
|
| 221443 | OARD1, C6orf130, TARG1, dJ34B21.3 | O-acyl-ADP-ribose deacylase 1 |
C00002514
|
| 4947 | OAZ2, AZ2 | ornithine decarboxylase antizyme 2 |
C00002514
|
| 23363 | OBSL1 | obscurin-like 1 (EC:2.7.11.18) |
C00002514
|
| 4957 | ODF2, CT134, ODF2/1, ODF2/2, ODF84 | outer dense fiber of sperm tails 2 |
C00002514
|
| 84798 | C19orf48 | chromosome 19 open reading frame 48 |
C00002514
|
| 11339 | OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18beta, hMIS18beta | Opa interacting protein 5 |
C00002514
|
| 497661 | C18orf32 | chromosome 18 open reading frame 32 |
C00002514
|
| 10133 | OPTN, ALS12, FIP2, GLC1E, HIP7, HYPL, NRP, TFIIIA-INTP | optineurin |
C00002514
|
| 4998 | ORC1, HSORC1, ORC1L, PARC1 | origin recognition complex, subunit 1 |
C00002514
|
| 716175 |
C00002514
|
||
| 9885 | OSBPL2, ORP-2, ORP2 | oxysterol binding protein-like 2 |
C00002514
|
| 51526 | OSER1, C20orf111, HSPC207, Osr1, Perit1, dJ1183I21.1 | oxidative stress responsive serine-rich 1 |
C00002514
|
| 28962 | OSTM1, GIPN, GL, OPTB5 | osteopetrosis associated transmembrane protein 1 |
C00002514
|
| 220213 | OTUD1, DUBA7, OTDC1 | OTU domain containing 1 (EC:3.4.19.12) |
C00002514
|
| 5019 | OXCT1, OXCT, SCOT | 3-oxoacid CoA transferase 1 (EC:2.8.3.5) |
C00002514
|
| 256302 | C17orf103, Gtlf3b | chromosome 17 open reading frame 103 |
C00002514
|
| 5034 | P4HB, DSI, ERBA2L, GIT, P4Hbeta, PDI, PDIA1, PHDB, PO4DB, PO4HB, PROHB | prolyl 4-hydroxylase, beta polypeptide (EC:5.3.4.1) |
C00002514
|
| 79447 | PAGR1, C16orf53, GAS, PA1 | PAXIP1 associated glutamate-rich protein 1 |
C00002514
|
| 10606 | PAICS, ADE2, ADE2H1, AIRC, PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase (EC:6.3.2.6 4.1.1.21) |
C00002514
|
| 10605 | PAIP1 | poly(A) binding protein interacting protein 1 |
C00002514
|
| 9924 | PAN2, USP52 | PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (EC:3.1.13.4) |
C00002514
|
| 11044 | PAPD7, LAK-1, LAK1, POLK, POLS, TRF4, TRF4-1, TRF41, TUTASE5 | PAP associated domain containing 7 (EC:2.7.7.7) |
C00002514
|
| 10914 | PAPOLA, PAP | poly(A) polymerase alpha (EC:2.7.7.19) |
C00002514
|
| 29105 | C16orf80, EVORF, GTL3, fSAP23 | chromosome 16 open reading frame 80 |
C00002514
|
| 54852 | PAQR5, MPRG | progestin and adipoQ receptor family member V |
C00002514
|
| 56288 | PARD3, ASIP, Baz, PAR3, PAR3alpha, PARD-3, PARD3A, SE2-5L16, SE2-5LT1, SE2-5T2 | par-3 family cell polarity regulator |
C00002514
|
| 283643 | C14orf80 | chromosome 14 open reading frame 80 |
C00002514
|
| 10038 | PARP2, ADPRT2, ADPRTL2, ADPRTL3, ARTD2, PARP-2, pADPRT-2 | poly (ADP-ribose) polymerase 2 (EC:2.4.2.30) |
C00002514
|
| 55010 | PARPBP, AROM, C12orf48, PARI | PARP1 binding protein |
C00002514
|
| 55742 | PARVA, CH-ILKBP, MXRA2 | parvin, alpha |
C00002514
|
| 5074 | PAWR, PAR4, Par-4 | PRKC, apoptosis, WT1, regulator |
C00002514
|
| 790955 | C11orf83, CCDS41658.1, UNQ655 | chromosome 11 open reading frame 83 |
C00002514
|
| 220042 | C11orf82, noxin | chromosome 11 open reading frame 82 |
C00002514
|
| 301164 |
C00002514
|
||
| 5094 | PCBP2, HNRNPE2, HNRPE2, hnRNP-E2 | poly(rC) binding protein 2 |
C00002514
|
| 57575 | PCDH10, OL-PCDH, PCDH19 | protocadherin 10 |
C00002514
|
| 100528825 |
C00002514
|
||
| 699 | BUB1, BUB1A, BUB1L, hBUB1 | BUB1 mitotic checkpoint serine/threonine kinase (EC:2.7.11.1) |
C00002514
|
| 115294 | PCMTD1 | protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
C00002514
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00002514
|
| 5116 | PCNT, KEN, MOPD2, PCN, PCNT2, PCNTB, PCTN2, SCKL4 | pericentrin |
C00002514
|
| 5121 | PCP4, PEP-19 | Purkinje cell protein 4 |
C00002514
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00002514
|
| 100380451 |
C00002514
|
||
| 58488 | PCTP, PC-TP, STARD2 | phosphatidylcholine transfer protein |
C00002514
|
| 55727 | BTBD7, FUP1 | BTB (POZ) domain containing 7 |
C00002514
|
| 10015 | PDCD6IP, AIP1, ALIX, DRIP4, HP95 | programmed cell death 6 interacting protein |
C00002514
|
| 2923 | PDIA3, ER60, ERp57, ERp60, ERp61, GRP57, GRP58, HsT17083, P58, PI-PLC | protein disulfide isomerase family A, member 3 (EC:5.3.4.1) |
C00002514
|
| 10130 | PDIA6, ERP5, P5, TXNDC7 | protein disulfide isomerase family A, member 6 (EC:5.3.4.1) |
C00002514
|
| 682 | BSG, 5F7, CD147, EMMPRIN, M6, OK, TCSF | basigin |
C00002514
|
| 27295 | PDLIM3, ALP | PDZ and LIM domain 3 |
C00002514
|
| 55108 | BSDC1 | BSD domain containing 1 |
C00002514
|
| 23590 | PDSS1, COQ1, COQ10D2, DPS, RP13-16H11.3, SPS, TPRT, TPT, TPT_1, hDPS1 | prenyl (decaprenyl) diphosphate synthase, subunit 1 (EC:2.5.1.91) |
C00002514
|
| 5174 | PDZK1, CAP70, CLAMP, NHERF-3, NHERF3, PDZD1 | PDZ domain containing 1 |
C00002514
|
| 64065 | PERP, KCP1, KRTCAP1, PIGPC1, RP3-496H19.1, THW, dJ496H19.1 | PERP, TP53 apoptosis effector |
C00002514
|
| 5211 | PFKL, PFK-B | phosphofructokinase, liver (EC:2.7.1.11) |
C00002514
|
| 5228 | PGF, D12S1900, PGFL, PLGF, PlGF-2, SHGC-10760 | placental growth factor |
C00002514
|
| 5229 | PGGT1B, BGGI, GGTI | protein geranylgeranyltransferase type I, beta subunit (EC:2.5.1.59) |
C00002514
|
| 5230 | PGK1, MIG10, PGKA | phosphoglycerate kinase 1 (EC:2.7.2.3) |
C00002514
|
| 54014 | BRWD1, C21orf107, N143, WDR9 | bromodomain and WD repeat domain containing 1 |
C00002514
|
| 140707 | BRI3BP, BNAS1, HCCR-1, HCCR-2, HCCRBP-1, KG19 | BRI3 binding protein |
C00002514
|
| 25798 | BRI3, I3 | brain protein I3 |
C00002514
|
| 10857 | PGRMC1, HPR6.6, MPR | progesterone receptor membrane component 1 |
C00002514
|
| 10424 | PGRMC2, DG6, PMBP | progesterone receptor membrane component 2 |
C00002514
|
| 9749 | PHACTR2, C6orf56 | phosphatase and actin regulator 2 |
C00002514
|
| 55274 | PHF10, BAF45A, XAP135 | PHD finger protein 10 |
C00002514
|
| 26147 | PHF19, MTF2L1, PCL3, TDRD19B | PHD finger protein 19 |
C00002514
|
| 51105 | PHF20L1, TDRD20B | PHD finger protein 20-like 1 |
C00002514
|
| 26227 | PHGDH, 3-PGDH, 3PGDH, PDG, PGAD, PGD, PGDH, SERA | phosphoglycerate dehydrogenase (EC:1.1.1.95) |
C00002514
|
| 8019 | BRD3, ORFX, RING3L | bromodomain containing 3 |
C00002514
|
| 90102 | PHLDB2, LL5b, LL5beta | pleckstrin homology-like domain, family B, member 2 |
C00002514
|
| 493911 | PHOSPHO2 | phosphatase, orphan 2 (EC:3.1.3.74) |
C00002514
|
| 10745 | PHTF1, PHTF | putative homeodomain transcription factor 1 |
C00002514
|
| 57157 | PHTF2 | putative homeodomain transcription factor 2 |
C00002514
|
| 85007 | PHYKPL, AGXT2L2, PHLU | 5-phosphohydroxy-L-lysine phospho-lyase (EC:4.2.3.134) |
C00002514
|
| 8301 | PICALM, CALM, CLTH, LAP | phosphatidylinositol binding clathrin assembly protein |
C00002514
|
| 80119 | PIF1, C15orf20, PIF | PIF1 5'-to-3' DNA helicase (EC:3.6.4.12) |
C00002514
|
| 55011 | PIH1D1, NOP17 | PIH1 domain containing 1 |
C00002514
|
| 5286 | PIK3C2A, CPK, PI3-K-C2(ALPHA), PI3-K-C2A | phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha (EC:2.7.1.154) |
C00002514
|
| 5289 | PIK3C3, VPS34, hVps34 | phosphatidylinositol 3-kinase, catalytic subunit type 3 (EC:2.7.1.137) |
C00002514
|
| 65018 | PINK1, BRPK, PARK6 | PTEN induced putative kinase 1 (EC:2.7.11.1) |
C00002514
|
| 8394 | PIP5K1A | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha (EC:2.7.1.68) |
C00002514
|
| 5570 | PKIB, PRKACN2 | protein kinase (cAMP-dependent, catalytic) inhibitor beta |
C00002514
|
| 396456 |
C00002514
|
||
| 9088 | PKMYT1, MYT1 | protein kinase, membrane associated tyrosine/threonine 1 (EC:2.7.11.1) |
C00002514
|
| 55344 | PLCXD1 | phosphatidylinositol-specific phospholipase C, X domain containing 1 |
C00002514
|
| 54477 | PLEKHA5, PEPP-2, PEPP2 | pleckstrin homology domain containing, family A member 5 |
C00002514
|
| 55041 | PLEKHB2, EVT2 | pleckstrin homology domain containing, family B (evectins) member 2 |
C00002514
|
| 423589 |
C00002514
|
||
| 79666 | PLEKHF2, EAPF, PHAFIN2, ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
C00002514
|
| 5347 | PLK1, PLK, STPK13 | polo-like kinase 1 (EC:2.7.11.21) |
C00002514
|
| 10733 | PLK4, SAK, STK18 | polo-like kinase 4 (EC:2.7.11.21) |
C00002514
|
| 675 | BRCA2, BRCC2, BROVCA2, FACD, FAD, FAD1, FANCB, FANCD, FANCD1, GLM3, PNCA2 | breast cancer 2, early onset |
C00002514
|
| 5352 | PLOD2, LH2, TLH | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (EC:1.14.11.4) |
C00002514
|
| 23654 | PLXNB2, MM1, Nbla00445, PLEXB2, dJ402G11.3 | plexin B2 |
C00002514
|
| 23129 | PLXND1, PLEXD1 | plexin D1 |
C00002514
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00002514
|
| 5376 | PMP22, CMT1A, CMT1E, DSS, GAS-3, HMSNIA, HNPP, Sp110 | peripheral myelin protein 22 |
C00002514
|
| 9512 | PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52 | peptidase (mitochondrial processing) beta (EC:3.4.24.64) |
C00002514
|
| 5378 | PMS1, HNPCC3, PMSL1, hPMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
C00002514
|
| 4860 | PNP, NP, PRO1837, PUNP | purine nucleoside phosphorylase (EC:2.4.2.1) |
C00002514
|
| 50640 | PNPLA8, IPLA2(GAMMA), IPLA2-2, IPLA2G, iPLA2gamma | patatin-like phospholipase domain containing 8 (EC:3.1.1.5) |
C00002514
|
| 10957 | PNRC1, B4-2, PNAS-145, PROL2, PRR2 | proline-rich nuclear receptor coactivator 1 |
C00002514
|
| 25886 | POC1A, PIX2, SOFT, WDR51A | POC1 centriolar protein A |
C00002514
|
| 5422 | POLA1, POLA, p180 | polymerase (DNA directed), alpha 1, catalytic subunit (EC:2.7.7.7) |
C00002514
|
| 23649 | POLA2 | polymerase (DNA directed), alpha 2, accessory subunit |
C00002514
|
| 5424 | POLD1, CDC2, CRCS10, MDPL, POLD | polymerase (DNA directed), delta 1, catalytic subunit (EC:2.7.7.7) |
C00002514
|
| 5425 | POLD2 | polymerase (DNA directed), delta 2, accessory subunit (EC:2.7.7.7) |
C00002514
|
| 10714 | POLD3, P66, P68 | polymerase (DNA-directed), delta 3, accessory subunit |
C00002514
|
| 26073 | POLDIP2, PDIP38, POLD4 | polymerase (DNA-directed), delta interacting protein 2 |
C00002514
|
| 5426 | POLE, CRCS12, FILS, POLE1 | polymerase (DNA directed), epsilon, catalytic subunit (EC:2.7.7.7) |
C00002514
|
| 5427 | POLE2, DPE2 | polymerase (DNA directed), epsilon 2, accessory subunit (EC:2.7.7.7) |
C00002514
|
| 54107 | POLE3, CHARAC17, CHRAC17, YBL1, p17 | polymerase (DNA directed), epsilon 3, accessory subunit (EC:2.7.7.7) |
C00002514
|
| 11201 | POLI, RAD30B, RAD3OB | polymerase (DNA directed) iota (EC:2.7.7.7) |
C00002514
|
| 10721 | POLQ, POLH, PRO0327 | polymerase (DNA directed), theta (EC:2.7.7.7) |
C00002514
|
| 221927 | BRAT1, BAAT1, C7orf27, RMFSL | BRCA1-associated ATM activator 1 |
C00002514
|
| 5433 | POLR2D, HSRBP4, HSRPB4, RBP4, RPB16 | polymerase (RNA) II (DNA directed) polypeptide D (EC:2.7.7.6) |
C00002514
|
| 5434 | POLR2E, RPABC1, RPB5, XAP4, hRPB25, hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa (EC:2.7.7.6) |
C00002514
|
| 5436 | POLR2G, RPB19, RPB7, hRPB19, hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G (EC:2.7.7.6) |
C00002514
|
| 5437 | POLR2H, RPABC3, RPB17, RPB8 | polymerase (RNA) II (DNA directed) polypeptide H (EC:2.7.7.6) |
C00002514
|
| 8612 | PPAP2C, LPP2, PAP-2c, PAP2-g | phosphatidic acid phosphatase type 2C (EC:3.1.3.4) |
C00002514
|
| 403313 | PPAPDC2, PDP1, PSDP, bA6J24.6 | phosphatidic acid phosphatase type 2 domain containing 2 |
C00002514
|
| 5471 | PPAT, ATASE, GPAT, PRAT | phosphoribosyl pyrophosphate amidotransferase (EC:2.4.2.14) |
C00002514
|
| 60490 | PPCDC | phosphopantothenoylcysteine decarboxylase (EC:4.1.1.36) |
C00002514
|
| 5479 | PPIB, CYP-S1, CYPB, OI9, SCYLP | peptidylprolyl isomerase B (cyclophilin B) (EC:5.2.1.8) |
C00002514
|
| 79866 | BORA, C13orf34 | bora, aurora kinase A activator |
C00002514
|
| 665 | BNIP3L, BNIP3a, NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like |
C00002514
|
| 5494 | PPM1A, PP2C-ALPHA, PP2CA, PP2Calpha | protein phosphatase, Mg2+/Mn2+ dependent, 1A (EC:3.1.3.16) |
C00002514
|
| 22843 | PPM1E, CaMKP-N, POPX1, PP2CH, caMKN | protein phosphatase, Mg2+/Mn2+ dependent, 1E (EC:3.1.3.16) |
C00002514
|
| 5514 | PPP1R10, CAT53, FB19, PNUTS, PP1R10, R111, p99 | protein phosphatase 1, regulatory subunit 10 |
C00002514
|
| 4659 | PPP1R12A, M130, MBS, MYPT1 | protein phosphatase 1, regulatory subunit 12A |
C00002514
|
| 10848 | PPP1R13L, IASPP, NKIP1, RAI, RAI4 | protein phosphatase 1, regulatory subunit 13 like |
C00002514
|
| 659 | BMPR2, BMPR-II, BMPR3, BMR2, BRK-3, PPH1, T-ALK | bone morphogenetic protein receptor, type II (serine/threonine kinase) (EC:2.7.11.30) |
C00002514
|
| 5504 | PPP1R2, IPP-2, IPP2 | protein phosphatase 1, regulatory (inhibitor) subunit 2 (EC:3.1.3.16) |
C00002514
|
| 5520 | PPP2R2A, B55A, B55ALPHA, PR52A, PR55A | protein phosphatase 2, regulatory subunit B, alpha (EC:3.1.3.16) |
C00002514
|
| 5523 | PPP2R3A, PPP2R3, PR130, PR72 | protein phosphatase 2, regulatory subunit B'', alpha (EC:3.1.3.16) |
C00002514
|
| 5529 | PPP2R5E | protein phosphatase 2, regulatory subunit B', epsilon isoform |
C00002514
|
| 5533 | PPP3CC, CALNA3, CNA3, PP2Bgamma | protein phosphatase 3, catalytic subunit, gamma isozyme (EC:3.1.3.16) |
C00002514
|
| 5536 | PPP5C, PP5, PPP5, PPT | protein phosphatase 5, catalytic subunit (EC:3.1.3.16) |
C00002514
|
| 10549 | PRDX4, AOE37-2, PRX-4 | peroxiredoxin 4 (EC:1.11.1.15) |
C00002514
|
| 57580 | PREX1, P-REX1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
C00002514
|
| 5557 | PRIM1, p49 | primase, DNA, polypeptide 1 (49kDa) (EC:2.7.7.7) |
C00002514
|
| 5558 | PRIM2, PRIM2A, p58 | primase, DNA, polypeptide 2 (58kDa) (EC:2.7.7.-) |
C00002514
|
| 5567 | PRKACB, PKACB | protein kinase, cAMP-dependent, catalytic, beta (EC:2.7.11.11) |
C00002514
|
| 5583 | PRKCH, PKC-L, PKCL, PRKCL, nPKC-eta | protein kinase C, eta (EC:2.7.11.13) |
C00002514
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00002514
|
| 10419 | PRMT5, HRMT1L5, IBP72, JBP1, SKB1, SKB1Hs | protein arginine methyltransferase 5 (EC:2.1.1.125) |
C00002514
|
| 150696 | PROM2, PROML2 | prominin 2 |
C00002514
|
| 9128 | PRPF4, HPRP4, HPRP4P, PRP4, Prp4p, SNRNP60 | pre-mRNA processing factor 4 |
C00002514
|
| 5634 | PRPS2, PRSII | phosphoribosyl pyrophosphate synthetase 2 (EC:2.7.6.1) |
C00002514
|
| 78994 | PRR14 | proline rich 14 |
C00002514
|
| 653 | BMP5 | bone morphogenetic protein 5 |
C00002514
|
| 90427 | BMF | Bcl2 modifying factor |
C00002514
|
| 23215 | PRRC2C, BAT2-iso, BAT2D1, BAT2L2, XTP2 | proline-rich coiled-coil 2C |
C00002514
|
| 285368 | PRRT3 | proline-rich transmembrane protein 3 |
C00002514
|
| 8548 | BLZF1, GOLGIN-45, JEM-1, JEM-1s, JEM1 | basic leucine zipper nuclear factor 1 |
C00002514
|
| 5652 | PRSS8, CAP1, PROSTASIN | protease, serine, 8 |
C00002514
|
| 645 | BLVRB, BVRB, FLR, SDR43U1 | biliverdin reductase B (flavin reductase (NADPH)) (EC:1.3.1.24 1.5.1.30) |
C00002514
|
| 11168 | PSIP1, DFS70, LEDGF, PAIP, PSIP2, p52, p75 | PC4 and SFRS1 interacting protein 1 |
C00002514
|
| 5702 | PSMC3, TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
C00002514
|
| 29893 | PSMC3IP, GT198, HOP2, HUMGT198A, ODG3, TBPIP | PSMC3 interacting protein |
C00002514
|
| 5721 | PSME2, PA28B, PA28beta, REGbeta | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
C00002514
|
| 84262 | PSMG3, C7orf48, PAC3 | proteasome (prosome, macropain) assembly chaperone 3 |
C00002514
|
| 84722 | PSRC1, DDA3, FP3214 | proline/serine-rich coiled-coil 1 |
C00002514
|
| 5725 | PTBP1, HNRNP-I, HNRNPI, HNRPI, PTB, PTB-1, PTB-T, PTB2, PTB3, PTB4, pPTB | polypyrimidine tract binding protein 1 |
C00002514
|
| 55037 | PTCD3, MRP-S39 | pentatricopeptide repeat domain 3 |
C00002514
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00002514
|
| 26258 | BLOC1S6, BLOS6, HPS9, PA, PALLID, PLDN | biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
C00002514
|
| 7803 | PTP4A1, HH72, PRL-1, PRL1, PTP(CAAX1), PTPCAAX1 | protein tyrosine phosphatase type IVA, member 1 (EC:3.1.3.48) |
C00002514
|
| 5770 | PTPN1, PTP1B | protein tyrosine phosphatase, non-receptor type 1 (EC:3.1.3.48) |
C00002514
|
| 11099 | PTPN21, PTPD1, PTPRL10 | protein tyrosine phosphatase, non-receptor type 21 (EC:3.1.3.48) |
C00002514
|
| 5792 | PTPRF, LAR | protein tyrosine phosphatase, receptor type, F (EC:3.1.3.48) |
C00002514
|
| 5796 | PTPRK, R-PTP-kappa | protein tyrosine phosphatase, receptor type, K (EC:3.1.3.48) |
C00002514
|
| 9232 | PTTG1, EAP1, HPTTG, PTTG, TUTR1 | pituitary tumor-transforming 1 |
C00002514
|
| 83480 | PUS3, 2610020J05Rik | pseudouridylate synthase 3 (EC:5.4.99.45) |
C00002514
|
| 5819 | PVRL2, CD112, HVEB, PRR2, PVRR2 | poliovirus receptor-related 2 (herpesvirus entry mediator B) |
C00002514
|
| 79912 | PYROXD1 | pyridine nucleotide-disulphide oxidoreductase domain 1 (EC:1.8.1.-) |
C00002514
|
| 79832 | QSER1 | glutamine and serine rich 1 |
C00002514
|
| 22864 | R3HDM2, PR01365 | R3H domain containing 2 |
C00002514
|
| 80223 | RAB11FIP1, NOEL1A, RCP, rab11-FIP1 | RAB11 family interacting protein 1 (class I) |
C00002514
|
| 51552 | RAB14, FBP, RAB-14 | RAB14, member RAS oncogene family |
C00002514
|
| 35577 |
C00002514
|
||
| 53917 | RAB24 | RAB24, member RAS oncogene family |
C00002514
|
| 29760 | BLNK, AGM4, BASH, BLNK-S, LY57, SLP-65, SLP65, bca | B-cell linker |
C00002514
|
| 10981 | RAB32 | RAB32, member RAS oncogene family |
C00002514
|
| 83452 | RAB33B, SMC2 | RAB33B, member RAS oncogene family |
C00002514
|
| 10966 | RAB40B, RAR, SEC4L | RAB40B, member RAS oncogene family |
C00002514
|
| 5870 | RAB6A, RAB6 | RAB6A, member RAS oncogene family |
C00002514
|
| 8934 | RAB7L1, RAB7L | RAB7, member RAS oncogene family-like 1 |
C00002514
|
| 4218 | RAB8A, MEL, RAB8 | RAB8A, member RAS oncogene family |
C00002514
|
| 55909 | BIN3 | bridging integrator 3 |
C00002514
|
| 5810 | RAD1, HRAD1, REC1 | RAD1 homolog (S. pombe) (EC:3.1.11.2) |
C00002514
|
| 56852 | RAD18, RNF73 | RAD18 homolog (S. cerevisiae) |
C00002514
|
| 5885 | RAD21, CDLS4, HR21, HRAD21, MCD1, NXP1, SCC1, hHR21 | RAD21 homolog (S. pombe) |
C00002514
|
| 5888 | RAD51, BRCC5, HRAD51, HsRad51, HsT16930, MRMV2, RAD51A, RECA | RAD51 recombinase |
C00002514
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00002514
|
| 5889 | RAD51C, BROVCA3, FANCO, R51H3, RAD51L2 | RAD51 paralog C |
C00002514
|
| 25788 | RAD54B, RDH54 | RAD54 homolog B (S. cerevisiae) |
C00002514
|
| 8438 | RAD54L, HR54, RAD54A, hHR54, hRAD54 | RAD54-like (S. cerevisiae) |
C00002514
|
| 8480 | RAE1, MIG14, MRNP41, Mnrp41, dJ481F12.3, dJ800J21.1 | ribonucleic acid export 1 |
C00002514
|
| 638 | BIK, BIP1, BP4, NBK | BCL2-interacting killer (apoptosis-inducing) |
C00002514
|
| 253959 | RALGAPA1, GARNL1, GRIPE, RalGAPalpha1, TULIP1, p240 | Ral GTPase activating protein, alpha subunit 1 (catalytic) |
C00002514
|
| 5902 | RANBP1, HTF9A | RAN binding protein 1 |
C00002514
|
| 5905 | RANGAP1, Fug1, RANGAP, SD | Ran GTPase activating protein 1 |
C00002514
|
| 9693 | RAPGEF2, CNrasGEF, NRAPGEP, PDZ-GEF1, PDZGEF1, RA-GEF, RA-GEF-1, Rap-GEP, nRap_GEP | Rap guanine nucleotide exchange factor (GEF) 2 |
C00002514
|
| 51195 | RAPGEFL1, Link-GEFII | Rap guanine nucleotide exchange factor (GEF)-like 1 |
C00002514
|
| 5921 | RASA1, CM-AVM, CMAVM, GAP, PKWS, RASA, RASGAP, p120GAP, p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 |
C00002514
|
| 100172339 |
C00002514
|
||
| 158158 | RASEF, RAB45 | RAS and EF-hand domain containing |
C00002514
|
| 84707 | BEX2, BEX1, DJ79P11.1 | brain expressed X-linked 2 |
C00002514
|
| 65997 | RASL11B | RAS-like, family 11, member B |
C00002514
|
| 283349 | RASSF3, RASSF5 | Ras association (RalGDS/AF-6) domain family member 3 |
C00002514
|
| 125950 | RAVER1 | ribonucleoprotein, PTB-binding 1 |
C00002514
|
| 617 | BCS1L, BCS, BCS1, BJS, FLNMS, GRACILE, Hs.6719, MC3DN1, PTD, h-BCS | BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
C00002514
|
| 5928 | RBBP4, NURF55, RBAP48 | retinoblastoma binding protein 4 |
C00002514
|
| 9774 | BCLAF1, BTF, bK211L9.1 | BCL2-associated transcription factor 1 |
C00002514
|
| 10741 | RBBP9, BOG, RBBP10 | retinoblastoma binding protein 9 |
C00002514
|
| 23543 | RBFOX2, FOX2, Fox-2, HNRBP2, HRNBP2, RBM9, RTA, dJ106I20.3, fxh | RNA binding protein, fox-1 homolog (C. elegans) 2 |
C00002514
|
| 5934 | RBL2, P130, Rb2 | retinoblastoma-like 2 (p130) |
C00002514
|
| 8241 | RBM10, DXS8237E, GPATC9, GPATCH9, S1-1, TARPS, ZRANB5 | RNA binding motif protein 10 |
C00002514
|
| 64783 | RBM15, OTT, OTT1, SPEN | RNA binding motif protein 15 |
C00002514
|
| 221662 | RBM24, RNPC6, dJ259A10.1 | RNA binding motif protein 24 |
C00002514
|
| 55285 | RBM41 | RNA binding motif protein 41 |
C00002514
|
| 5937 | RBMS1, C2orf12, HCC-4, MSSP, MSSP-1, MSSP-2, MSSP-3, SCR2, YC1 | RNA binding motif, single stranded interacting protein 1 |
C00002514
|
| 27316 | RBMX, HNRNPG, HNRPG, RBMXP1, RBMXRT, RNMX, hnRNP-G | RNA binding motif protein, X-linked |
C00002514
|
| 55213 | RCBTB1, CLLD7, CLLL7, GLP, RP11-185C18.1 | regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
C00002514
|
| 91433 | RCCD1 | RCC1 domain containing 1 |
C00002514
|
| 55758 | RCOR3, RP11-318L16.1 | REST corepressor 3 |
C00002514
|
| 112724 | RDH13, SDR7C3 | retinol dehydrogenase 13 (all-trans/9-cis) (EC:1.1.1.-) |
C00002514
|
| 201299 | RDM1, RAD52B | RAD52 motif 1 |
C00002514
|
| 9401 | RECQL4, RECQ4 | RecQ protein-like 4 (EC:3.6.4.12) |
C00002514
|
| 221035 | REEP3, C10orf74 | receptor accessory protein 3 |
C00002514
|
| 80346 | REEP4, C8orf20 | receptor accessory protein 4 |
C00002514
|
| 7905 | REEP5, C5orf18, D5S346, DP1, TB2, YOP1 | receptor accessory protein 5 |
C00002514
|
| 85004 | RERG | RAS-like, estrogen-regulated, growth inhibitor |
C00002514
|
| 5979 | RET, CDHF12, CDHR16, HSCR1, MEN2A, MEN2B, MTC1, PTC, RET-ELE1, RET51 | ret proto-oncogene (EC:2.7.10.1) |
C00002514
|
| 5982 | RFC2, RFC40 | replication factor C (activator 1) 2, 40kDa |
C00002514
|
| 5983 | RFC3, RFC38 | replication factor C (activator 1) 3, 38kDa |
C00002514
|
| 5984 | RFC4, A1, RFC37 | replication factor C (activator 1) 4, 37kDa |
C00002514
|
| 5985 | RFC5, RFC36 | replication factor C (activator 1) 5, 36.5kDa |
C00002514
|
| 23180 | RFTN1, PIB10, PIG9, RAFTLIN | raftlin, lipid raft linker 1 |
C00002514
|
| 283149 | BCL9L, BCL9-2, DLNB11 | B-cell CLL/lymphoma 9-like |
C00002514
|
| 64285 | RHBDF1, C16orf8, Dist1, EGFR-RS, gene-89, gene-90, hDist1 | rhomboid 5 homolog 1 (Drosophila) |
C00002514
|
| 83695 | RHNO1, C12orf32, RHINO | RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
C00002514
|
| 9886 | RHOBTB1 | Rho-related BTB domain containing 1 |
C00002514
|
| 23433 | RHOQ, ARHQ, RASL7A, TC10, TC10A | ras homolog family member Q |
C00002514
|
| 55288 | RHOT1, ARHT1, MIRO-1, MIRO1 | ras homolog family member T1 |
C00002514
|
| 26150 | RIBC2, C22orf11 | RIB43A domain with coiled-coils 2 |
C00002514
|
| 607 | BCL9, LGS | B-cell CLL/lymphoma 9 |
C00002514
|
| 151393 | RMDN2, FAM82A, FAM82A1, PRO34163, PYST9371, RMD-2, RMD2, RMD4 | regulator of microtubule dynamics 2 |
C00002514
|
| 80010 | RMI1, BLAP75, C9orf76, FAAP75, RP11-346I8.1 | RecQ mediated genome instability 1 |
C00002514
|
| 116028 | RMI2, BLAP18, C16orf75 | RecQ mediated genome instability 2 |
C00002514
|
| 10535 | RNASEH2A, AGS4, JUNB, RNASEHI, RNHIA, RNHL | ribonuclease H2, subunit A (EC:3.1.26.4) |
C00002514
|
| 602 | BCL3, BCL4, D19S37 | B-cell CLL/lymphoma 3 |
C00002514
|
| 83596 | BCL2L12 | BCL2-like 12 (proline rich) |
C00002514
|
| 149603 | RNF187, RACO-1, RACO1 | ring finger protein 187 |
C00002514
|
| 551328 |
C00002514
|
||
| 79102 | RNF26 | ring finger protein 26 |
C00002514
|
| 9475 | ROCK2, ROCK-II | Rho-associated, coiled-coil containing protein kinase 2 (EC:2.7.11.1) |
C00002514
|
| 6117 | RPA1, HSSB, MST075, REPA1, RF-A, RP-A, RPA70 | replication protein A1, 70kDa |
C00002514
|
| 6119 | RPA3, REPA3 | replication protein A3, 14kDa |
C00002514
|
| 6160 | RPL31, L31 | ribosomal protein L31 |
C00002514
|
| 6168 | RPL37A, L37A | ribosomal protein L37a |
C00002514
|
| 116832 | RPL39L, RPL39L1 | ribosomal protein L39-like |
C00002514
|
| 6184 | RPN1, OST1, RBPH1 | ribophorin I (EC:2.4.99.18) |
C00002514
|
| 56475 | RPRM, REPRIMO | reprimo, TP53 dependent G2 arrest mediator candidate |
C00002514
|
| 64121 | RRAGC, GTR2, RAGC, TIB929 | Ras-related GTP binding C |
C00002514
|
| 6240 | RRM1, R1, RIR1, RR1 | ribonucleotide reductase M1 (EC:1.17.4.1) |
C00002514
|
| 594 | BCKDHB, E1B, dJ279A18.1 | branched chain keto acid dehydrogenase E1, beta polypeptide (EC:1.2.4.4) |
C00002514
|
| 51018 | RRP15, KIAA0507 | ribosomal RNA processing 15 homolog (S. cerevisiae) |
C00002514
|
| 51773 | RSF1, HBXAP, RSF-1, XAP8, p325 | remodeling and spacing factor 1 |
C00002514
|
| 51319 | RSRC1, SFRS21, SRrp53 | arginine/serine-rich coiled-coil 1 |
C00002514
|
| 6251 | RSU1, RSP-1 | Ras suppressor protein 1 |
C00002514
|
| 6253 | RTN2, NSP2, NSPL1, NSPLI, SPG12 | reticulon 2 |
C00002514
|
| 10900 | RUNDC3A, RAP2IP, RPIP-8, RPIP8 | RUN domain containing 3A |
C00002514
|
| 154661 | RUNDC3B, RPIB9, RPIP9 | RUN domain containing 3B |
C00002514
|
| 10856 | RUVBL2, ECP51, INO80J, REPTIN, RVB2, TIH2, TIP48, TIP49B | RuvB-like AAA ATPase 2 (EC:3.6.4.12) |
C00002514
|
| 6259 | RYK, D3S3195, JTK5, JTK5A, RYK1 | receptor-like tyrosine kinase (EC:2.7.10.1) |
C00002514
|
| 54828 | BCAS3, GAOB1, MAAB | breast carcinoma amplified sequence 3 |
C00002514
|
| 6277 | S100A6, 2A9, 5B10, CABP, CACY, PRA | S100 calcium binding protein A6 |
C00002514
|
| 1903 | S1PR3, EDG-3, EDG3, LPB3, S1P3 | sphingosine-1-phosphate receptor 3 |
C00002514
|
| 113174 | SAAL1, SPACIA1 | serum amyloid A-like 1 |
C00002514
|
| 29901 | SAC3D1, HSU79266, SHD1 | SAC3 domain containing 1 |
C00002514
|
| 26278 | SACS, ARSACS, DNAJC29 | spastic ataxia of Charlevoix-Saguenay (sacsin) |
C00002514
|
| 10055 | SAE1, AOS1, HSPC140, SUA1, UBLE1A | SUMO1 activating enzyme subunit 1 |
C00002514
|
| 6294 | SAFB, HAP, HET, SAF-B1, SAFB1 | scaffold attachment factor B |
C00002514
|
| 57167 | SALL4, DRRS, HSAL4, ZNF797, dJ1112F19.1 | spalt-like transcription factor 4 |
C00002514
|
| 23034 | SAMD4A, SAMD4, SMAUG, SMAUG1, SMG, SMGA | sterile alpha motif domain containing 4A |
C00002514
|
| 582 | BBS1, BBS2L2 | Bardet-Biedl syndrome 1 |
C00002514
|
| 6303 | SAT1, DC21, KFSD, KFSDX, SAT, SSAT, SSAT-1 | spermidine/spermine N1-acetyltransferase 1 (EC:2.3.1.57) |
C00002514
|
| 23314 | SATB2 | SATB homeobox 2 |
C00002514
|
| 949 | SCARB1, CD36L1, CLA-1, CLA1, HDLQTL6, SR-BI, SRB1 | scavenger receptor class B, member 1 |
C00002514
|
| 950 | SCARB2, AMRF, CD36L2, EPM4, HLGP85, LGP85, LIMP-2, LIMPII, SR-BII | scavenger receptor class B, member 2 |
C00002514
|
| 51097 | SCCPDH, NET11, RP11-439E19.2 | saccharopine dehydrogenase (putative) (EC:1.5.1.9) |
C00002514
|
| 11176 | BAZ2A, TIP5, WALp3 | bromodomain adjacent to zinc finger domain, 2A |
C00002514
|
| 6383 | SDC2, CD362, HSPG, HSPG1, SYND2 | syndecan 2 |
C00002514
|
| 10483 | SEC23B, CDA-II, CDAII, CDAN2, HEMPAS | Sec23 homolog B (S. cerevisiae) |
C00002514
|
| 9871 | SEC24D | SEC24 family member D |
C00002514
|
| 29927 | SEC61A1, HSEC61, SEC61, SEC61A | Sec61 alpha 1 subunit (S. cerevisiae) |
C00002514
|
| 8991 | SELENBP1, LPSB, SBP56, SP56, hSBP | selenium binding protein 1 |
C00002514
|
| 362713 |
C00002514
|
||
| 51714 | SELT | selenoprotein T |
C00002514
|
| 100155979 |
C00002514
|
||
| 29843 | SENP1, SuPr-2 | SUMO1/sentrin specific peptidase 1 (EC:3.4.22.68) |
C00002514
|
| 22929 | SEPHS1, SELD, SPS, SPS1 | selenophosphate synthetase 1 (EC:2.7.9.3) |
C00002514
|
| 6414 | SEPP1, SELP, SeP | selenoprotein P, plasma, 1 |
C00002514
|
| 23176 | SEPT8, SEP2 | septin 8 |
C00002514
|
| 10801 | SEPT9, AF17q25, MSF, MSF1, NAPB, PNUTL4, SINT1, SeptD1 | septin 9 |
C00002514
|
| 94009 | SERHL, BK126B4.1, BK126B4.2, HS126B42, dJ222E13.1 | serine hydrolase-like |
C00002514
|
| 57515 | SERINC1, TDE1L, TDE2, TMS-2, TMS2 | serine incorporator 1 |
C00002514
|
| 256987 | SERINC5, C5orf12, TPO1 | serine incorporator 5 |
C00002514
|
| 1992 | SERPINB1, EI, ELANH2, LEI, M/NEI, MNEI, PI-2, PI2 | serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
C00002514
|
| 393917 |
C00002514
|
||
| 7536 | SF1, BBP, D11S636, MBBP, ZCCHC25, ZFM1, ZNF162 | splicing factor 1 |
C00002514
|
| 23450 | SF3B3, RSE1, SAP130, SF3b130, STAF130 | splicing factor 3b, subunit 3, 130kDa |
C00002514
|
| 2810 | SFN, YWHAS | stratifin |
C00002514
|
| 119392 | SFR1, C10orf78, MEI5, MEIR5, bA373N18.1 | SWI5-dependent recombination repair 1 |
C00002514
|
| 100380358 |
C00002514
|
||
| 100049528 |
C00002514
|
||
| 100305297 |
C00002514
|
||
| 94081 | SFXN1 | sideroflexin 1 |
C00002514
|
| 118980 | SFXN2 | sideroflexin 2 |
C00002514
|
| 6445 | SGCG, A4, DAGA4, DMDA, DMDA1, LGMD2C, MAM, SCARMD2, SCG3, TYPE | sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
C00002514
|
| 580 | BARD1 | BRCA1 associated RING domain 1 |
C00002514
|
| 151648 | SGOL1, NY-BR-85, SGO, Sgo1 | shugoshin-like 1 (S. pombe) |
C00002514
|
| 8815 | BANF1, BAF, BCRP1, D14S1460, NGPS | barrier to autointegration factor 1 |
C00002514
|
| 6451 | SH3BGRL, SH3BGR | SH3 domain binding glutamic acid-rich protein like |
C00002514
|
| 9532 | BAG2, BAG-2, dJ417I1.2 | BCL2-associated athanogene 2 |
C00002514
|
| 152503 | SH3D19, EBP, EVE1, Kryn, SH3P19 | SH3 domain containing 19 |
C00002514
|
| 30011 | SH3KBP1, CD2BP3, CIN85, GIG10, HSB-1, HSB1, MIG18 | SH3-domain kinase binding protein 1 |
C00002514
|
| 57630 | SH3RF1, POSH, RNF142, SH3MD2 | SH3 domain containing ring finger 1 |
C00002514
|
| 26751 | SH3YL1, RAY | SH3 and SYLF domain containing 1 |
C00002514
|
| 25825 | BACE2, AEPLC, ALP56, ASP1, ASP21, BAE2, CEAP1, DRAP | beta-site APP-cleaving enzyme 2 (EC:3.4.23.45) |
C00002514
|
| 387914 | SHISA2, C13orf13, PRO28631, TMEM46, WGAR9166, bA398O19.2, hShisa | shisa family member 2 |
C00002514
|
| 23621 | BACE1, ASP2, BACE, HSPC104 | beta-site APP-cleaving enzyme 1 (EC:3.4.23.46) |
C00002514
|
| 6472 | SHMT2, GLYA, SHMT | serine hydroxymethyltransferase 2 (mitochondrial) (EC:2.1.2.1) |
C00002514
|
| 57619 | SHROOM3, APXL3, SHRM, ShrmL | shroom family member 3 |
C00002514
|
| 8706 | B3GALNT1, B3GALT3, GLCT3, GLOB, Gb4Cer, P, P1, beta3Gal-T3, galT3 | beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) (EC:2.4.1.79) |
C00002514
|
| 220134 | SKA1, C18orf24 | spindle and kinetochore associated complex subunit 1 |
C00002514
|
| 348235 | SKA2, FAM33A | spindle and kinetochore associated complex subunit 2 |
C00002514
|
| 221150 | SKA3, C13orf3, RAMA1 | spindle and kinetochore associated complex subunit 3 |
C00002514
|
| 8935 | SKAP2, PRAP, RA70, SAPS, SCAP2, SKAP-HOM, SKAP55R | src kinase associated phosphoprotein 2 |
C00002514
|
| 6498 | SKIL, SNO, SnoA, SnoI, SnoN | SKI-like oncogene |
C00002514
|
| 567 | B2M | beta-2-microglobulin |
C00002514
|
| 7884 | SLBP, HBP | stem-loop binding protein |
C00002514
|
| 6558 | SLC12A2, BSC, BSC2, NKCC1 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
C00002514
|
| 6560 | SLC12A4, KCC1 | solute carrier family 12 (potassium/chloride transporter), member 4 |
C00002514
|
| 84561 | SLC12A8, CCC9 | solute carrier family 12, member 8 |
C00002514
|
| 26503 | SLC17A5, AST, ISSD, NSD, SD, SIALIN, SIASD, SLD | solute carrier family 17 (acidic sugar transporter), member 5 |
C00002514
|
| 33419 |
C00002514
|
||
| 63027 | SLC22A23, C6orf85 | solute carrier family 22, member 23 |
C00002514
|
| 8402 | SLC25A11, OGC, SLC20A4 | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
C00002514
|
| 10166 | SLC25A15, D13S327, HHH, ORC1, ORNT1 | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
C00002514
|
| 26053 | AUTS2, FBRSL2 | autism susceptibility candidate 2 |
C00002514
|
| 51312 | SLC25A37, MFRN, MFRN1, MSC, MSCP, PRO1278, PRO1584, PRO2217 | solute carrier family 25 (mitochondrial iron transporter), member 37 |
C00002514
|
| 51629 | SLC25A39, CGI69 | solute carrier family 25, member 39 |
C00002514
|
| 284129 | SLC26A11 | solute carrier family 26 (anion exchanger), member 11 |
C00002514
|
| 9212 | AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, PPP1R48, STK12, STK5, aurkb-sv1, aurkb-sv2 | aurora kinase B (EC:2.7.11.1) |
C00002514
|
| 11001 | SLC27A2, ACSVL1, FACVL1, FATP2, HsT17226, VLACS, VLCS, hFACVL1 | solute carrier family 27 (fatty acid transporter), member 2 (EC:6.2.1.3) |
C00002514
|
| 11000 | SLC27A3, ACSVL3, FATP3, VLCS-3 | solute carrier family 27 (fatty acid transporter), member 3 (EC:6.2.1.-) |
C00002514
|
| 2030 | SLC29A1, ENT1 | solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
C00002514
|
| 56731 | SLC2A4RG, GEF, HDBP-1, HDBP1, Si-1-2, Si-1-2-19 | SLC2A4 regulator |
C00002514
|
| 7779 | SLC30A1, ZNT1, ZRC1 | solute carrier family 30 (zinc transporter), member 1 |
C00002514
|
| 1317 | SLC31A1, COPT1, CTR1 | solute carrier family 31 (copper transporter), member 1 |
C00002514
|
| 23443 | SLC35A3 | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
C00002514
|
| 11046 | SLC35D2, HFRC1, SQV7L, UGTrel8, hfrc | solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
C00002514
|
| 79939 | SLC35E1 | solute carrier family 35, member E1 |
C00002514
|
| 6310 | ATXN1, ATX1, D6S504E, SCA1 | ataxin 1 |
C00002514
|
| 55630 | SLC39A4, AEZ, AWMS2, ZIP4 | solute carrier family 39 (zinc transporter), member 4 |
C00002514
|
| 10079 | ATP9A, ATPIIA | ATPase, class II, type 9A (EC:3.6.3.1) |
C00002514
|
| 57198 | ATP8B2, ATPID | ATPase, aminophospholipid transporter, class I, type 8B, member 2 (EC:3.6.3.1) |
C00002514
|
| 11254 | SLC6A14, BMIQ11 | solute carrier family 6 (amino acid transporter), member 14 |
C00002514
|
| 6541 | SLC7A1, ATRC1, CAT-1, ERR, HCAT1, REC1L | solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
C00002514
|
| 8992 | ATP6V0E1, ATP6H, ATP6V0E, M9.2, Vma21, Vma21p | ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (EC:3.6.3.14) |
C00002514
|
| 516 | ATP5G1, ATP5A, ATP5G | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
C00002514
|
| 27032 | ATP2C1, ATP2C1A, BCPM, HHD, PMR1, SPCA1, hSPCA1 | ATPase, Ca++ transporting, type 2C, member 1 (EC:3.6.3.8) |
C00002514
|
| 490 | ATP2B1, PMCA1, PMCA1kb | ATPase, Ca++ transporting, plasma membrane 1 (EC:3.6.3.8) |
C00002514
|
| 489 | ATP2A3, SERCA3 | ATPase, Ca++ transporting, ubiquitous (EC:3.6.3.8) |
C00002514
|
| 8243 | SMC1A, CDLS2, DXS423E, SB1.8, SMC1, SMC1L1, SMC1alpha, SMCB | structural maintenance of chromosomes 1A |
C00002514
|
| 22863 | ATG14, ATG14L, BARKOR, KIAA0831 | autophagy related 14 |
C00002514
|
| 22809 | ATF5, ATFX, HMFN0395 | activating transcription factor 5 |
C00002514
|
| 23137 | SMC5, SMC5L1 | structural maintenance of chromosomes 5 |
C00002514
|
| 201895 | SMIM14, C4orf34 | small integral membrane protein 14 |
C00002514
|
| 56950 | SMYD2, HSKM-B, KMT3C, ZMYND14 | SET and MYND domain containing 2 (EC:2.1.1.43) |
C00002514
|
| 8773 | SNAP23, HsT17016, SNAP-23, SNAP23A, SNAP23B | synaptosomal-associated protein, 23kDa |
C00002514
|
| 6619 | SNAPC3, PTFbeta, SNAP50 | small nuclear RNA activating complex, polypeptide 3, 50kDa |
C00002514
|
| 84973 | SNHG7, NCRNA00061 | small nucleolar RNA host gene 7 (non-protein coding) |
C00002514
|
| 8303 | SNN | stannin |
C00002514
|
| 677809 | SNORA24, ACA24 | small nucleolar RNA, H/ACA box 24 |
C00002514
|
| 26797 | SNORD52, RNU52, U52 | small nucleolar RNA, C/D box 52 |
C00002514
|
| 79622 | SNRNP25, C16orf33 | small nuclear ribonucleoprotein 25kDa (U11/U12) |
C00002514
|
| 6626 | SNRPA, Mud1, U1-A, U1A | small nuclear ribonucleoprotein polypeptide A |
C00002514
|
| 6627 | SNRPA1, Lea1 | small nuclear ribonucleoprotein polypeptide A' |
C00002514
|
| 6628 | SNRPB, COD, SNRPB1, Sm-B/B', SmB/B', SmB/SmB', snRNP-B | small nuclear ribonucleoprotein polypeptides B and B1 |
C00002514
|
| 6632 | SNRPD1, HsT2456, SMD1, SNRPD, Sm-D1 | small nuclear ribonucleoprotein D1 polypeptide 16kDa |
C00002514
|
| 1386 | ATF2, CRE-BP1, CREB2, HB16, TREB7 | activating transcription factor 2 (EC:2.3.1.48) |
C00002514
|
| 23161 | SNX13, RGS-PX1 | sorting nexin 13 |
C00002514
|
| 28966 | SNX24, PRO1284 | sorting nexin 24 |
C00002514
|
| 83891 | SNX25, SBBI31 | sorting nexin 25 |
C00002514
|
| 92017 | SNX29, A-388D4.1, RUNDC2A | sorting nexin 29 |
C00002514
|
| 58533 | SNX6, MSTP010, TFAF2 | sorting nexin 6 |
C00002514
|
| 51429 | SNX9, SDP1, SH3PX1, SH3PXD3A, WISP | sorting nexin 9 |
C00002514
|
| 55723 | ASF1B, CIA-II | anti-silencing function 1B histone chaperone |
C00002514
|
| 9655 | SOCS5, CIS6, CISH6, Cish5, SOCS-5 | suppressor of cytokine signaling 5 |
C00002514
|
| 6658 | SOX3, GHDX, MRGH, PHP, PHPX, SOXB | SRY (sex determining region Y)-box 3 |
C00002514
|
| 170302 | ARX, CT121, EIEE1, ISSX, MRX29, MRX32, MRX33, MRX36, MRX38, MRX43, MRX54, MRX76, MRX87, MRXS1, PRTS | aristaless related homeobox |
C00002514
|
| 414 | ARSD, ASD | arylsulfatase D (EC:3.1.6.1) |
C00002514
|
| 10615 | SPAG5, DEEPEST, MAP126, hMAP126 | sperm associated antigen 5 |
C00002514
|
| 64847 | SPATA20, SSP411, Tisp78 | spermatogenesis associated 20 |
C00002514
|
| 124044 | SPATA2L, C16orf76, tamo | spermatogenesis associated 2-like |
C00002514
|
| 55812 | SPATA7, HSD-3.1, HSD3, LCA3 | spermatogenesis associated 7 |
C00002514
|
| 147841 | SPC24, SPBC24 | SPC24, NDC80 kinetochore complex component |
C00002514
|
| 35539 |
C00002514
|
||
| 25803 | SPDEF, PDEF, RP11-375E1__A.3, bA375E1.3 | SAM pointed domain containing ETS transcription factor |
C00002514
|
| 54908 | SPDL1, CCDC99 | spindle apparatus coiled-coil protein 1 |
C00002514
|
| 23111 | SPG20, SPARTIN, TAHCCP1 | spastic paraplegia 20 (Troyer syndrome) |
C00002514
|
| 139886 | SPIN4 | spindlin family, member 4 |
C00002514
|
| 84501 | SPIRE2, Spir-2 | spire-type actin nucleation factor 2 |
C00002514
|
| 200734 | SPRED2, Spred-2 | sprouty-related, EVH1 domain containing 2 |
C00002514
|
| 10558 | SPTLC1, HSAN1, HSN1, LBC1, LCB1, SPT1, SPTI | serine palmitoyltransferase, long chain base subunit 1 (EC:2.3.1.50) |
C00002514
|
| 57561 | ARRDC3, TLIMP | arrestin domain containing 3 |
C00002514
|
| 403 | ARL3, ARFL3 | ADP-ribosylation factor-like 3 |
C00002514
|
| 79644 | SRD5A3, CDG1P, CDG1Q, KRIZI, SRD5A2L, SRD5A2L1 | steroid 5 alpha-reductase 3 (EC:1.3.1.22 1.3.1.94) |
C00002514
|
| 6717 | SRI, CP-22, CP22, SCN, V19 | sorcin |
C00002514
|
| 6723 | SRM, PAPT, SPDSY, SPS1, SRML1 | spermidine synthase (EC:2.5.1.16) |
C00002514
|
| 58477 | SRPRB, APMCF1 | signal recognition particle receptor, B subunit |
C00002514
|
| 63826 | SRR, ILV1, ISO1 | serine racemase (EC:4.3.1.17 5.1.1.18 4.3.1.18) |
C00002514
|
| 23524 | SRRM2, 300-KD, CWF21, Cwc21, SRL300, SRm300 | serine/arginine repetitive matrix 2 |
C00002514
|
| 10772 | SRSF10, FUSIP1, FUSIP2, NSSR, SFRS13, SFRS13A, SRp38, SRrp40, TASR, TASR1, TASR2 | serine/arginine-rich splicing factor 10 |
C00002514
|
| 23635 | SSBP2, HSPC116, SOSS-B2 | single-stranded DNA binding protein 2 |
C00002514
|
| 6749 | SSRP1, FACT, FACT80, T160 | structure specific recognition protein 1 |
C00002514
|
| 117178 | SSX2IP, ADIP | synovial sarcoma, X breakpoint 2 interacting protein |
C00002514
|
| 51742 | ARID4B, BCAA, BRCAA1, RBBP1L1, RBP1L1, SAP180 | AT rich interactive domain 4B (RBP1-like) |
C00002514
|
| 7903 | ST8SIA4, PST, PST1, SIAT8D, ST8SIA-IV | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (EC:2.4.99.-) |
C00002514
|
| 338596 | ST8SIA6, SIA8F, SIAT8F, ST8SIA-VI | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 (EC:2.4.99.-) |
C00002514
|
| 10274 | STAG1, SA1, SCC3A | stromal antigen 1 |
C00002514
|
| 84904 | ARHGEF39, C9orf100, RP11-331F9.7 | Rho guanine nucleotide exchange factor (GEF) 39 |
C00002514
|
| 389337 | ARHGEF37 | Rho guanine nucleotide exchange factor (GEF) 37 |
C00002514
|
| 26084 | ARHGEF26, CSGEF, HMFN1864, SGEF | Rho guanine nucleotide exchange factor (GEF) 26 |
C00002514
|
| 6491 | STIL, MCPH7, SIL | SCL/TAL1 interrupting locus |
C00002514
|
| 10963 | STIP1, HOP, IEF-SSP-3521, P60, STI1, STI1L | stress-induced-phosphoprotein 1 |
C00002514
|
| 23012 | STK38L, NDR2 | serine/threonine kinase 38 like (EC:2.7.11.1) |
C00002514
|
| 6789 | STK4, KRS2, MST1, TIIAC, YSK3 | serine/threonine kinase 4 (EC:2.7.11.1) |
C00002514
|
| 412271 |
C00002514
|
||
| 3925 | STMN1, C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | stathmin 1 |
C00002514
|
| 85439 | STON2, STN2, STNB, STNB2 | stonin 2 |
C00002514
|
| 92335 | STRADA, LYK5, NY-BR-96, PMSE, STRAD, Stlk | STE20-related kinase adaptor alpha |
C00002514
|
| 29966 | STRN3, SG2NA | striatin, calmodulin binding protein 3 |
C00002514
|
| 55014 | STX17 | syntaxin 17 |
C00002514
|
| 80728 | ARHGAP39, CrGAP, Vilse | Rho GTPase activating protein 39 |
C00002514
|
| 8417 | STX7 | syntaxin 7 |
C00002514
|
| 23213 | SULF1, HSULF-1, SULF-1 | sulfatase 1 |
C00002514
|
| 84986 | ARHGAP19 | Rho GTPase activating protein 19 |
C00002514
|
| 6820 | SULT2B1, HSST2 | sulfotransferase family, cytosolic, 2B, member 1 (EC:2.8.2.2) |
C00002514
|
| 11198 | SUPT16H, CDC68, FACTP140, SPT16/CDC68 | suppressor of Ty 16 homolog (S. cerevisiae) |
C00002514
|
| 6839 | SUV39H1, H3-K9-HMTase_1, KMT1A, MG44, SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00002514
|
| 79723 | SUV39H2, KMT1B | suppressor of variegation 3-9 homolog 2 (Drosophila) (EC:2.1.1.43) |
C00002514
|
| 51111 | SUV420H1, CGI85, KMT5B | suppressor of variegation 4-20 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00002514
|
| 23075 | SWAP70, SWAP-70 | SWAP switching B-cell complex 70kDa subunit |
C00002514
|
| 10564 | ARFGEF2, BIG2, PVNH2, dJ1164I10.1 | ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
C00002514
|
| 256126 | SYCE2, CESC1 | synaptonemal complex central element protein 2 |
C00002514
|
| 84144 | SYDE2, RP11-33E12.1 | synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
C00002514
|
| 10492 | SYNCRIP, GRY-RBP, GRYRBP, HNRNPQ, HNRPQ1, NSAP1, PP68, hnRNP-Q | synaptotagmin binding, cytoplasmic RNA interacting protein |
C00002514
|
| 23224 | SYNE2, EDMD5, NUA, NUANCE, Nesp2, Nesprin-2, SYNE-2, TROPH | spectrin repeat containing, nuclear envelope 2 |
C00002514
|
| 9143 | SYNGR3 | synaptogyrin 3 |
C00002514
|
| 55333 | SYNJ2BP, ARIP2, OMP25 | synaptojanin 2 binding protein |
C00002514
|
| 51760 | SYT17 | synaptotagmin XVII |
C00002514
|
| 54843 | SYTL2, CHR11SYT, EXO4, SGA72M, SLP2, SLP2A | synaptotagmin-like 2 |
C00002514
|
| 351 | APP, AAA, ABETA, ABPP, AD1, APPI, CTFgamma, CVAP, PN-II, PN2 | amyloid beta (A4) precursor protein |
C00002514
|
| 378708 | APITD1, CENP-S, CENPS, FAAP16, MHF1 | apoptosis-inducing, TAF9-like domain 1 |
C00002514
|
| 23118 | TAB2, CHTD2, MAP3K7IP2 | TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
C00002514
|
| 10460 | TACC3, ERIC-1, ERIC1 | transforming, acidic coiled-coil containing protein 3 |
C00002514
|
| 323 | APBB2, FE65L, FE65L1 | amyloid beta (A4) precursor protein-binding, family B, member 2 |
C00002514
|
| 6883 | TAF12, TAF2J, TAFII20 | TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
C00002514
|
| 6877 | TAF5, TAF2D, TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
C00002514
|
| 26115 | TANC2, ROLSA, rols | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
C00002514
|
| 57551 | TAOK1, KFC-B, MAP3K16, MARKK, PSK-2, PSK2, TAO1, hKFC-B, hTAOK1 | TAO kinase 1 (EC:2.7.11.1) |
C00002514
|
| 51347 | TAOK3, DPK, JIK, MAP3K18 | TAO kinase 3 (EC:2.7.11.1) |
C00002514
|
| 80222 | TARS2, TARSL1, thrRS | threonyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.3) |
C00002514
|
| 9797 | TATDN2 | TatD DNase domain containing 2 |
C00002514
|
| 57465 | TBC1D24, EIEE16, FIME, TLDC6 | TBC1 domain family, member 24 |
C00002514
|
| 6904 | TBCD, SSD-1, tfcD | tubulin folding cofactor D |
C00002514
|
| 6907 | TBL1X, EBI, SMAP55, TBL1 | transducin (beta)-like 1X-linked |
C00002514
|
| 1175 | AP2S1, AP17, AP17-DELTA, CLAPS2, HHC3 | adaptor-related protein complex 2, sigma 1 subunit |
C00002514
|
| 6941 | TCF19, SC1, TCF-19 | transcription factor 19 |
C00002514
|
| 83439 | TCF7L1, TCF-3, TCF3 | transcription factor 7-like 1 (T-cell specific, HMG-box) |
C00002514
|
| 55775 | TDP1 | tyrosyl-DNA phosphodiesterase 1 (EC:3.1.4.-) |
C00002514
|
| 26136 | TES, TESS, TESS-2 | testis derived transcript (3 LIM domains) |
C00002514
|
| 93081 | TEX30, C13orf27 | testis expressed 30 |
C00002514
|
| 7022 | TFAP2C, AP2-GAMMA, ERF1, TFAP2G, hAP-2g | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
C00002514
|
| 7027 | TFDP1, DP1, DRTF1, Dp-1 | transcription factor Dp-1 |
C00002514
|
| 8907 | AP1M1, AP47, CLAPM2, CLTNM, MU-1A | adaptor-related protein complex 1, mu 1 subunit |
C00002514
|
| 162 | AP1B1, ADTB1, AP105A, BAM22, CLAPB2 | adaptor-related protein complex 1, beta 1 subunit |
C00002514
|
| 8416 | ANXA9, ANX31 | annexin A9 |
C00002514
|
| 7037 | TFRC, CD71, T9, TFR, TFR1, TR, TRFR, p90 | transferrin receptor |
C00002514
|
| 304 | ANXA2P2, ANX2L2, ANX2P2, LPC2B | annexin A2 pseudogene 2 |
C00002514
|
| 7046 | TGFBR1, AAT5, ACVRLK4, ALK-5, ALK5, LDS1A, LDS2A, MSSE, SKR4, TGFR-1 | transforming growth factor, beta receptor 1 (EC:2.7.11.30) |
C00002514
|
| 302 | ANXA2, ANX2, ANX2L4, CAL1H, LIP2, LPC2, LPC2D, P36, PAP-IV | annexin A2 |
C00002514
|
| 60436 | TGIF2 | TGFB-induced factor homeobox 2 |
C00002514
|
| 81611 | ANP32E, LANP-L, LANPL | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
C00002514
|
| 51337 | THEM6, C8orf55, DSCD75 | thioesterase superfamily member 6 |
C00002514
|
| 8563 | THOC5, C22orf19, Fmip, PK1.3, fSAP79 | THO complex 5 |
C00002514
|
| 7064 | THOP1, EP24.15, MEPD_HUMAN, MP78, TOP | thimet oligopeptidase 1 (EC:3.4.24.15) |
C00002514
|
| 8125 | ANP32A, C15orf1, HPPCn, I1PP2A, LANP, MAPM, PHAP1, PHAPI, PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
C00002514
|
| 90381 | TICRR, C15orf42, SLD3, Treslin | TOPBP1-interacting checkpoint and replication regulator |
C00002514
|
| 8914 | TIMELESS, TIM, TIM1, hTIM | timeless circadian clock |
C00002514
|
| 29090 | TIMM21, C18orf55, TIM21 | translocase of inner mitochondrial membrane 21 homolog (yeast) |
C00002514
|
| 7077 | TIMP2, CSC-21K, DDC8 | TIMP metallopeptidase inhibitor 2 |
C00002514
|
| 196527 | ANO6, BDPLT7, SCTS, TMEM16F | anoctamin 6 |
C00002514
|
| 54962 | TIPIN | TIMELESS interacting protein |
C00002514
|
| 84250 | ANKRD32, BRCTD1, BRCTx | ankyrin repeat domain 32 |
C00002514
|
| 116238 | TLCD1 | TLC domain containing 1 |
C00002514
|
| 7088 | TLE1, ESG, ESG1, GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
C00002514
|
| 11011 | TLK2, HsHPK, PKU-ALPHA | tousled-like kinase 2 (EC:2.7.11.1) |
C00002514
|
| 4071 | TM4SF1, H-L6, L6, M3S1, TAAL6 | transmembrane 4 L six family member 1 |
C00002514
|
| 253650 | ANKRD18A | ankyrin repeat domain 18A |
C00002514
|
| 147798 | TMC4 | transmembrane channel-like 4 |
C00002514
|
| 23023 | TMCC1 | transmembrane and coiled-coil domain family 1 |
C00002514
|
| 51434 | ANAPC7, APC7 | anaphase promoting complex subunit 7 |
C00002514
|
| 54664 | TMEM106B | transmembrane protein 106B |
C00002514
|
| 79022 | TMEM106C | transmembrane protein 106C |
C00002514
|
| 84314 | TMEM107, GRVS638, PRO1268 | transmembrane protein 107 |
C00002514
|
| 129303 | TMEM150A, TM6P1, TMEM150 | transmembrane protein 150A |
C00002514
|
| 441027 | TMEM150C | transmembrane protein 150C |
C00002514
|
| 51433 | ANAPC5, APC5 | anaphase promoting complex subunit 5 |
C00002514
|
| 64418 | TMEM168 | transmembrane protein 168 |
C00002514
|
| 57583 | TMEM181, GPR178, KIAA1423 | transmembrane protein 181 |
C00002514
|
| 55266 | TMEM19 | transmembrane protein 19 |
C00002514
|
| 23306 | TMEM194A, TMEM194 | transmembrane protein 194A |
C00002514
|
| 29058 | TMEM230, C20orf30, HSPC274, dJ1116H23.2.1 | transmembrane protein 230 |
C00002514
|
| 57393 | TMEM27, NX-17, NX17 | transmembrane protein 27 |
C00002514
|
| 55754 | TMEM30A, C6orf67, CDC50A | transmembrane protein 30A |
C00002514
|
| 55151 | TMEM38B, C9orf87, D4Ertd89e, OI14, TRIC-B, TRICB, bA219P18.1 | transmembrane protein 38B |
C00002514
|
| 120224 | TMEM45B | transmembrane protein 45B |
C00002514
|
| 51421 | AMOTL2, LCCP | angiomotin like 2 |
C00002514
|
| 169200 | TMEM64 | transmembrane protein 64 |
C00002514
|
| 157378 | TMEM65 | transmembrane protein 65 |
C00002514
|
| 267 | AMFR, GP78, RNF45 | autocrine motility factor receptor, E3 ubiquitin protein ligase (EC:6.3.2.19) |
C00002514
|
| 7112 | TMPO, CMD1T, LAP2, LEMD4, PRO0868, TP | thymopoietin |
C00002514
|
| 262 | AMD1, ADOMETDC, AMD, SAMDC | adenosylmethionine decarboxylase 1 (EC:4.1.1.50) |
C00002514
|
| 10189 | ALYREF, ALY, ALY/REF, BEF, REF, THOC4 | Aly/REF export factor |
C00002514
|
| 160335 | TMTC2, IBDBP1 | transmembrane and tetratricopeptide repeat containing 2 |
C00002514
|
| 81542 | TMX1, PDIA11, TMX, TXNDC, TXNDC1 | thioredoxin-related transmembrane protein 1 |
C00002514
|
| 56255 | TMX4, DJ971N18.2, PDIA14, TXNDC13 | thioredoxin-related transmembrane protein 4 |
C00002514
|
| 7126 | TNFAIP1, B12, B61, BTBD34, EDP1, hBACURD2 | tumor necrosis factor, alpha-induced protein 1 (endothelial) |
C00002514
|
| 51330 | TNFRSF12A, CD266, FN14, TWEAKR | tumor necrosis factor receptor superfamily, member 12A |
C00002514
|
| 702262 |
C00002514
|
||
| 57679 | ALS2, ALS2CR6, ALSJ, IAHSP, PLSJ | amyotrophic lateral sclerosis 2 (juvenile) |
C00002514
|
| 114034 | TOE1 | target of EGR1, member 1 (nuclear) |
C00002514
|
| 84266 | ALKBH7, ABH7, SPATA11, UNQ6002 | alkB, alkylation repair homolog 7 (E. coli) |
C00002514
|
| 11073 | TOPBP1, TOP2BP1 | topoisomerase (DNA) II binding protein 1 |
C00002514
|
| 94241 | TP53INP1, SIP, TP53DINP1, TP53INP1A, TP53INP1B, Teap, p53DINP1 | tumor protein p53 inducible nuclear protein 1 |
C00002514
|
| 58476 | TP53INP2, C20orf110, DOR, PINH, dJ1181N3.1 | tumor protein p53 inducible nuclear protein 2 |
C00002514
|
| 7163 | TPD52, D52, N8L, PC-1, PrLZ, hD52 | tumor protein D52 |
C00002514
|
| 79053 | ALG8, CDG1H | ALG8, alpha-1,3-glucosyltransferase (EC:2.4.1.265) |
C00002514
|
| 7167 | TPI1, TIM, TPI, TPID | triosephosphate isomerase 1 (EC:5.3.1.1) |
C00002514
|
| 29929 | ALG6, CDG1C | ALG6, alpha-1,3-glucosyltransferase (EC:2.4.1.267) |
C00002514
|
| 127262 | TPRG1L, FAM79A, RP11-46F15.3, h-mover, mover | tumor protein p63 regulated 1-like |
C00002514
|
| 7179 | TPTE, CT44, PTEN2 | transmembrane phosphatase with tensin homology (EC:3.1.3.48) |
C00002514
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00002514
|
| 6434 | TRA2B, Htra2-beta, SFRS10, SRFS10, TRA2-BETA, TRAN2B | transformer 2 beta homolog (Drosophila) |
C00002514
|
| 9618 | TRAF4, CART1, MLN62, RNF83 | TNF receptor-associated factor 4 |
C00002514
|
| 6399 | TRAPPC2, MIP2A, SEDL, SEDT, TRAPPC2P1, TRS20, ZNF547L, hYP38334 | trafficking protein particle complex 2 |
C00002514
|
| 22878 | TRAPPC8, GSG1, HsT2706, KIAA1012, TRS85 | trafficking protein particle complex 8 |
C00002514
|
| 126133 | ALDH16A1 | aldehyde dehydrogenase 16 family, member A1 |
C00002514
|
| 10221 | TRIB1, C8FW, GIG-2, GIG2, SKIP1, TRB-1, TRB1 | tribbles pseudokinase 1 |
C00002514
|
| 286827 | TRIM59, MRF1, RNF104, TRIM57, TSBF1 | tripartite motif containing 59 |
C00002514
|
| 11078 | TRIOBP, DFNB28, TARA, dJ37E16.4 | TRIO and F-actin binding protein |
C00002514
|
| 9319 | TRIP13, 16E1BP | thyroid hormone receptor interactor 13 |
C00002514
|
| 93587 | TRMT10A, RG9MTD2, TRM10 | tRNA methyltransferase 10 homolog A (S. cerevisiae) |
C00002514
|
| 57570 | TRMT5, KIAA1393, TRM5 | tRNA methyltransferase 5 (EC:2.1.1.228) |
C00002514
|
| 51605 | TRMT6, GCD10, Gcd10p | tRNA methyltransferase 6 homolog (S. cerevisiae) |
C00002514
|
| 10024 | TROAP, TASTIN | trophinin associated protein |
C00002514
|
| 7249 | TSC2, LAM, TSC4 | tuberous sclerosis 2 |
C00002514
|
| 116461 | TSEN15, C1orf19, sen15 | TSEN15 tRNA splicing endonuclease subunit |
C00002514
|
| 7247 | TSN, BCLF-1, C3PO, RCHF1, REHF-1, TBRBP, TRSLN | translin |
C00002514
|
| 10103 | TSPAN1, NET1, TM4C, TM4SF | tetraspanin 1 |
C00002514
|
| 6302 | TSPAN31, SAS | tetraspanin 31 |
C00002514
|
| 7259 | TSPYL1, TSPYL | TSPY-like 1 |
C00002514
|
| 158219 | TTC39B, C9orf52 | tetratricopeptide repeat domain 39B |
C00002514
|
| 23508 | TTC9, TTC9A | tetratricopeptide repeat domain 9 |
C00002514
|
| 84962 | AJUBA, JUB | ajuba LIM protein |
C00002514
|
| 7272 | TTK, CT96, ESK, MPH1, MPS1, MPS1L1, PYT | TTK protein kinase (EC:2.7.12.1) |
C00002514
|
| 406303 |
C00002514
|
||
| 10598 | AHSA1, AHA1, C14orf3, p38 | AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
C00002514
|
| 10768 | AHCYL1, DCAL, IRBIT, PRO0233, XPVKONA | adenosylhomocysteinase-like 1 (EC:3.3.1.1) |
C00002514
|
| 100691897 |
C00002514
|
||
| 51174 | TUBD1, TUBD | tubulin, delta 1 |
C00002514
|
| 7283 | TUBG1, CDCBM4, GCP-1, TUBG, TUBGCP1 | tubulin, gamma 1 |
C00002514
|
| 27229 | TUBGCP4, 76P, GCP-4, GCP4, Grip76 | tubulin, gamma complex associated protein 4 |
C00002514
|
| 7286 | TUFT1 | tuftelin 1 |
C00002514
|
| 55000 | TUG1, LINC00080, NCRNA00080, TI-227H | taurine up-regulated 1 (non-protein coding) |
C00002514
|
| 56995 | TULP4, TUSP | tubby like protein 4 |
C00002514
|
| 57085 | AGTRAP, ATRAP | angiotensin II receptor-associated protein |
C00002514
|
| 155465 | AGR3, AG3, BCMP11, HAG3, PDIA18, hAG-3 | anterior gradient 3 |
C00002514
|
| 283991 | UBALD2, FAM100B | UBA-like domain containing 2 |
C00002514
|
| 7320 | UBE2B, E2-17kDa, HHR6B, HR6B, RAD6B, UBC2 | ubiquitin-conjugating enzyme E2B (EC:6.3.2.19) |
C00002514
|
| 7321 | UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A | ubiquitin-conjugating enzyme E2D 1 (EC:6.3.2.19) |
C00002514
|
| 7322 | UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B | ubiquitin-conjugating enzyme E2D 2 (EC:6.3.2.19) |
C00002514
|
| 10551 | AGR2, AG2, GOB-4, HAG-2, PDIA17, XAG-2 | anterior gradient 2 |
C00002514
|
| 51465 | UBE2J1, HSPC153, HSPC205, HSU93243, NCUBE-1, NCUBE1, UBC6, Ubc6p | ubiquitin-conjugating enzyme E2, J1 (EC:6.3.2.19) |
C00002514
|
| 7334 | UBE2N, UBC13, UbcH-ben, UbcH13 | ubiquitin-conjugating enzyme E2N (EC:6.3.2.19) |
C00002514
|
| 134111 | UBE2QL1 | ubiquitin-conjugating enzyme E2Q family-like 1 (EC:6.3.2.19) |
C00002514
|
| 29089 | UBE2T, PIG50 | ubiquitin-conjugating enzyme E2T (putative) (EC:6.3.2.19) |
C00002514
|
| 55284 | UBE2W, UBC-16, UBC16 | ubiquitin-conjugating enzyme E2W (putative) (EC:6.3.2.19) |
C00002514
|
| 55148 | UBR7, C14orf130 | ubiquitin protein ligase E3 component n-recognin 7 (putative) |
C00002514
|
| 51377 | UCHL5, CGI-70, INO80R, UCH-L5, UCH37 | ubiquitin carboxyl-terminal hydrolase L5 (EC:3.4.19.12) |
C00002514
|
| 7371 | UCK2, TSA903, UK, UMPK | uridine-cytidine kinase 2 (EC:2.7.1.48) |
C00002514
|
| 7358 | UGDH, GDH, UDP-GlcDH, UDPGDH, UGD | UDP-glucose 6-dehydrogenase (EC:1.1.1.22) |
C00002514
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00002514
|
| 127933 | UHMK1, KIS, KIST, P-CIP2 | U2AF homology motif (UHM) kinase 1 (EC:2.7.11.1) |
C00002514
|
| 29128 | UHRF1, ICBP90, Np95, RNF106, hNP95, hUHRF1 | ubiquitin-like with PHD and ring finger domains 1 (EC:6.3.2.19) |
C00002514
|
| 137964 | AGPAT6, 1-AGPAT_6, LPAAT-zeta, LPAATZ, TSARG7 | 1-acylglycerol-3-phosphate O-acyltransferase 6 (EC:2.3.1.15) |
C00002514
|
| 56894 | AGPAT3, LPAAT-GAMMA1, LPAAT3 | 1-acylglycerol-3-phosphate O-acyltransferase 3 (EC:2.3.1.51) |
C00002514
|
| 65109 | UPF3B, HUPF3B, MRXS14, RENT3B, UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) |
C00002514
|
| 84300 | UQCC2, C6orf125, Cbp6, M19, MNF1, bA6B20.2 | ubiquinol-cytochrome c reductase complex assembly factor 2 (EC:2.7.4.21) |
C00002514
|
| 7398 | USP1, UBP | ubiquitin specific peptidase 1 (EC:3.4.19.12) |
C00002514
|
| 9960 | USP3, SIH003, UBP | ubiquitin specific peptidase 3 (EC:3.4.19.12) |
C00002514
|
| 7375 | USP4, UNP, Unph | ubiquitin specific peptidase 4 (proto-oncogene) (EC:3.4.19.12) |
C00002514
|
| 55230 | USP40 | ubiquitin specific peptidase 40 (EC:3.4.19.12) |
C00002514
|
| 8078 | USP5, ISOT | ubiquitin specific peptidase 5 (isopeptidase T) (EC:3.4.19.12) |
C00002514
|
| 159195 | USP54, C10orf29, bA137L10.3, bA137L10.4 | ubiquitin specific peptidase 54 |
C00002514
|
| 27161 | AGO2, EIF2C2, Q10 | argonaute RISC catalytic component 2 |
C00002514
|
| 7407 | VARS, G7A, VARS1, VARS2 | valyl-tRNA synthetase (EC:6.1.1.9) |
C00002514
|
| 57176 | VARS2, G7a, VARS2L, VARSL | valyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.9) |
C00002514
|
| 10493 | VAT1, VATI | vesicle amine transport 1 |
C00002514
|
| 7416 | VDAC1, PORIN, VDAC-1 | voltage-dependent anion channel 1 |
C00002514
|
| 116987 | AGAP1, AGAP-1, CENTG2, GGAP1, cnt-g2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
C00002514
|
| 9686 | VGLL4, VGL-4 | vestigial like 4 (Drosophila) |
C00002514
|
| 79001 | VKORC1, EDTP308, IMAGE3455200, MST134, MST576, VKCFD2, VKOR | vitamin K epoxide reductase complex, subunit 1 (EC:1.1.4.1) |
C00002514
|
| 54832 | VPS13C | vacuolar protein sorting 13 homolog C (S. cerevisiae) |
C00002514
|
| 40542 |
C00002514
|
||
| 27072 | VPS41, HVPS41, HVSP41, hVps41p | vacuolar protein sorting 41 homolog (S. cerevisiae) |
C00002514
|
| 11311 | VPS45, H1, H1VPS45, SCN5, VPS45A, VPS45B, VPS54A, VSP45, VSP45A | vacuolar protein sorting 45 homolog (S. cerevisiae) |
C00002514
|
| 7443 | VRK1, PCH1, PCH1A | vaccinia related kinase 1 (EC:2.7.11.1) |
C00002514
|
| 374666 | WASH3P, FAM39DP | WAS protein family homolog 3 pseudogene |
C00002514
|
| 23559 | WBP1, WBP-1 | WW domain binding protein 1 |
C00002514
|
| 27125 | AFF4, AF5Q31, MCEF | AF4/FMR2 family, member 4 |
C00002514
|
| 89891 | WDR34, DIC5, FAP133, bA216B9.3 | WD repeat domain 34 |
C00002514
|
| 60312 | AFAP1, AFAP, AFAP-110, AFAP110 | actin filament associated protein 1 |
C00002514
|
| 84058 | WDR54 | WD repeat domain 54 |
C00002514
|
| 79968 | WDR76, CDW14 | WD repeat domain 76 |
C00002514
|
| 33865 |
C00002514
|
||
| 7468 | WHSC1, MMSET, NSD2, REIIBP, TRX5, WHS | Wolf-Hirschhorn syndrome candidate 1 (EC:2.1.1.43) |
C00002514
|
| 55062 | WIPI1, ATG18, ATG18A, WIPI49 | WD repeat domain, phosphoinositide interacting 1 |
C00002514
|
| 26100 | WIPI2, ATG18B, Atg21, WIPI-2 | WD repeat domain, phosphoinositide interacting 2 |
C00002514
|
| 132 | ADK, AK | adenosine kinase (EC:2.7.1.20) |
C00002514
|
| 58525 | WIZ, ZNF803 | widely interspaced zinc finger motifs |
C00002514
|
| 55884 | WSB2, SBA2 | WD repeat and SOCS box containing 2 |
C00002514
|
| 80014 | WWC2, BOMB | WW and C2 domain containing 2 |
C00002514
|
| 55841 | WWC3, BM042 | WWC family member 3 |
C00002514
|
| 11059 | WWP1, AIP5, Tiul1, hSDRP1 | WW domain containing E3 ubiquitin protein ligase 1 |
C00002514
|
| 23039 | XPO7, EXP7, RANBP16 | exportin 7 |
C00002514
|
| 2547 | XRCC6, CTC75, CTCBF, G22P1, KU70, ML8, TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 |
C00002514
|
| 91419 | XRCC6BP1, KUB3 | XRCC6 binding protein 1 (EC:3.4.24.-) |
C00002514
|
| 4904 | YBX1, BP-8, CSDA2, CSDB, DBPB, MDR-NF1, NSEP-1, NSEP1, YB-1, YB1 | Y box binding protein 1 |
C00002514
|
| 388403 | YPEL2 | yippee-like 2 (Drosophila) |
C00002514
|
| 51646 | YPEL5 | yippee-like 5 (Drosophila) |
C00002514
|
| 7529 | YWHAB, GW128, HS1, KCIP-1, YWHAA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
C00002514
|
| 7531 | YWHAE, 14-3-3E, KCIP-1, MDCR, MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
C00002514
|
| 22890 | ZBTB1, ZNF909 | zinc finger and BTB domain containing 1 |
C00002514
|
| 65986 | ZBTB10, RINZF | zinc finger and BTB domain containing 10 |
C00002514
|
| 57684 | ZBTB26, ZNF481, bioref | zinc finger and BTB domain containing 26 |
C00002514
|
| 253461 | ZBTB38, CIBZ, ZNF921 | zinc finger and BTB domain containing 38 |
C00002514
|
| 57659 | ZBTB4, KAISO-L1, ZNF903 | zinc finger and BTB domain containing 4 |
C00002514
|
| 140685 | ZBTB46, BTBD4, BZEL, RINZF, ZNF340, dJ583P15.7, dJ583P15.8 | zinc finger and BTB domain containing 46 |
C00002514
|
| 339487 | ZBTB8OS, ARCH, ARCH2 | zinc finger and BTB domain containing 8 opposite strand |
C00002514
|
| 51101 | ZC2HC1A, C8orf70, FAM164A | zinc finger, C2HC-type containing 1A |
C00002514
|
| 80149 | ZC3H12A, MCPIP, MCPIP1, RP3-423B22.1, dJ423B22.1 | zinc finger CCCH-type containing 12A |
C00002514
|
| 120 | ADD3, ADDL | adducin 3 (gamma) |
C00002514
|
| 23390 | ZDHHC17, HIP14, HIP3, HYPH | zinc finger, DHHC-type containing 17 (EC:2.3.1.225) |
C00002514
|
| 64429 | ZDHHC6, ZNF376 | zinc finger, DHHC-type containing 6 (EC:2.3.1.225) |
C00002514
|
| 55625 | ZDHHC7, DHHC7, SERZ-B, SERZ1, ZNF370 | zinc finger, DHHC-type containing 7 (EC:2.3.1.225) |
C00002514
|
| 90637 | ZFAND2A, AIRAP | zinc finger, AN1-type domain 2A |
C00002514
|
| 161882 | ZFPM1, FOG, FOG1, ZC2HC11A, ZNF408, ZNF89A | zinc finger protein, FOG family member 1 |
C00002514
|
| 53349 | ZFYVE1, DFCP1, SR3, TAFF1, ZNFN2A1 | zinc finger, FYVE domain containing 1 |
C00002514
|
| 124220 | ZG16B, EECP, HRPE773, JCLN2, PAUF, PRO1567 | zymogen granule protein 16B |
C00002514
|
| 7545 | ZIC1, ZIC, ZNF201 | Zic family member 1 |
C00002514
|
| 7586 | ZKSCAN1, 9130423L19Rik, KOX18, PHZ-37, ZNF139, ZNF36, ZSCAN33 | zinc finger with KRAB and SCAN domains 1 |
C00002514
|
| 57178 | ZMIZ1, MIZ, RAI17, TRAFIP10, ZIMP10, hZIMP10 | zinc finger, MIZ-type containing 1 |
C00002514
|
| 9205 | ZMYM5, HSPC050, MYM, ZNF198L1, ZNF237 | zinc finger, MYM-type 5 |
C00002514
|
| 51351 | ZNF117, H-plk, HPF9 | zinc finger protein 117 |
C00002514
|
| 7559 | ZNF12, GIOT-3, HZF11, KOX3, ZNF325 | zinc finger protein 12 |
C00002514
|
| 7678 | ZNF124, HZF-16, HZF16, ZK7 | zinc finger protein 124 |
C00002514
|
| 90338 | ZNF160, F11, HKr18, HZF5, KR18 | zinc finger protein 160 |
C00002514
|
| 7769 | ZNF226 | zinc finger protein 226 |
C00002514
|
| 339324 | ZNF260, OZRF1, PEX1, ZFP260 | zinc finger protein 260 |
C00002514
|
| 11179 | ZNF277, NRIF4, ZNF277P | zinc finger protein 277 |
C00002514
|
| 23036 | ZNF292, Nbla00365, ZFP292, ZN-16, Zn-15, bA393I2.3 | zinc finger protein 292 |
C00002514
|
| 59348 | ZNF350, ZBRK1, ZFQR | zinc finger protein 350 |
C00002514
|
| 6940 | ZNF354A, EZNF, HKL1, KID-1, KID1, TCF17 | zinc finger protein 354A |
C00002514
|
| 23536 | ADAT1, HADAT1 | adenosine deaminase, tRNA-specific 1 (EC:3.5.4.34) |
C00002514
|
| 151126 | ZNF385B, ZNF533 | zinc finger protein 385B |
C00002514
|
| 55893 | ZNF395, HDBP-2, HDBP2, HDRF-2, PBF, PRF-1, PRF1, Si-1-8-14 | zinc finger protein 395 |
C00002514
|
| 51710 | ZNF44, GIOT-2, KOX7, ZNF, ZNF504, ZNF55, ZNF58 | zinc finger protein 44 |
C00002514
|
| 168544 | ZNF467, EZI, Zfp467 | zinc finger protein 467 |
C00002514
|
| 84838 | ZNF496, NIZP1, ZFP496, ZKSCAN17, ZSCAN49 | zinc finger protein 496 |
C00002514
|
| 93134 | ZNF561 | zinc finger protein 561 |
C00002514
|
| 84914 | ZNF587, ZF6 | zinc finger protein 587 |
C00002514
|
| 9831 | ZNF623 | zinc finger protein 623 |
C00002514
|
| 199692 | ZNF627 | zinc finger protein 627 |
C00002514
|
| 121274 | ZNF641 | zinc finger protein 641 |
C00002514
|
| 55279 | ZNF654 | zinc finger protein 654 |
C00002514
|
| 339500 | ZNF678 | zinc finger protein 678 |
C00002514
|
| 80139 | ZNF703, ZEPPO1, ZNF503L, ZPO1 | zinc finger protein 703 |
C00002514
|
| 255403 | ZNF718 | zinc finger protein 718 |
C00002514
|
| 79755 | ZNF750, ZFP750 | zinc finger protein 750 |
C00002514
|
| 284309 | ZNF776 | zinc finger protein 776 |
C00002514
|
| 146540 | ZNF785 | zinc finger protein 785 |
C00002514
|
| 7637 | ZNF84, HPF2 | zinc finger protein 84 |
C00002514
|
| 7644 | ZNF91, HPF7, HTF10 | zinc finger protein 91 |
C00002514
|
| 30834 | ZNRD1, HTEX-6, Rpa12, TEX6, ZR14, hZR14, tctex-6 | zinc ribbon domain containing 1 (EC:2.7.7.6) |
C00002514
|
| 23053 | ZSWIM8, KIAA0913 | zinc finger, SWIM-type containing 8 |
C00002514
|
| 55055 | ZWILCH, KNTC1AP, hZwilch | zwilch kinetochore protein |
C00002514
|
| 11130 | ZWINT, HZwint-1, KNTC2AP, ZWINT1 | ZW10 interacting kinetochore protein |
C00002514
|
| 158586 | ZXDB, ZNF905, dJ83L6.1 | zinc finger, X-linked, duplicated B |
C00002514
|
| 7791 | ZYX, ESP-2, HED-2 | zyxin |
C00002514
|
| 148 | ADRA1A, ADRA1C, ADRA1L1, ALPHA1AAR | adrenoceptor alpha 1A |
C00001424
|
| 147 | ADRA1B, ADRA1, ALPHA1BAR | adrenoceptor alpha 1B |
C00001424
|
| 146 | ADRA1D, ADRA1, ADRA1A, ADRA1R, ALPHA1, DAR, dJ779E11.2 | adrenoceptor alpha 1D |
C00001424
|
| 151 | ADRA2B, ADRA2L1, ADRA2RL1, ADRARL1, ALPHA2BAR, alpha-2BAR | adrenoceptor alpha 2B |
C00001424
|
| 97 | ACYP1, ACYPE | acylphosphatase 1, erythrocyte (common) type (EC:3.6.1.7) |
C00002514
|
| 153 | ADRB1, ADRB1R, B1AR, BETA1AR, RHR | adrenoceptor beta 1 |
C00001424
|
| 154 | ADRB2, ADRB2R, ADRBR, B2AR, BAR, BETA2AR | adrenoceptor beta 2, surface |
C00001424
|
| 155 | ADRB3, BETA3AR | adrenoceptor beta 3 |
C00001424
|
| 79611 | ACSS3 | acyl-CoA synthetase short-chain family member 3 (EC:6.2.1.1) |
C00002514
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00001424
|
| 2181 | ACSL3, ACS3, FACL3, PRO2194 | acyl-CoA synthetase long-chain family member 3 (EC:6.2.1.3) |
C00002514
|
| 1813 | DRD2, D2DR, D2R | dopamine receptor D2 |
C00001424
|
| 11332 | ACOT7, ACH1, ACT, BACH, CTE-II, LACH, LACH1, RP1-120G22.10, hBACH | acyl-CoA thioesterase 7 (EC:3.1.2.2) |
C00002514
|
| 122970 | ACOT4, PTE-Ib, PTE1B, PTE2B | acyl-CoA thioesterase 4 (EC:3.1.2.2) |
C00002514
|
| 47 | ACLY, ACL, ATPCL, CLATP | ATP citrate lyase (EC:2.3.3.8) |
C00002514
|
| 57007 | ACKR3, CMKOR1, CXC-R7, CXCR-7, CXCR7, GPR159, RDC-1, RDC1 | atypical chemokine receptor 3 |
C00002514
|
| 64746 | ACBD3, GCP60, GOCAP1, GOLPH1, PAP7 | acyl-CoA binding domain containing 3 |
C00002514
|
| 10449 | ACAA2, DSAEC | acetyl-CoA acyltransferase 2 (EC:2.3.1.16) |
C00002514
|
| 10006 | ABI1, ABI-1, ABLBP4, E3B1, NAP1BP, SSH3BP, SSH3BP1 | abl-interactor 1 |
C00002514
|
| 4879 | NPPB, BNP | natriuretic peptide B |
C00001424
|
| 23 | ABCF1, ABC27, ABC50 | ATP-binding cassette, sub-family F (GCN20), member 1 |
C00002514
|
| 6059 | ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I | ATP-binding cassette, sub-family E (OABP), member 1 |
C00002514
|
| 6279 | S100A8, 60B8AG, CAGA, CFAG, CGLA, CP-10, L1Ag, MA387, MIF, MRP8, NIF, P8 | S100 calcium binding protein A8 |
C00001424
|
| 6280 | S100A9, 60B8AG, CAGB, CFAG, CGLB, L1AG, LIAG, MAC387, MIF, MRP14, NIF, P14 | S100 calcium binding protein A9 |
C00001424
|
| 6285 | S100B, NEF, S100, S100-B, S100beta | S100 calcium binding protein B |
C00001424
|
| 10057 | ABCC5, ABC33, EST277145, MOAT-C, MOATC, MRP5, SMRP, pABC11 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
C00002514
|
| 16 | AARS, CMT2N | alanyl-tRNA synthetase (EC:6.1.1.7) |
C00002514
|
| 8086 | AAAS, AAA, AAASb, ADRACALA, ADRACALIN, ALADIN | achalasia, adrenocortical insufficiency, alacrimia |
C00002514
|
| 2741 | GLRA1, HKPX1, STHE | glycine receptor, alpha 1 |
C00001267
|
| 5446 | PON3 | paraoxonase 3 (EC:3.1.1.2 3.1.8.1 3.1.1.81) |
C00000750
|
| 5445 | PON2 | paraoxonase 2 (EC:3.1.1.2 3.1.1.81) |
C00000750
|
| 5444 | PON1, ESA, MVCD5, PON | paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81) |
C00000750
|
| 7391 | USF1, FCHL, FCHL1, HYPLIP1, MLTF, MLTFI, UEF, bHLHb11 | upstream transcription factor 1 |
C00001424
|
| 14 | AAMP | angio-associated, migratory cell protein |
C00034649
|
| 23460 | ABCA6, EST155051 | ATP-binding cassette, sub-family A (ABC1), member 6 |
C00034649
|
| 10351 | ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 |
C00034649
|
| 23457 | ABCB9, EST122234, TAPL | ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
C00034649
|
| 1244 | ABCC2, ABC30, CMOAT, DJS, MRP2, cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
C00034649
|
| 8714 | ABCC3, ABC31, EST90757, MLP2, MOAT-D, MRP3, cMOAT2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
C00034649
|
| 10257 | ABCC4, EST170205, MOAT-B, MOATB, MRP4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
C00034649
|
| 368 | ABCC6, ABC34, ARA, EST349056, GACI2, MLP1, MOAT-E, MOATE, MRP6, PXE, PXE1, URG7 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
C00034649
|
| 10060 | ABCC9, ABC37, ATFB12, CANTU, CMD1O, SUR2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
C00034649
|
| 9429 | ABCG2, ABC15, ABCP, BCRP, BCRP1, BMDP, CD338, CDw338, EST157481, GOUT1, MRX, MXR, MXR1, UAQTL1 | ATP-binding cassette, sub-family G (WHITE), member 2 |
C00034649
|
| 64240 | ABCG5, STSL | ATP-binding cassette, sub-family G (WHITE), member 5 |
C00034649
|
| 64241 | ABCG8, GBD4, STSL | ATP-binding cassette, sub-family G (WHITE), member 8 |
C00034649
|
| 51099 | ABHD5, CDS, CGI58, IECN2, NCIE2 | abhydrolase domain containing 5 (EC:2.3.1.51) |
C00034649
|
| 25890 | ABI3BP, NESHBP, TARSH | ABI family, member 3 (NESH) binding protein |
C00034649
|
| 3983 | ABLIM1, ABLIM, LIMAB1, LIMATIN, abLIM-1 | actin binding LIM protein 1 |
C00034649
|
| 84448 | ABLIM2 | actin binding LIM protein family, member 2 |
C00034649
|
| 176 | ACAN, AGC1, AGCAN, CSPG1, CSPGCP, MSK16, SEDK | aggrecan |
C00034649
|
| 51554 | ACKR4, CC-CKR-11, CCBP2, CCR-11, CCR10, CCR11, CCRL1, CCX_CKR, CCX-CKR, CKR-11, PPR1, VSHK1 | atypical chemokine receptor 4 |
C00034649
|
| 55 | ACPP, 5'-NT, ACP-3, ACP3, PAP | acid phosphatase, prostate (EC:3.1.3.2 3.1.3.5) |
C00034649
|
| 2180 | ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2 | acyl-CoA synthetase long-chain family member 1 (EC:6.2.1.3) |
C00034649
|
| 6296 | ACSM3, SA, SAH | acyl-CoA synthetase medium-chain family member 3 (EC:6.2.1.2) |
C00034649
|
| 59 | ACTA2, AAT6, ACTSA, MYMY5 | actin, alpha 2, smooth muscle, aorta |
C00034649
|
| 70 | ACTC1, ACTC, ASD5, CMD1R, CMH11, LVNC4 | actin, alpha, cardiac muscle 1 |
C00034649
|
| 72 | ACTG2, ACT, ACTA3, ACTE, ACTL3, ACTSG | actin, gamma 2, smooth muscle, enteric |
C00034649
|
| 87 | ACTN1, BDPLT15 | actinin, alpha 1 |
C00034649
|
| 100 | ADA | adenosine deaminase (EC:3.5.4.4) |
C00034649
|
| 8038 | ADAM12, MCMP, MCMPMltna, MLTN, MLTNA | ADAM metallopeptidase domain 12 |
C00034649
|
| 6868 | ADAM17, ADAM18, CD156B, CSVP, NISBD, TACE | ADAM metallopeptidase domain 17 (EC:3.4.24.86) |
C00034649
|
| 10863 | ADAM28, ADAM_28, ADAM23, MDC-L, MDC-Lm, MDC-Ls, MDCL, eMDC_II, eMDCII | ADAM metallopeptidase domain 28 |
C00034649
|
| 8754 | ADAM9, CORD9, MCMP, MDC9, Mltng | ADAM metallopeptidase domain 9 |
C00034649
|
| 9510 | ADAMTS1, C3-C5, METH1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
C00034649
|
| 9509 | ADAMTS2, ADAM-TS2, ADAMTS-2, ADAMTS-3, NPI, PC_I-NP, PCI-NP, PCINP, PCPNI, PNPI | ADAM metallopeptidase with thrombospondin type 1 motif, 2 (EC:3.4.24.14) |
C00034649
|
| 80070 | ADAMTS20, ADAM-TS20, ADAMTS-20, GON-1 | ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
C00034649
|
| 11095 | ADAMTS8, ADAM-TS8, METH2 | ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
C00034649
|
| 56999 | ADAMTS9 | ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
C00034649
|
| 55803 | ADAP2, CENTA2, HSA272195, cent-b | ArfGAP with dual PH domains 2 |
C00034649
|
| 107 | ADCY1, AC1 | adenylate cyclase 1 (brain) (EC:4.6.1.1) |
C00034649
|
| 116 | ADCYAP1, PACAP | adenylate cyclase activating polypeptide 1 (pituitary) |
C00034649
|
| 117 | ADCYAP1R1, PAC1, PAC1R, PACAPR, PACAPRI | adenylate cyclase activating polypeptide 1 (pituitary) receptor type I |
C00034649
|
| 125 | ADH1B, ADH2 | alcohol dehydrogenase 1B (class I), beta polypeptide (EC:1.1.1.1) |
C00034649
|
| 126 | ADH1C, ADH3 | alcohol dehydrogenase 1C (class I), gamma polypeptide (EC:1.1.1.1) |
C00034649
|
| 55256 | ADI1, APL1, ARD, Fe-ARD, MTCBP1, Ni-ARD, SIPL, mtnD | acireductone dioxygenase 1 (EC:1.13.11.54) |
C00034649
|
| 9370 | ADIPOQ, ACDC, ACRP30, ADIPQTL1, ADPN, APM-1, APM1, GBP28 | adiponectin, C1Q and collagen domain containing |
C00034649
|
| 79602 | ADIPOR2, ACDCR2, PAQR2 | adiponectin receptor 2 |
C00034649
|
| 10974 | ADIRF, AFRO, APM2, C10orf116, apM-2 | adipogenesis regulatory factor |
C00034649
|
| 133 | ADM, AM | adrenomedullin |
C00034649
|
| 84632 | AFAP1L2, KIAA1914, XB130 | actin filament associated protein 1-like 2 |
C00034649
|
| 174 | AFP, FETA, HPAFP | alpha-fetoprotein |
C00034649
|
| 116988 | AGAP3, AGAP-3, CENTG3, CRAG, MRIP-1, cnt-g3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
C00034649
|
| 56895 | AGPAT4, 1-AGPAT4, LPAAT-delta, dJ473J16.2 | 1-acylglycerol-3-phosphate O-acyltransferase 4 (EC:2.3.1.51) |
C00034649
|
| 183 | AGT, ANHU, SERPINA8 | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
C00034649
|
| 55057 | AIM1L, CRYBG2 | absent in melanoma 1-like |
C00034649
|
| 9590 | AKAP12, AKAP250, SSeCKS | A kinase (PRKA) anchor protein 12 |
C00034649
|
| 10270 | AKAP8, AKAP_95, AKAP-8, AKAP-95, AKAP95 | A kinase (PRKA) anchor protein 8 |
C00034649
|
| 10142 | AKAP9, AKAP-9, AKAP350, AKAP450, CG-NAP, HYPERION, LQT11, MU-RMS-40.16A, PPP1R45, PRKA9, YOTIAO | A kinase (PRKA) anchor protein 9 |
C00034649
|
| 57016 | AKR1B10, AKR1B11, AKR1B12, ALDRLn, ARL-1, ARL1, HIS, HSI | aldo-keto reductase family 1, member B10 (aldose reductase) (EC:1.1.1.2) |
C00034649
|
| 1645 | AKR1C1, 2-ALPHA-HSD, 20-ALPHA-HSD, C9, DD1, DD1/DD2, DDH, DDH1, H-37, HAKRC, HBAB, MBAB | aldo-keto reductase family 1, member C1 (EC:1.3.1.20 1.1.1.149 1.1.1.112) |
C00034649
|
| 1646 | AKR1C2, AKR1C-pseudo, BABP, DD, DD2, DDH2, HAKRD, HBAB, MCDR2, SRXY8 | aldo-keto reductase family 1, member C2 (EC:1.3.1.20 1.1.1.357) |
C00034649
|
| 8644 | AKR1C3, DD3, DDX, HA1753, HAKRB, HAKRe, HSD17B5, PGFS, hluPGFS | aldo-keto reductase family 1, member C3 (EC:1.3.1.20 1.1.1.188 1.1.1.239 1.1.1.64 1.1.1.112 1.1.1.357) |
C00034649
|
| 1109 | AKR1C4, 3-alpha-HSD, C11, CDR, CHDR, DD-4, DD4, HAKRA | aldo-keto reductase family 1, member C4 (EC:1.1.1.225 1.1.1.357) |
C00034649
|
| 6718 | AKR1D1, 3o5bred, CBAS2, SRD5B1 | aldo-keto reductase family 1, member D1 (EC:1.3.1.3) |
C00034649
|
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00034649
|
| 210 | ALAD, ALADH, PBGS | aminolevulinate dehydratase (EC:4.2.1.24) |
C00034649
|
| 213 | ALB, PRO0883, PRO0903, PRO1341 | albumin |
C00034649
|
| 220 | ALDH1A3, ALDH1A6, ALDH6, MCOP8, RALDH3 | aldehyde dehydrogenase 1 family, member A3 (EC:1.2.1.5) |
C00034649
|
| 218 | ALDH3A1, ALDH3, ALDHIII | aldehyde dehydrogenase 3 family, member A1 (EC:1.2.1.5) |
C00034649
|
| 85365 | ALG2, CDGIi, NET38, hALPG2 | ALG2, alpha-1,3/1,6-mannosyltransferase (EC:2.4.1.132 2.4.1.257) |
C00034649
|
| 247 | ALOX15B, 15-LOX-2 | arachidonate 15-lipoxygenase, type B (EC:1.13.11.33) |
C00034649
|
| 250 | ALPP, ALP, PALP, PLAP, PLAP-1 | alkaline phosphatase, placental (EC:3.1.3.1) |
C00034649
|
| 347902 | AMIGO2, ALI1, AMIGO-2, DEGA | adhesion molecule with Ig-like domain 2 |
C00034649
|
| 276 | AMY1A, AMY1 | amylase, alpha 1A (salivary) (EC:3.2.1.1) |
C00034649
|
| 283 | ANG, ALS9, HEL168, RNASE4, RNASE5 | angiogenin, ribonuclease, RNase A family, 5 (EC:3.1.27.-) |
C00034649
|
| 284 | ANGPT1, AGP1, AGPT, ANG1 | angiopoietin 1 |
C00034649
|
| 285 | ANGPT2, AGPT2, ANG2 | angiopoietin 2 |
C00034649
|
| 23452 | ANGPTL2, ARP2, HARP | angiopoietin-like 2 |
C00034649
|
| 51129 | ANGPTL4, ANGPTL2, ARP4, FIAF, HARP, HFARP, NL2, PGAR, UNQ171, pp1158 | angiopoietin-like 4 |
C00034649
|
| 288 | ANK3, ANKYRIN-G, MRT37 | ankyrin 3, node of Ranvier (ankyrin G) |
C00034649
|
| 84210 | ANKRD20A1, ANKRD20A | ankyrin repeat domain 20 family, member A1 |
C00034649
|
| 118932 | ANKRD22 | ankyrin repeat domain 22 |
C00034649
|
| 148741 | ANKRD35 | ankyrin repeat domain 35 |
C00034649
|
| 57182 | ANKRD50 | ankyrin repeat domain 50 |
C00034649
|
| 79722 | ANKRD55 | ankyrin repeat domain 55 |
C00034649
|
| 84168 | ANTXR1, ATR, GAPO, TEM8 | anthrax toxin receptor 1 |
C00034649
|
| 118429 | ANTXR2, CMG-2, CMG2, HFS, ISH, JHF | anthrax toxin receptor 2 |
C00034649
|
| 316 | AOX1, AO, AOH1 | aldehyde oxidase 1 (EC:1.2.3.1) |
C00034649
|
| 10947 | AP3M2, AP47B, CLA20, P47B | adaptor-related protein complex 3, mu 2 subunit |
C00034649
|
| 55745 | AP5M1, C14orf108, MUDENG, Mu5, MuD | adaptor-related protein complex 5, mu 1 subunit |
C00034649
|
| 147495 | APCDD1, B7323, DRAPC1, HHS, HTS | adenomatosis polyposis coli down-regulated 1 |
C00034649
|
| 328 | APEX1, APE, APE1, APEN, APEX, APX, HAP1, REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 (EC:4.2.99.18) |
C00034649
|
| 346 | APOC4, APO-CIV, APOC-IV | apolipoprotein C-IV |
C00034649
|
| 55937 | APOM, G3a, HSPC336, NG20, apo-M | apolipoprotein M |
C00034649
|
| 359 | AQP2, AQP-CD, WCH-CD | aquaporin 2 (collecting duct) |
C00034649
|
| 64411 | ARAP3, CENTD3, DRAG1 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
C00034649
|
| 374 | AREG, AR, CRDGF, SDGF | amphiregulin |
C00034649
|
| 384 | ARG2 | arginase 2 (EC:3.5.3.1) |
C00034649
|
| 93663 | ARHGAP18, MacGAP, SENEX, bA307O14.2 | Rho GTPase activating protein 18 |
C00034649
|
| 79822 | ARHGAP28 | Rho GTPase activating protein 28 |
C00034649
|
| 9828 | ARHGEF17, P164RHOGEF, TEM4, p164-RhoGEF | Rho guanine nucleotide exchange factor (GEF) 17 |
C00034649
|
| 50650 | ARHGEF3, GEF3, STA3, XPLN | Rho guanine nucleotide exchange factor (GEF) 3 |
C00034649
|
| 10124 | ARL4A, ARL4 | ADP-ribosylation factor-like 4A |
C00034649
|
| 10123 | ARL4C, ARL7, LAK | ADP-ribosylation factor-like 4C |
C00034649
|
| 80210 | ARMC9, ARM, KU-MEL-1 | armadillo repeat containing 9 |
C00034649
|
| 406 | ARNTL, BMAL1, BMAL1c, JAP3, MOP3, PASD3, TIC, bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like |
C00034649
|
| 419 | ART3, ARTC3 | ADP-ribosyltransferase 3 (EC:2.4.2.31) |
C00034649
|
| 8853 | ASAP2, AMAP2, CENTB3, DDEF2, PAG3, PAP, Pap-alpha, SHAG1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
C00034649
|
| 51676 | ASB2, ASB-2 | ankyrin repeat and SOCS box containing 2 |
C00034649
|
| 444 | ASPH, AAH, BAH, CASQ2BP1, HAAH, JCTN, junctin | aspartate beta-hydroxylase (EC:1.14.11.16) |
C00034649
|
| 57168 | ASPHD2 | aspartate beta-hydroxylase domain containing 2 |
C00034649
|
| 54829 | ASPN, OS3, PLAP-1, PLAP1, SLRR1C | asporin |
C00034649
|
| 80150 | ASRGL1, ALP, ALP1, CRASH | asparaginase like 1 (EC:3.5.1.1 3.4.19.5) |
C00034649
|
| 468 | ATF4, CREB-2, CREB2, TAXREB67, TXREB | activating transcription factor 4 |
C00034649
|
| 55102 | ATG2B, C14orf103 | autophagy related 2B |
C00034649
|
| 57194 | ATP10A, ATP10C, ATPVA, ATPVC | ATPase, class V, type 10A (EC:3.6.3.1) |
C00034649
|
| 476 | ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide (EC:3.6.3.9) |
C00034649
|
| 493 | ATP2B4, ATP2B2, MXRA1, PMCA4, PMCA4b, PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 (EC:3.6.3.8) |
C00034649
|
| 523 | ATP6V1A, ATP6A1, ATP6V1A1, HO68, VA68, VPP2, Vma1 | ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (EC:3.6.3.14) |
C00034649
|
| 540 | ATP7B, PWD, WC1, WD, WND | ATPase, Cu++ transporting, beta polypeptide (EC:3.6.3.54) |
C00034649
|
| 51761 | ATP8A2, ATP, ATPIB, CAMRQ4, IB, ML-1 | ATPase, aminophospholipid transporter, class I, type 8A, member 2 (EC:3.6.3.1) |
C00034649
|
| 60370 | AVPI1, PP5395, RP11-548K23.7, VIP32, VIT32 | arginine vasopressin-induced 1 |
C00034649
|
| 552 | AVPR1A, AVPR_V1a, AVPR1, V1aR | arginine vasopressin receptor 1A |
C00034649
|
| 8313 | AXIN2, AXIL, ODCRCS | axin 2 |
C00034649
|
| 22994 | AZI1, AZ1, Cep131, ZA1 | 5-azacytidine induced 1 |
C00034649
|
| 8704 | B4GALT2, B4Gal-T2, B4Gal-T3, beta4Gal-T2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (EC:2.4.1.22 2.4.1.90 2.4.1.38) |
C00034649
|
| 79870 | BAALC | brain and acute leukemia, cytoplasmic |
C00034649
|
| 573 | BAG1, BAG-1, HAP, RAP46 | BCL2-associated athanogene |
C00034649
|
| 10458 | BAIAP2, BAP2, FLAF3, IRSP53 | BAI1-associated protein 2 |
C00034649
|
| 79738 | BBS10, C12orf58 | Bardet-Biedl syndrome 10 |
C00034649
|
| 55212 | BBS7, BBS2L1 | Bardet-Biedl syndrome 7 |
C00034649
|
| 586 | BCAT1, BCATC, BCT1, ECA39, MECA39, PNAS121, PP18 | branched chain amino-acid transaminase 1, cytosolic (EC:2.6.1.42) |
C00034649
|
| 587 | BCAT2, BCAM, BCATM, BCT2, PP18 | branched chain amino-acid transaminase 2, mitochondrial (EC:2.6.1.42) |
C00034649
|
| 53335 | BCL11A, BCL11A-L, BCL11A-S, BCL11A-XL, BCL11a-M, CTIP1, EVI9, HBFQTL5, ZNF856 | B-cell CLL/lymphoma 11A (zinc finger protein) |
C00034649
|
| 623 | BDKRB1, B1BKR, B1R, BKB1R, BKR1, BRADYB1 | bradykinin receptor B1 |
C00034649
|
| 627 | BDNF, ANON2, BULN2 | brain-derived neurotrophic factor |
C00034649
|
| 221336 | BEND6, C6orf65, bA203B9.1 | BEN domain containing 6 |
C00034649
|
| 10282 | BET1, HBET1 | Bet1 golgi vesicular membrane trafficking protein |
C00034649
|
| 8553 | BHLHE40, BHLHB2, DEC1, HLHB2, SHARP-2, STRA13, Stra14 | basic helix-loop-helix family, member e40 |
C00034649
|
| 79365 | BHLHE41, BHLHB3, DEC2, SHARP1, hDEC2 | basic helix-loop-helix family, member e41 |
C00034649
|
| 45827 |
C00034649
|
||
| 636 | BICD1, BICD | bicaudal D homolog 1 (Drosophila) |
C00034649
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00034649
|
| 274 | BIN1, AMPH2, AMPHL, SH3P9 | bridging integrator 1 |
C00034649
|
| 330 | BIRC3, AIP1, API2, CIAP2, HAIP1, HIAP1, MALT2, MIHC, RNF49, c-IAP2 | baculoviral IAP repeat containing 3 |
C00034649
|
| 650 | BMP2, BDA2, BMP2A | bone morphogenetic protein 2 |
C00034649
|
| 654 | BMP6, VGR, VGR1 | bone morphogenetic protein 6 |
C00034649
|
| 669 | BPGM, DPGM | 2,3-bisphosphoglycerate mutase (EC:3.1.3.13 5.4.2.4 5.4.2.11) |
C00034649
|
| 55299 | BRIX1, BRIX, BXDC2 | BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
C00034649
|
| 22903 | BTBD3, dJ742J24.1 | BTB (POZ) domain containing 3 |
C00034649
|
| 153579 | BTNL9, BTN3, VDLS1900 | butyrophilin-like 9 |
C00034649
|
| 11067 | C10orf10, DEPP, FIG | chromosome 10 open reading frame 10 |
C00034649
|
| 348110 | C15orf38, Arpin | chromosome 15 open reading frame 38 |
C00034649
|
| 388135 | C15orf59 | chromosome 15 open reading frame 59 |
C00034649
|
| 708 | C1QBP, GC1QBP, HABP1, SF2p32, gC1Q-R, gC1qR, p32 | complement component 1, q subcomponent binding protein |
C00034649
|
| 114905 | C1QTNF7, CTRP7, ZACRP7 | C1q and tumor necrosis factor related protein 7 |
C00034649
|
| 717 | C2, CO2 | complement component 2 (EC:3.4.21.43) |
C00034649
|
| 145741 | C2CD4A, FAM148A, NLF1 | C2 calcium-dependent domain containing 4A |
C00034649
|
| 8913 | CACNA1G, Ca(V)T.1, Cav3.1, NBR13 | calcium channel, voltage-dependent, T type, alpha 1G subunit |
C00034649
|
| 23705 | CADM1, BL2, IGSF4, IGSF4A, NECL2, Necl-2, RA175, ST17, SYNCAM, TSLC1, sTSLC-1, sgIGSF, synCAM1 | cell adhesion molecule 1 |
C00034649
|
| 93664 | CADPS2 | Ca++-dependent secretion activator 2 |
C00034649
|
| 811 | CALR, CRT, RO, SSA, cC1qR | calreticulin |
C00034649
|
| 816 | CAMK2B, CAM2, CAMK2, CAMKB | calcium/calmodulin-dependent protein kinase II beta (EC:2.7.11.17) |
C00034649
|
| 820 | CAMP, CAP-18, CAP18, CRAMP, FALL-39, FALL39, LL37 | cathelicidin antimicrobial peptide |
C00034649
|
| 827 | CAPN6, CANPX, CAPNX, CalpM, DJ914P14.1 | calpain 6 |
C00034649
|
| 79820 | CATSPERB, C14orf161, CatSper(beta) | catsper channel auxiliary subunit beta |
C00034649
|
| 859 | CAV3, LGMD1C, LQT9, VIP-21, VIP21 | caveolin 3 |
C00034649
|
| 23624 | CBLC, CBL-3, CBL-SL, RNF57 | Cbl proto-oncogene C, E3 ubiquitin protein ligase |
C00034649
|
| 874 | CBR3, SDR21C2, hCBR3 | carbonyl reductase 3 (EC:1.1.1.184) |
C00034649
|
| 875 | CBS, HIP4 | cystathionine-beta-synthase (EC:4.2.1.22) |
C00034649
|
| 79839 | CCDC102B, ACY1L, AN, C18orf14, HsT1731 | coiled-coil domain containing 102B |
C00034649
|
| 83643 | CCDC3 | coiled-coil domain containing 3 |
C00034649
|
| 80323 | CCDC68, SE57-1 | coiled-coil domain containing 68 |
C00034649
|
| 168455 | CCDC71L, C7orf74 | coiled-coil domain containing 71-like |
C00034649
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00034649
|
| 8900 | CCNA1 | cyclin A1 |
C00034649
|
| 10666 | CD226, DNAM-1, DNAM1, PTA1, TLiSA1 | CD226 molecule |
C00034649
|
| 9936 | CD302, BIMLEC, CLEC13A, DCL-1, DCL1 | CD302 molecule |
C00034649
|
| 952 | CD38, T10 | CD38 molecule (EC:3.2.2.5) |
C00034649
|
| 9308 | CD83, BL11, HB15 | CD83 molecule |
C00034649
|
| 10225 | CD96, TACTILE | CD96 molecule |
C00034649
|
| 978 | CDA, CDD | cytidine deaminase (EC:3.5.4.5) |
C00034649
|
| 991 | CDC20, CDC20A, bA276H19.3, p55CDC | cell division cycle 20 |
C00034649
|
| 994 | CDC25B | cell division cycle 25B (EC:3.1.3.48) |
C00034649
|
| 10602 | CDC42EP3, BORG2, CEP3, UB1 | CDC42 effector protein (Rho GTPase binding) 3 |
C00034649
|
| 83879 | CDCA7, JPO1 | cell division cycle associated 7 |
C00034649
|
| 1003 | CDH5, 7B4, CD144 | cadherin 5, type 2 (vascular endothelium) |
C00034649
|
| 1004 | CDH6, CAD6, KCAD | cadherin 6, type 2, K-cadherin (fetal kidney) |
C00034649
|
| 23097 | CDK19, CDC2L6, CDK11, bA346C16.3 | cyclin-dependent kinase 19 (EC:2.7.11.22) |
C00034649
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00034649
|
| 1024 | CDK8, K35 | cyclin-dependent kinase 8 (EC:2.7.11.23 2.7.11.22) |
C00034649
|
| 1025 | CDK9, C-2k, CDC2L4, CTK1, PITALRE, TAK | cyclin-dependent kinase 9 (EC:2.7.11.23 2.7.11.22) |
C00034649
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00034649
|
| 634 | CEACAM1, BGP, BGP1, BGPI | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
C00034649
|
| 1048 | CEACAM5, CD66e, CEA | carcinoembryonic antigen-related cell adhesion molecule 5 |
C00034649
|
| 4680 | CEACAM6, CD66c, CEAL, NCA | carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
C00034649
|
| 1054 | CEBPG, GPE1BP, IG/EBP-1 | CCAAT/enhancer binding protein (C/EBP), gamma |
C00034649
|
| 201161 | CENPV, 3110013H01Rik, CENP-V, PRR6, p30 | centromere protein V |
C00034649
|
| 79959 | CEP76, C18orf9, HsT1705 | centrosomal protein 76kDa |
C00034649
|
| 64781 | CERK, LK4, dA59H18.2, dA59H18.3, hCERK | ceramide kinase (EC:2.7.1.138) |
C00034649
|
| 253782 | CERS6, CERS5, LASS6 | ceramide synthase 6 |
C00034649
|
| 283848 | CES4A, CES6, CES8 | carboxylesterase 4A |
C00034649
|
| 3075 | CFH, AHUS1, AMBP1, ARMD4, ARMS1, CFHL3, FH, FHL1, HF, HF1, HF2, HUS | complement factor H (EC:4.2.1.2) |
C00034649
|
| 3078 | CFHR1, CFHL, CFHL1, CFHL1P, CFHR1P, FHR1, H36-1, H36-2, HFL1, HFL2 | complement factor H-related 1 |
C00034649
|
| 379258 |
C00034649
|
||
| 3091 | HIF1A, HIF-1A, HIF-1alpha, HIF1, HIF1-ALPHA, MOP1, PASD8, bHLHe78 | hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
C00002525
|
| 1081 | CGA, CG-ALPHA, FSHA, GPHA1, GPHa, HCG, LHA, TSHA | glycoprotein hormones, alpha polypeptide |
C00034649
|
| 1082 | CGB, CGB3, CGB5, CGB7, CGB8, hCGB | chorionic gonadotropin, beta polypeptide |
C00034649
|
| 9023 | CH25H, C25H | cholesterol 25-hydroxylase (EC:1.14.99.38) |
C00034649
|
| 79094 | CHAC1 | ChaC, cation transport regulator homolog 1 (E. coli) |
C00034649
|
| 1113 | CHGA, CGA | chromogranin A (parathyroid secretory protein 1) |
C00034649
|
| 1116 | CHI3L1, ASRT7, CGP-39, GP-39, GP39, HC-gp39, HCGP-3P, YKL-40, YKL40, YYL-40, hCGP-39 | chitinase 3-like 1 (cartilage glycoprotein-39) |
C00034649
|
| 1124 | CHN2, ARHGAP3, BCH, CHN2-3, RHOGAP3 | chimerin 2 |
C00034649
|
| 26973 | CHORDC1, CHP1 | cysteine and histidine-rich domain (CHORD) containing 1 |
C00034649
|
| 1137 | CHRNA4, BFNC, EBN, EBN1, NACHR, NACHRA4, NACRA4 | cholinergic receptor, nicotinic, alpha 4 (neuronal) |
C00034649
|
| 1141 | CHRNB2, EFNL3, nAChRB2 | cholinergic receptor, nicotinic, beta 2 (neuronal) |
C00034649
|
| 1145 | CHRNE, ACHRE, CMS1D, CMS1E, CMS2A, FCCMS, SCCMS | cholinergic receptor, nicotinic, epsilon (muscle) |
C00034649
|
| 50515 | CHST11, C4ST, C4ST-1, C4ST1, HSA269537 | carbohydrate (chondroitin 4) sulfotransferase 11 (EC:2.8.2.5) |
C00034649
|
| 51363 | CHST15, BRAG, GALNAC4S-6ST, RP11-47G11.1 | carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 (EC:2.8.2.33) |
C00034649
|
| 10164 | CHST4, GST3, GlcNAc6ST2, HECGLCNAC6ST, LSST | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
C00034649
|
| 56548 | CHST7, C6ST-2, GST-5 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (EC:2.8.2.17) |
C00034649
|
| 337876 | CHSY3, CHSY2, CSS3 | chondroitin sulfate synthase 3 (EC:2.4.1.175 2.4.1.226) |
C00034649
|
| 8483 | CILP, CILP-1, HsT18872 | cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
C00034649
|
| 1152 | CKB, B-CK, CKBB | creatine kinase, brain (EC:2.7.3.2) |
C00034649
|
| 22802 | CLCA4, CaCC, CaCC2 | chloride channel accessory 4 |
C00034649
|
| 1183 | CLCN4, CLC4, ClC-4, ClC-4A | chloride channel, voltage-sensitive 4 |
C00034649
|
| 7122 | CLDN5, AWAL, BEC1, CPETRL1, TMVCF | claudin 5 |
C00034649
|
| 9073 | CLDN8 | claudin 8 |
C00034649
|
| 9976 | CLEC2B, AICL, CLECSF2, HP10085, IFNRG1 | C-type lectin domain family 2, member B |
C00034649
|
| 7123 | CLEC3B, TN, TNA | C-type lectin domain family 3, member B |
C00034649
|
| 1047 | CLGN | calmegin |
C00034649
|
| 1193 | CLIC2, CLIC2b, MRXS32, XAP121 | chloride intracellular channel 2 |
C00034649
|
| 9022 | CLIC3 | chloride intracellular channel 3 |
C00034649
|
| 25932 | CLIC4, CLIC4L, H1, MTCLIC, huH1, p64H1 | chloride intracellular channel 4 |
C00034649
|
| 134147 | CMBL, JS-1 | carboxymethylenebutenolidase homolog (Pseudomonas) (EC:3.1.-.-) |
C00034649
|
| 129607 | CMPK2, TMPK2, TYKi, UMP-CMPK2 | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial (EC:2.7.4.6 2.7.4.14) |
C00034649
|
| 123920 | CMTM3, BNAS2, CKLFSF3 | CKLF-like MARVEL transmembrane domain containing 3 |
C00034649
|
| 55748 | CNDP2, CN2, CPGL, HsT2298, PEPA | CNDP dipeptidase 2 (metallopeptidase M20 family) (EC:3.4.13.18) |
C00034649
|
| 1259 | CNGA1, CNCG, CNCG1, CNG-1, CNG1, RCNC1, RCNCa, RCNCalpha, RP49 | cyclic nucleotide gated channel alpha 1 |
C00034649
|
| 149111 | CNIH3, CNIH-3 | cornichon family AMPA receptor auxiliary protein 3 |
C00034649
|
| 1264 | CNN1, SMCC, Sm-Calp | calponin 1, basic, smooth muscle |
C00034649
|
| 29883 | CNOT7, CAF1, Caf1a, hCAF-1 | CCR4-NOT transcription complex, subunit 7 (EC:3.1.13.4) |
C00034649
|
| 1268 | CNR1, CANN6, CB-R, CB1, CB1A, CB1K5, CB1R, CNR | cannabinoid receptor 1 (brain) |
C00034649
|
| 1272 | CNTN1, F3, GP135 | contactin 1 |
C00034649
|
| 5067 | CNTN3, BIG-1, PANG, PCS | contactin 3 (plasmacytoma associated) |
C00034649
|
| 79937 | CNTNAP3, CASPR3, CNTNAP3A, RP11-138L21.1, RP11-290L7.1 | contactin associated protein-like 3 |
C00034649
|
| 1301 | COL11A1, CO11A1, COLL6, STL2 | collagen, type XI, alpha 1 |
C00034649
|
| 1303 | COL12A1, BA209D8.1, COL12A1L, DJ234P15.1 | collagen, type XII, alpha 1 |
C00034649
|
| 1305 | COL13A1, COLXIIIA1 | collagen, type XIII, alpha 1 |
C00034649
|
| 7373 | COL14A1, UND | collagen, type XIV, alpha 1 |
C00034649
|
| 1306 | COL15A1 | collagen, type XV, alpha 1 |
C00034649
|
| 1307 | COL16A1, 447AA | collagen, type XVI, alpha 1 |
C00034649
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00034649
|
| 1278 | COL1A2, OI4 | collagen, type I, alpha 2 |
C00034649
|
| 1282 | COL4A1, HANAC, ICH, POREN1, arresten | collagen, type IV, alpha 1 |
C00034649
|
| 1290 | COL5A2 | collagen, type V, alpha 2 |
C00034649
|
| 1291 | COL6A1, OPLL | collagen, type VI, alpha 1 |
C00034649
|
| 1296 | COL8A2, FECD, FECD1, PPCD, PPCD2 | collagen, type VIII, alpha 2 |
C00034649
|
| 10699 | CORIN, ATC2, CRN, Lrp4, PEE5, TMPRSS10 | corin, serine peptidase |
C00034649
|
| 23406 | COTL1, CLP | coactosin-like 1 (Dictyostelium) |
C00034649
|
| 1346 | COX7A1, COX7A, COX7AH, COX7AM | cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) (EC:1.9.3.1) |
C00034649
|
| 1356 | CP, CP-2 | ceruloplasmin (ferroxidase) (EC:1.16.3.1) |
C00034649
|
| 1368 | CPM | carboxypeptidase M (EC:3.4.17.12) |
C00034649
|
| 8532 | CPZ | carboxypeptidase Z |
C00034649
|
| 1382 | CRABP2, CRABP-II, RBP6 | cellular retinoic acid binding protein 2 |
C00034649
|
| 9586 | CREB5, CRE-BPA | cAMP responsive element binding protein 5 |
C00034649
|
| 8804 | CREG1, CREG | cellular repressor of E1A-stimulated genes 1 |
C00034649
|
| 1392 | CRH, CRF | corticotropin releasing hormone |
C00034649
|
| 1394 | CRHR1, CRF-R, CRF-R-1, CRF-R1, CRF1, CRFR-1, CRFR1, CRH-R-1, CRH-R1, CRH-R1h, CRHR, CRHR1L, CRHR1f | corticotropin releasing hormone receptor 1 |
C00034649
|
| 1396 | CRIP1, CRHP, CRIP, CRP-1, CRP1 | cysteine-rich protein 1 (intestinal) |
C00034649
|
| 83716 | CRISPLD2, CRISP11, LCRISP2 | cysteine-rich secretory protein LCCL domain containing 2 |
C00034649
|
| 9244 | CRLF1, CISS, CISS1, CLF, CLF-1, NR6, zcytor5 | cytokine receptor-like factor 1 |
C00034649
|
| 56253 | CRTAM, CD355 | cytotoxic and regulatory T cell molecule |
C00034649
|
| 1410 | CRYAB, CMD1II, CRYA2, CTPP2, CTRCT16, HSPB5, MFM2 | crystallin, alpha B |
C00034649
|
| 1438 | CSF2RA, CD116, CDw116, CSF2R, CSF2RAX, CSF2RAY, CSF2RX, CSF2RY, GM-CSF-R-alpha, GMCSFR, GMR, SMDP4 | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
C00034649
|
| 1440 | CSF3, C17orf33, CSF3OS, GCSF, G-CSF | colony stimulating factor 3 (granulocyte) |
C00034649
|
| 1445 | CSK | c-src tyrosine kinase (EC:2.7.10.2) |
C00034649
|
| 114784 | CSMD2, dJ1007G16.1, dJ1007G16.2, dJ947L8.1 | CUB and Sushi multiple domains 2 |
C00034649
|
| 1466 | CSRP2, CRP2, LMO5, SmLIM | cysteine and glycine-rich protein 2 |
C00034649
|
| 1472 | CST4 | cystatin S |
C00034649
|
| 10217 | CTDSPL, C3orf8, HYA22, PSR1, RBSP3, SCP3 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (EC:3.1.3.16) |
C00034649
|
| 1491 | CTH | cystathionase (cystathionine gamma-lyase) (EC:4.4.1.1) |
C00034649
|
| 1496 | CTNNA2, CAP-R, CAPR, CT114, CTNR | catenin (cadherin-associated protein), alpha 2 |
C00034649
|
| 1509 | CTSD, CLN10, CPSD | cathepsin D (EC:3.4.23.5) |
C00034649
|
| 1510 | CTSE, CATE | cathepsin E (EC:3.4.23.34) |
C00034649
|
| 1521 | CTSW, LYPN | cathepsin W |
C00034649
|
| 23316 | CUX2, CDP2, CUTL2 | cut-like homeobox 2 |
C00034649
|
| 80157 | CWH43, CWH43-C | cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) |
C00034649
|
| 6376 | CX3CL1, ABCD-3, C3Xkine, CXC3, CXC3C, NTN, NTT, SCYD1, fractalkine, neurotactin | chemokine (C-X3-C motif) ligand 1 |
C00034649
|
| 653108 |
C00034649
|
||
| 2919 | CXCL1, FSP, GRO1, GROa, MGSA, MGSA-a, NAP-3, SCYB1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
C00034649
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00034649
|
| 6373 | CXCL11, H174, I-TAC, IP-9, IP9, SCYB11, SCYB9B, b-R1 | chemokine (C-X-C motif) ligand 11 |
C00034649
|
| 9547 | CXCL14, BMAC, BRAK, KEC, KS1, MIP-2g, MIP2G, NJAC, SCYB14 | chemokine (C-X-C motif) ligand 14 |
C00034649
|
| 2920 | CXCL2, CINC-2a, GRO2, GROb, MGSA-b, MIP-2a, MIP2, MIP2A, SCYB2 | chemokine (C-X-C motif) ligand 2 |
C00034649
|
| 6374 | CXCL5, ENA-78, SCYB5 | chemokine (C-X-C motif) ligand 5 |
C00034649
|
| 4283 | CXCL9, CMK, Humig, MIG, SCYB9, crg-10 | chemokine (C-X-C motif) ligand 9 |
C00034649
|
| 7852 | CXCR4, CD184, D2S201E, FB22, HM89, HSY3RR, LAP3, LCR1, LESTR, NPY3R, NPYR, NPYRL, NPYY3R, WHIM | chemokine (C-X-C motif) receptor 4 |
C00034649
|
| 1528 | CYB5A, CYB5, MCB5 | cytochrome b5 type A (microsomal) |
C00034649
|
| 1583 | CYP11A1, CYP11A, CYPXIA1, P450SCC | cytochrome P450, family 11, subfamily A, polypeptide 1 (EC:1.14.15.6) |
C00034649
|
| 2056 | EPO, EP, MVCD2 | erythropoietin |
C00002525
|
| 1588 | CYP19A1, ARO, ARO1, CPV1, CYAR, CYP19, CYPXIX, P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00034649
|
| 1589 | CYP21A2, CA21H, CAH1, CPS1, CYP21, CYP21B, P450c21B | cytochrome P450, family 21, subfamily A, polypeptide 2 (EC:1.14.99.10) |
C00034649
|
| 1591 | CYP24A1, CP24, CYP24, HCAI, P450-CC24 | cytochrome P450, family 24, subfamily A, polypeptide 1 (EC:1.14.13.126) |
C00034649
|
| 1592 | CYP26A1, CP26, CYP26, P450RAI, P450RAI1 | cytochrome P450, family 26, subfamily A, polypeptide 1 (EC:1.14.-.-) |
C00034649
|
| 1593 | CYP27A1, CP27, CTX, CYP27 | cytochrome P450, family 27, subfamily A, polypeptide 1 (EC:1.14.13.15) |
C00034649
|
| 1594 | CYP27B1, CP2B, CYP1, CYP1alpha, CYP27B, P450c1, PDDR, VDD1, VDDR, VDDRI, VDR | cytochrome P450, family 27, subfamily B, polypeptide 1 (EC:1.14.13.13) |
C00034649
|
| 1548 | CYP2A6, CPA6, CYP2A, CYP2A3, CYPIIA6, P450C2A, P450PB | cytochrome P450, family 2, subfamily A, polypeptide 6 (EC:1.14.14.1) |
C00034649
|
| 1555 | CYP2B6, CPB6, CYP2B, CYP2B7, CYP2B7P, CYPIIB6, EFVM, IIB1, P450 | cytochrome P450, family 2, subfamily B, polypeptide 6 (EC:1.14.14.1) |
C00034649
|
| 1562 | CYP2C18, CPCI, CYP2C, CYP2C17, P450-6B/29C, P450IIC17 | cytochrome P450, family 2, subfamily C, polypeptide 18 (EC:1.14.14.1) |
C00034649
|
| 1557 | CYP2C19, CPCJ, CYP2C, P450C2C, P450IIC19 | cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00034649
|
| 1558 | CYP2C8, CPC8, CYPIIC8, MP-12/MP-20 | cytochrome P450, family 2, subfamily C, polypeptide 8 (EC:1.14.14.1) |
C00034649
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00034649
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00034649
|
| 1577 | CYP3A5, CP35, CYPIIIA5, P450PCN3, PCN3 | cytochrome P450, family 3, subfamily A, polypeptide 5 (EC:1.14.14.1) |
C00034649
|
| 10858 | CYP46A1, CP46, CYP46 | cytochrome P450, family 46, subfamily A, polypeptide 1 (EC:1.14.13.98) |
C00034649
|
| 1580 | CYP4B1, CYPIVB1, P-450HP | cytochrome P450, family 4, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00034649
|
| 199974 | CYP4Z1, CYP4A20 | cytochrome P450, family 4, subfamily Z, polypeptide 1 (EC:1.14.14.1) |
C00034649
|
| 1602 | DACH1, DACH | dachshund homolog 1 (Drosophila) |
C00034649
|
| 51339 | DACT1, DAPPER, DAPPER1, DPR1, FRODO, HDPR1, THYEX3 | dishevelled-binding antagonist of beta-catenin 1 |
C00034649
|
| 55861 | DBNDD2, C20orf35, CK1BP, HSMNP1 | dysbindin (dystrobrevin binding protein 1) domain containing 2 |
C00034649
|
| 131566 | DCBLD2, CLCP1, ESDN | discoidin, CUB and LCCL domain containing 2 |
C00034649
|
| 8642 | DCHS1, CDH25, CDHR6, FIB1, PCDH16 | dachsous cadherin-related 1 |
C00034649
|
| 23576 | DDAH1, DDAH | dimethylarginine dimethylaminohydrolase 1 (EC:3.5.3.18) |
C00034649
|
| 100286728 |
C00034649
|
||
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00034649
|
| 55510 | DDX43, CT13, HAGE | DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 (EC:3.6.4.13) |
C00034649
|
| 1667 | DEFA1, DEF1, DEFA2, HNP-1, HP-1, HP1, MRS | defensin, alpha 1 |
C00034649
|
| 1668 | DEFA3, DEF3, HNP-3, HNP3, HP-3, HP3 | defensin, alpha 3, neutrophil-specific |
C00034649
|
| 1672 | DEFB1, BD1, DEFB-1, DEFB101, HBD1 | defensin, beta 1 |
C00034649
|
| 1673 | DEFB4A, BD-2, DEFB-2, DEFB102, DEFB2, DEFB4, HBD-2, SAP1 | defensin, beta 4A |
C00034649
|
| 27147 | DENND2A, FAM31D, KIAA1277 | DENN/MADD domain containing 2A |
C00034649
|
| 64798 | DEPTOR, DEP.6, DEPDC6 | DEP domain containing MTOR-interacting protein |
C00034649
|
| 1674 | DES, CSM1, CSM2, LGMD2R | desmin |
C00034649
|
| 1687 | DFNA5, ICERE-1 | deafness, autosomal dominant 5 |
C00034649
|
| 1607 | DGKB, DAGK2, DGK, DGK-BETA | diacylglycerol kinase, beta 90kDa (EC:2.7.1.107) |
C00034649
|
| 8527 | DGKD, DGKdelta, dgkd-2 | diacylglycerol kinase, delta 130kDa (EC:2.7.1.107) |
C00034649
|
| 9249 | DHRS3, DD83.1, RDH17, Rsdr1, SDR1, SDR16C1, retSDR1 | dehydrogenase/reductase (SDR family) member 3 (EC:1.1.1.300) |
C00034649
|
| 1665 | DHX15, DBP1, DDX15, HRH2, PRP43, PRPF43, PrPp43p | DEAH (Asp-Glu-Ala-His) box helicase 15 (EC:3.6.4.13) |
C00034649
|
| 1729 | DIAPH1, DFNA1, DIA1, DRF1, LFHL1, hDIA1 | diaphanous-related formin 1 |
C00034649
|
| 1730 | DIAPH2, DIA, DIA2, DRF2, POF, POF2 | diaphanous-related formin 2 |
C00034649
|
| 1734 | DIO2, 5DII, D2, DIOII, SelY, TXDI2 | deiodinase, iodothyronine, type II (EC:1.97.1.10) |
C00034649
|
| 729582 | DIRC3 | disrupted in renal carcinoma 3 |
C00034649
|
| 22943 | DKK1, DKK-1, SK | dickkopf WNT signaling pathway inhibitor 1 |
C00034649
|
| 27122 | DKK3, REIC, RIG | dickkopf WNT signaling pathway inhibitor 3 |
C00034649
|
| 10395 | DLC1, ARHGAP7, HP, STARD12, p122-RhoGAP | deleted in liver cancer 1 |
C00034649
|
| 9231 | DLG5, LP-DLG, P-DLG5, PDLG | discs, large homolog 5 (Drosophila) |
C00034649
|
| 1748 | DLX4, BP1, DLX7, DLX8, DLX9 | distal-less homeobox 4 |
C00034649
|
| 1755 | DMBT1, GP340, muclin | deleted in malignant brain tumors 1 |
C00034649
|
| 1756 | DMD, BMD, CMD3B, DXS142, DXS164, DXS206, DXS230, DXS239, DXS268, DXS269, DXS270, DXS272 | dystrophin |
C00034649
|
| 55172 | DNAAF2, C14orf104, CILD10, KTU, PF13 | dynein, axonemal, assembly factor 2 |
C00034649
|
| 3337 | DNAJB1, HSPF1, Hdj1, Hsp40, RSPH16B, Sis1 | DnaJ (Hsp40) homolog, subfamily B, member 1 |
C00034649
|
| 29103 | DNAJC15, DNAJD1, HSD18, MCJ | DnaJ (Hsp40) homolog, subfamily C, member 15 |
C00034649
|
| 92737 | DNER, UNQ26, bet | delta/notch-like EGF repeat containing |
C00034649
|
| 26052 | DNM3, Dyna_III | dynamin 3 (EC:3.6.5.5) |
C00034649
|
| 9732 | DOCK4 | dedicator of cytokinesis 4 |
C00034649
|
| 1803 | DPP4, ADABP, ADCP2, CD26, DPPIV, TP103 | dipeptidyl-peptidase 4 (EC:3.4.14.5) |
C00034649
|
| 667 | DST, BP240, BPA, BPAG1, CATX-15, CATX15, D6S1101, DMH, DT, EBSB2, HSAN6, MACF2 | dystonin |
C00034649
|
| 51514 | DTL, CDT2, DCAF2, L2DTL, RAMP | denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
C00034649
|
| 53905 | DUOX1, LNOX1, NOXEF1, THOX1 | dual oxidase 1 (EC:1.6.3.1) |
C00034649
|
| 56931 | DUS3L, DUS3 | dihydrouridine synthase 3-like (S. cerevisiae) |
C00034649
|
| 1847 | DUSP5, DUSP, HVH3 | dual specificity phosphatase 5 (EC:3.1.3.16 3.1.3.48) |
C00034649
|
| 1848 | DUSP6, HH19, MKP3, PYST1 | dual specificity phosphatase 6 (EC:3.1.3.16 3.1.3.48) |
C00034649
|
| 1797 | DXO, DOM3L, DOM3Z, NG6, RAI1 | decapping exoribonuclease |
C00034649
|
| 8445 | DYRK2 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 (EC:2.7.12.1) |
C00034649
|
| 55840 | EAF2, BM040, TRAITS, U19 | ELL associated factor 2 |
C00034649
|
| 10148 | EBI3, IL-27B, IL27B | Epstein-Barr virus induced 3 |
C00034649
|
| 9718 | ECE2 | endothelin converting enzyme 2 (EC:3.4.24.71) |
C00034649
|
| 55268 | ECHDC2 | enoyl CoA hydratase domain containing 2 |
C00034649
|
| 1893 | ECM1, URBWD | extracellular matrix protein 1 |
C00034649
|
| 1908 | EDN3, ET-3, ET3, HSCR4, PPET3, WS4B | endothelin 3 |
C00034649
|
| 1909 | EDNRA, ET-A, ETA, ETA-R, ETAR, ETRA, hET-AR | endothelin receptor type A |
C00034649
|
| 1910 | EDNRB, ABCDS, ET-B, ET-BR, ETB, ETBR, ETRB, HSCR, HSCR2, WS4A | endothelin receptor type B |
C00034649
|
| 29904 | EEF2K, HSU93850, eEF-2K | eukaryotic elongation factor-2 kinase (EC:2.7.11.20) |
C00034649
|
| 10278 | EFS, CAS3, CASS3, EFS1, EFS2, HEFS, SIN | embryonal Fyn-associated substrate |
C00034649
|
| 1950 | EGF, HOMG4, URG | epidermal growth factor |
C00034649
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00034649
|
| 1959 | EGR2, AT591, CMT1D, CMT4E, KROX20 | early growth response 2 |
C00034649
|
| 23301 | EHBP1, HPC12, NACSIN | EH domain binding protein 1 |
C00034649
|
| 10938 | EHD1, H-PAST, HPAST1, PAST, PAST1 | EH-domain containing 1 |
C00034649
|
| 30846 | EHD2, PAST2 | EH-domain containing 2 |
C00034649
|
| 26298 | EHF, ESE3, ESE3B, ESEJ | ets homologous factor |
C00034649
|
| 10209 | EIF1, A121, EIF-1, EIF1A, ISO1, SUI1 | eukaryotic translation initiation factor 1 |
C00034649
|
| 1968 | EIF2S3, EIF2, EIF2G, EIF2gamma, eIF-2gA | eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
C00034649
|
| 1983 | EIF5, EIF-5, EIF-5A | eukaryotic translation initiation factor 5 |
C00034649
|
| 1999 | ELF3, EPR-1, ERT, ESE-1, ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
C00034649
|
| 64834 | ELOVL1, Ssc1 | ELOVL fatty acid elongase 1 (EC:2.3.1.199) |
C00034649
|
| 2013 | EMP2, XMP | epithelial membrane protein 2 |
C00034649
|
| 2028 | ENPEP, APA, CD249, gp160 | glutamyl aminopeptidase (aminopeptidase A) (EC:3.4.11.7) |
C00034649
|
| 5167 | ENPP1, ARHR2, M6S1, NPP1, NPPS, PC-1, PCA1, PDNP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 (EC:3.1.4.1 3.6.1.9) |
C00034649
|
| 22875 | ENPP4, NPP4 | ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) (EC:3.6.1.29) |
C00034649
|
| 64097 | EPB41L4A, EPB41L4, NBL4 | erythrocyte membrane protein band 4.1 like 4A |
C00034649
|
| 54566 | EPB41L4B, CG1, EHM2 | erythrocyte membrane protein band 4.1 like 4B |
C00034649
|
| 2042 | EPHA3, EK4, ETK, ETK1, HEK, HEK4, TYRO4 | EPH receptor A3 (EC:2.7.10.1) |
C00034649
|
| 2045 | EPHA7, EHK3, HEK11 | EPH receptor A7 (EC:2.7.10.1) |
C00034649
|
| 55914 | ERBB2IP, ERBIN, LAP2 | erbb2 interacting protein |
C00034649
|
| 121506 | ERP27, C12orf46, PDIA8 | endoplasmic reticulum protein 27 |
C00034649
|
| 30816 | ERVW-1, ENV, ENVW, ERVWE1, HERV-7q, HERV-W-ENV, HERV7Q, HERVW, HERVWENV | endogenous retrovirus group W, member 1 |
C00034649
|
| 2101 | ESRRA, ERR1, ERRa, ERRalpha, ESRL1, NR3B1 | estrogen-related receptor alpha |
C00034649
|
| 2115 | ETV1, ER81 | ets variant 1 |
C00034649
|
| 2119 | ETV5, ERM | ets variant 5 |
C00034649
|
| 2123 | EVI2A, EVDA, EVI-2A, EVI2 | ecotropic viral integration site 2A |
C00034649
|
| 2159 | F10, FX, FXA | coagulation factor X (EC:3.4.21.6) |
C00034649
|
| 2162 | F13A1, F13A | coagulation factor XIII, A1 polypeptide (EC:2.3.2.13) |
C00034649
|
| 2147 | F2, PT, RPRGL2, THPH1 | coagulation factor II (thrombin) (EC:3.4.21.5) |
C00034649
|
| 2151 | F2RL2, PAR-3, PAR3 | coagulation factor II (thrombin) receptor-like 2 |
C00034649
|
| 9002 | F2RL3, PAR4 | coagulation factor II (thrombin) receptor-like 3 |
C00034649
|
| 2152 | F3, CD142, TF, TFA | coagulation factor III (thromboplastin, tissue factor) |
C00034649
|
| 2166 | FAAH, FAAH-1, PSAB | fatty acid amide hydrolase (EC:3.5.1.99) |
C00034649
|
| 9415 | FADS2, D6D, DES6, FADSD6, LLCDL2, SLL0262, TU13 | fatty acid desaturase 2 (EC:1.14.19.-) |
C00034649
|
| 3995 | FADS3, CYB5RP, LLCDL3 | fatty acid desaturase 3 (EC:1.14.19.-) |
C00034649
|
| 55179 | FAIM, FAIM1 | Fas apoptotic inhibitory molecule |
C00034649
|
| 359845 | FAM101B | family with sequence similarity 101, member B |
C00034649
|
| 54491 | FAM105A, NET20 | family with sequence similarity 105, member A |
C00034649
|
| 11170 | FAM107A, DRR1, TU3A | family with sequence similarity 107, member A |
C00034649
|
| 54463 | FAM134B, JK-1, JK1 | family with sequence similarity 134, member B |
C00034649
|
| 285596 | FAM153A, NY-REN-7 | family with sequence similarity 153, member A |
C00034649
|
| 220382 | FAM181B | family with sequence similarity 181, member B |
C00034649
|
| 51313 | FAM198B, AD036, C4orf18 | family with sequence similarity 198, member B |
C00034649
|
| 338811 | FAM19A2, TAFA-2, TAFA2 | family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
C00034649
|
| 387680 | FAM21A, bA56A21.1 | family with sequence similarity 21, member A |
C00034649
|
| 441168 | FAM26F, C6orf187, dJ93H18.5 | family with sequence similarity 26, member F |
C00034649
|
| 421052 |
C00034649
|
||
| 55603 | FAM46A, C6orf37, XTP11 | family with sequence similarity 46, member A |
C00034649
|
| 2196 | FAT2, CDHF8, CDHR9, HFAT2, MEGF1 | FAT atypical cadherin 2 |
C00034649
|
| 10826 | FAXDC2, C5orf4 | fatty acid hydroxylase domain containing 2 |
C00034649
|
| 2192 | FBLN1, FBLN, FIBL1 | fibulin 1 |
C00034649
|
| 2199 | FBLN2 | fibulin 2 |
C00034649
|
| 10516 | FBLN5, ADCL2, ARCL1A, ARMD3, DANCE, EVEC, FIBL-5, UP50 | fibulin 5 |
C00034649
|
| 23194 | FBXL7, FBL6, FBL7 | F-box and leucine-rich repeat protein 7 |
C00034649
|
| 80204 | FBXO11, FBX11, PRMT9, UBR6, VIT1 | F-box protein 11 |
C00034649
|
| 114907 | FBXO32, Fbx32, MAFbx | F-box protein 32 |
C00034649
|
| 2214 | FCGR3A, CD16, CD16A, FCG3, FCGR3, FCGRIII, FCR-10, FCRIII, FCRIIIA, IGFR3 | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
C00034649
|
| 55612 | FERMT1, C20orf42, DTGCU2, KIND1, UNC112A, URP1 | fermitin family member 1 |
C00034649
|
| 10979 | FERMT2, KIND2, MIG2, PLEKHC1, UNC112, UNC112B, mig-2 | fermitin family member 2 |
C00034649
|
| 2243 | FGA, Fib2 | fibrinogen alpha chain |
C00034649
|
| 2244 | FGB | fibrinogen beta chain |
C00034649
|
| 121512 | FGD4, CMT4H, FRABP, ZFYVE6 | FYVE, RhoGEF and PH domain containing 4 |
C00034649
|
| 2258 | FGF13, FGF-13, FGF2, FHF-2, FHF2 | fibroblast growth factor 13 |
C00034649
|
| 8817 | FGF18, FGF-18, ZFGF5 | fibroblast growth factor 18 |
C00034649
|
| 2247 | FGF2, BFGF, FGF-2, FGFB, HBGF-2 | fibroblast growth factor 2 (basic) |
C00034649
|
| 2254 | FGF9, FGF-9, GAF, HBFG-9, HBGF-9, SYNS3 | fibroblast growth factor 9 |
C00034649
|
| 9982 | FGFBP1, FGF-BP, FGF-BP1, FGFBP, FGFBP-1, HBP17 | fibroblast growth factor binding protein 1 |
C00034649
|
| 2263 | FGFR2, BBDS, BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 | fibroblast growth factor receptor 2 (EC:2.7.10.1) |
C00034649
|
| 2267 | FGL1, HFREP1, HP-041, LFIRE-1, LFIRE1 | fibrinogen-like 1 |
C00034649
|
| 10875 | FGL2, T49, pT49 | fibrinogen-like 2 |
C00034649
|
| 80206 | FHOD3, FHOS2, Formactin2 | formin homology 2 domain containing 3 |
C00034649
|
| 24147 | FJX1 | four jointed box 1 (Drosophila) |
C00034649
|
| 23768 | FLRT2 | fibronectin leucine rich transmembrane protein 2 |
C00034649
|
| 342184 | FMN1, FMN, LD | formin 1 |
C00034649
|
| 84624 | FNDC1, AGS8, FNDC2, MEL4B3, RP11-243O10.2, bA243O10.1, dJ322A24.1 | fibronectin type III domain containing 1 |
C00034649
|
| 22862 | FNDC3A, FNDC3, HUGO, bA203I16.1, bA203I16.5 | fibronectin type III domain containing 3A |
C00034649
|
| 2348 | FOLR1, FBP, FOLR | folate receptor 1 (adult) |
C00034649
|
| 2354 | FOSB, AP-1, G0S3, GOS3, GOSB | FBJ murine osteosarcoma viral oncogene homolog B |
C00034649
|
| 2308 | FOXO1, FKH1, FKHR, FOXO1A | forkhead box O1 |
C00034649
|
| 768121 |
C00034649
|
||
| 50943 | FOXP3, AIID, DIETER, IPEX, PIDX, XPID | forkhead box P3 |
C00034649
|
| 55691 | FRMD4A, FRMD4, bA295P9.4 | FERM domain containing 4A |
C00034649
|
| 10129 | FRY, 13CDNA73, 214K23.2, C13orf14, CG003, bA207N4.2, bA37E23.1 | furry homolog (Drosophila) |
C00034649
|
| 2488 | FSHB | follicle stimulating hormone, beta polypeptide |
C00034649
|
| 10468 | FST, FS | follistatin |
C00034649
|
| 5349 | FXYD3, MAT8, PLML | FXYD domain containing ion transport regulator 3 |
C00034649
|
| 53828 | FXYD4, CHIF | FXYD domain containing ion transport regulator 4 |
C00034649
|
| 11211 | FZD10, CD350, FZ-10, Fz10, FzE7, hFz10 | frizzled family receptor 10 |
C00034649
|
| 7855 | FZD5, C2orf31, HFZ5 | frizzled family receptor 5 |
C00034649
|
| 8324 | FZD7, FzE3 | frizzled family receptor 7 |
C00034649
|
| 51343 | FZR1, CDC20C, CDH1, FZR, FZR2, HCDH, HCDH1 | fizzy/cell division cycle 20 related 1 (Drosophila) |
C00034649
|
| 50486 | G0S2, RP1-28O10.2 | G0/G1switch 2 |
C00034649
|
| 2538 | G6PC, G6PC1, G6PT, GSD1, GSD1a | glucose-6-phosphatase, catalytic subunit (EC:3.1.3.9) |
C00034649
|
| 2550 | GABBR1, GABABR1, GABBR1-3, GB1, GPRC3A, dJ271M21.1.1, dJ271M21.1.2 | gamma-aminobutyric acid (GABA) B receptor, 1 |
C00034649
|
| 2555 | GABRA2 | gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
C00034649
|
| 2558 | GABRA5 | gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
C00034649
|
| 2564 | GABRE | gamma-aminobutyric acid (GABA) A receptor, epsilon |
C00034649
|
| 2567 | GABRG3 | gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
C00034649
|
| 2568 | GABRP | gamma-aminobutyric acid (GABA) A receptor, pi |
C00034649
|
| 2585 | GALK2, GK2 | galactokinase 2 (EC:2.7.1.157) |
C00034649
|
| 2596 | GAP43, B-50, PP46 | growth associated protein 43 |
C00034649
|
| 54433 | GAR1, NOLA1 | GAR1 ribonucleoprotein |
C00034649
|
| 2617 | GARS, CMT2D, DSMAV, GlyRS, HMN5, SMAD1 | glycyl-tRNA synthetase (EC:6.1.1.14) |
C00034649
|
| 2619 | GAS1 | growth arrest-specific 1 |
C00034649
|
| 2621 | GAS6, AXLLG, AXSF | growth arrest-specific 6 |
C00034649
|
| 2520 | GAST, GAS | gastrin |
C00034649
|
| 2627 | GATA6 | GATA binding protein 6 |
C00034649
|
| 115361 | GBP4, Mpa2 | guanylate binding protein 4 |
C00034649
|
| 2650 | GCNT1, C2GNT, C2GNT-L, C2GNT1, G6NT, NACGT2, NAGCT2 | glucosaminyl (N-acetyl) transferase 1, core 2 (EC:2.4.1.102) |
C00034649
|
| 2661 | GDF9 | growth differentiation factor 9 |
C00034649
|
| 2672 | GFI1, GFI-1, GFI1A, SCN2, ZNF163 | growth factor independent 1 transcription repressor |
C00034649
|
| 9945 | GFPT2, GFAT2 | glutamine-fructose-6-phosphate transaminase 2 (EC:2.6.1.16) |
C00034649
|
| 2687 | GGT5, GGT-REL, GGTLA1 | gamma-glutamyltransferase 5 (EC:2.3.2.2 3.4.19.13 3.4.19.14) |
C00034649
|
| 14594 |
C00034649
|
||
| 2681 | GGTA1P, GGTA, GGTA1, GLYT2, a1/3GTP | glycoprotein, alpha-galactosyltransferase 1 pseudogene |
C00034649
|
| 2688 | GH1, GH, GH-N, GHN, IGHD1B, hGH-N | growth hormone 1 |
C00034649
|
| 2697 | GJA1, AVSD3, CMDR, CX43, DFNB38, GJAL, HLHS1, HSS, ODDD | gap junction protein, alpha 1, 43kDa |
C00034649
|
| 2701 | GJA4, CX37 | gap junction protein, alpha 4, 37kDa |
C00034649
|
| 2706 | GJB2, CX26, DFNA3, DFNA3A, DFNB1, DFNB1A, HID, KID, NSRD1, PPK | gap junction protein, beta 2, 26kDa |
C00034649
|
| 2710 | GK, GK1, GKD | glycerol kinase (EC:2.7.1.30) |
C00034649
|
| 89944 | GLB1L2, MST114, MSTP114 | galactosidase, beta 1-like 2 (EC:3.2.1.23) |
C00034649
|
| 11010 | GLIPR1, CRISP7, GLIPR, RTVP1 | GLI pathogenesis-related 1 |
C00034649
|
| 84662 | GLIS2, NKL, NPHP7 | GLIS family zinc finger 2 |
C00034649
|
| 144423 | GLT1D1 | glycosyltransferase 1 domain containing 1 (EC:2.4.-.-) |
C00034649
|
| 51228 | GLTP | glycolipid transfer protein |
C00034649
|
| 10249 | GLYAT, ACGNAT, CAT, GAT | glycine-N-acyltransferase (EC:2.3.1.13 2.3.1.71) |
C00034649
|
| 2766 | GMPR, GMPR1 | guanosine monophosphate reductase (EC:1.7.1.7) |
C00034649
|
| 2773 | GNAI3, 87U6, ARCND1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
C00034649
|
| 2791 | GNG11, GNGT11 | guanine nucleotide binding protein (G protein), gamma 11 |
C00034649
|
| 54331 | GNG2 | guanine nucleotide binding protein (G protein), gamma 2 |
C00034649
|
| 2786 | GNG4 | guanine nucleotide binding protein (G protein), gamma 4 |
C00034649
|
| 29889 | GNL2, HUMAUANTIG, NGP1, Ngp-1 | guanine nucleotide binding protein-like 2 (nucleolar) |
C00034649
|
| 2796 | GNRH1, GNRH, GRH, HH12, LHRH, LNRH | gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
C00034649
|
| 283768 | GOLGA8G | golgin A8 family, member G |
C00034649
|
| 785067 |
C00034649
|
||
| 23171 | GPD1L, GPD1-L | glycerol-3-phosphate dehydrogenase 1-like (EC:1.1.1.8) |
C00034649
|
| 57211 | GPR126, APG1, DREG, PS1TP2, VIGR | G protein-coupled receptor 126 |
C00034649
|
| 283383 | GPR133, PGR25 | G protein-coupled receptor 133 |
C00034649
|
| 151556 | GPR155, DEP.7, DEPDC3, PGR22 | G protein-coupled receptor 155 |
C00034649
|
| 2840 | GPR17 | G protein-coupled receptor 17 |
C00034649
|
| 51704 | GPRC5B, RAIG-2, RAIG2 | G protein-coupled receptor, family C, group 5, member B |
C00034649
|
| 2877 | GPX2, GI-GPx, GPRP, GPRP-2, GPx-2, GPx-GI, GSHPX-GI, GSHPx-2 | glutathione peroxidase 2 (gastrointestinal) (EC:1.11.1.9) |
C00034649
|
| 2878 | GPX3, GPx-P, GSHPx-3, GSHPx-P | glutathione peroxidase 3 (plasma) (EC:1.11.1.9) |
C00034649
|
| 26585 | GREM1, CKTSF1B1, DAND2, DRM, GREMLIN, IHG-2 | gremlin 1, DAN family BMP antagonist |
C00034649
|
| 29841 | GRHL1, LBP32, MGR, NH32, TFCP2L2 | grainyhead-like 1 (Drosophila) |
C00034649
|
| 2890 | GRIA1, GLUH1, GLUR1, GLURA, GluA1, HBGR1 | glutamate receptor, ionotropic, AMPA 1 |
C00034649
|
| 83743 | GRWD1, CDW4, GRWD, RRB1, WDR28 | glutamate-rich WD repeat containing 1 |
C00034649
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00000094
|
| 2938 | GSTA1, GST2, GSTA1-1, GTH1 | glutathione S-transferase alpha 1 (EC:2.5.1.18) |
C00034649
|
| 2940 | GSTA3, GSTA3-3, GTA3 | glutathione S-transferase alpha 3 (EC:2.5.1.18) |
C00034649
|
| 2952 | GSTT1 | glutathione S-transferase theta 1 (EC:2.5.1.18) |
C00034649
|
| 219409 | GSX1, GSH1, Gsh-1 | GS homeobox 1 |
C00034649
|
| 120071 | GYLTL1B, LARGE2 | glycosyltransferase-like 1B |
C00034649
|
| 2996 | GYPE, GPE, MNS, MiIX | glycophorin E (MNS blood group) |
C00034649
|
| 286436 | H2BFM | H2B histone family, member M |
C00034649
|
| 54145 |
C00034649
|
||
| 3026 | HABP2, FSAP, HABP, HGFAL, PHBP | hyaluronan binding protein 2 |
C00034649
|
| 9464 | HAND2, DHAND2, Hed, Thing2, bHLHa26, dHand | heart and neural crest derivatives expressed 2 |
C00034649
|
| 79804 | HAND2-AS1, DEIN, NBLA00301 | HAND2 antisense RNA 1 (head to head) |
C00034649
|
| 1404 | HAPLN1, CRTL1 | hyaluronan and proteoglycan link protein 1 |
C00034649
|
| 3036 | HAS1, HAS | hyaluronan synthase 1 (EC:2.4.1.212) |
C00034649
|
| 3038 | HAS3 | hyaluronan synthase 3 (EC:2.4.1.212) |
C00034649
|
| 3039 | HBA1, CD31, HBH | hemoglobin, alpha 1 |
C00034649
|
| 3040 | HBA2, HBH | hemoglobin, alpha 2 |
C00034649
|
| 3043 | HBB, CD113t-C, beta-globin | hemoglobin, beta |
C00034649
|
| 3045 | HBD | hemoglobin, delta |
C00034649
|
| 3047 | HBG1, HBGA, HBGR, HSGGL1 | hemoglobin, gamma A |
C00034649
|
| 3048 | HBG2, TNCY | hemoglobin, gamma G |
C00034649
|
| 10767 | HBS1L, EF-1a, ERFS, HBS1, HSPC276 | HBS1-like (S. cerevisiae) |
C00034649
|
| 8843 | HCAR3, GPR109B, HCA3, HM74, PUMAG, Puma-g | hydroxycarboxylic acid receptor 3 |
C00034649
|
| 3059 | HCLS1, CTTNL, HS1 | hematopoietic cell-specific Lyn substrate 1 |
C00034649
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00034649
|
| 57493 | HEG1, HEG, MST112, MSTP112 | heart development protein with EGF-like domains 1 |
C00034649
|
| 51191 | HERC5, CEB1, CEBP1 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
C00034649
|
| 9709 | HERPUD1, HERP, Mif1, SUP | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
C00034649
|
| 388585 | HES5, bHLHb38 | hes family bHLH transcription factor 5 |
C00034649
|
| 3081 | HGD, AKU, HGO | homogentisate 1,2-dioxygenase (EC:1.13.11.5) |
C00034649
|
| 3082 | HGF, DFNB39, F-TCF, HGFB, HPTA, SF | hepatocyte growth factor (hepapoietin A; scatter factor) |
C00034649
|
| 26275 | HIBCH, HIBYLCOAH | 3-hydroxyisobutyryl-CoA hydrolase (EC:3.1.2.4) |
C00034649
|
| 3090 | HIC1, ZBTB29, ZNF901, hic-1 | hypermethylated in cancer 1 |
C00034649
|
| 121504 | HIST4H4, H4/p | histone cluster 4, H4 |
C00034649
|
| 3122 | HLA-DRA, HLA-DRA1, MLRW | major histocompatibility complex, class II, DR alpha |
C00034649
|
| 3123 | HLA-DRB1, DRB1, DRw10, HLA-DR1B, HLA-DRB, SS1 | major histocompatibility complex, class II, DR beta 1 |
C00034649
|
| 3125 | HLA-DRB3, HLA-DR3B | major histocompatibility complex, class II, DR beta 3 |
C00034649
|
| 3133 | HLA-E, EA1.2, EA2.1, HLA-6.2, MHC, QA1 | major histocompatibility complex, class I, E |
C00034649
|
| 3134 | HLA-F, CDA12, HLA-5.4, HLA-CDA12, HLAF | major histocompatibility complex, class I, F |
C00034649
|
| 3135 | HLA-G, MHC-G | major histocompatibility complex, class I, G |
C00034649
|
| 83872 | HMCN1, ARMD1, FBLN6, FIBL-6, FIBL6 | hemicentin 1 |
C00034649
|
| 3156 | HMGCR, LDLCQ3 | 3-hydroxy-3-methylglutaryl-CoA reductase (EC:1.1.1.34) |
C00034649
|
| 3157 | HMGCS1, HMGCS | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) (EC:2.3.3.10) |
C00034649
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00034649
|
| 3172 | HNF4A, HNF4, HNF4a7, HNF4a8, HNF4a9, HNF4alpha, MODY, MODY1, NR2A1, NR2A21, TCF, TCF14 | hepatocyte nuclear factor 4, alpha |
C00034649
|
| 9454 | HOMER3, HOMER-3, VESL3 | homer homolog 3 (Drosophila) |
C00034649
|
| 3206 | HOXA10, HOX1, HOX1.8, HOX1H, PL | homeobox A10 |
C00034649
|
| 3207 | HOXA11, HOX1, HOX1I | homeobox A11 |
C00034649
|
| 3240 | HP, BP, HP2ALPHA2, HPA1S | haptoglobin |
C00034649
|
| 9953 | HS3ST3B1, 30ST3B1, 3OST3B1 | heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 (EC:2.8.2.30) |
C00034649
|
| 9394 | HS6ST1, HH15, HS6ST | heparan sulfate 6-O-sulfotransferase 1 (EC:2.8.2.-) |
C00034649
|
| 3290 | HSD11B1, 11-DH, 11-beta-HSD1, CORTRD2, HDL, HSD11, HSD11B, HSD11L, SDR26C1 | hydroxysteroid (11-beta) dehydrogenase 1 (EC:1.1.1.146) |
C00034649
|
| 3291 | HSD11B2, AME, AME1, HSD11K, HSD2, SDR9C3 | hydroxysteroid (11-beta) dehydrogenase 2 (EC:1.1.1.146) |
C00034649
|
| 51170 | HSD17B11, 17BHSD11, DHRS8, PAN1B, RETSDR2, SDR16C2 | hydroxysteroid (17-beta) dehydrogenase 11 (EC:1.1.1.62) |
C00034649
|
| 3294 | HSD17B2, EDH17B2, HSD17, SDR9C2 | hydroxysteroid (17-beta) dehydrogenase 2 (EC:1.1.1.62 1.1.1.239) |
C00034649
|
| 3283 | HSD3B1, 3BETAHSD, HSD3B, HSDB3, HSDB3A, I, SDR11E1 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 (EC:1.1.1.145 5.3.3.1) |
C00034649
|
| 3284 | HSD3B2, HSD3B, HSDB, SDR11E2 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 (EC:1.1.1.145 5.3.3.1) |
C00034649
|
| 3304 | HSPA1B, HSP70-1B, HSP70-2 | heat shock 70kDa protein 1B |
C00034649
|
| 3310 | HSPA6 | heat shock 70kDa protein 6 (HSP70B') |
C00034649
|
| 26353 | HSPB8, CMT2L, DHMN2, E2IG1, H11, HMN2, HMN2A, HSP22 | heat shock 22kDa protein 8 |
C00034649
|
| 7127 | TNFAIP2, B94, EXOC3L3 | tumor necrosis factor, alpha-induced protein 2 |
C00000094
|
| 3357 | HTR2B, 5-HT(2B), 5-HT2B | 5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
C00034649
|
| 8692 | HYAL2, LUCA2 | hyaluronoglucosaminidase 2 (EC:3.2.1.35) |
C00034649
|
| 57061 | HYMAI, NCRNA00020 | hydatidiform mole associated and imprinted (non-protein coding) |
C00034649
|
| 10525 | HYOU1, GRP-170, Grp170, HSP12A, ORP-150, ORP150 | hypoxia up-regulated 1 |
C00034649
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00034649
|
| 3397 | ID1, ID, bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
C00034649
|
| 3398 | ID2, GIG8, ID2A, ID2H, bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
C00034649
|
| 3400 | ID4, IDB4, bHLHb27 | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
C00034649
|
| 3417 | IDH1, IDCD, IDH, IDP, IDPC, PICD | isocitrate dehydrogenase 1 (NADP+), soluble (EC:1.1.1.42) |
C00034649
|
| 55853 | IDI2-AS1, C10orf110, IDI2-AS | IDI2 antisense RNA 1 |
C00034649
|
| 3434 | IFIT1, C56, G10P1, IFI-56, IFI-56K, IFI56, IFIT-1, IFNAI1, ISG56, P56, RNM561 | interferon-induced protein with tetratricopeptide repeats 1 |
C00034649
|
| 3437 | IFIT3, CIG-49, GARG-49, IFI60, IFIT4, IRG2, ISG60, P60, RIG-G | interferon-induced protein with tetratricopeptide repeats 3 |
C00034649
|
| 3440 | IFNA2, IFN-alphaA, IFNA, IFNA2B, INFA2 | interferon, alpha 2 |
C00034649
|
| 3455 | IFNAR2, IFN-R, IFN-alpha-REC, IFNABR, IFNARB | interferon (alpha, beta and omega) receptor 2 |
C00034649
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00034649
|
| 3459 | IFNGR1, CD119, IFNGR | interferon gamma receptor 1 |
C00034649
|
| 3475 | IFRD1, PC4, TIS7 | interferon-related developmental regulator 1 |
C00034649
|
| 80173 | IFT74, CCDC2, CMG-1, CMG1 | intraflagellar transport 74 homolog (Chlamydomonas) |
C00034649
|
| 3479 | IGF1, IGF-I, IGF1A, IGFI | insulin-like growth factor 1 (somatomedin C) |
C00034649
|
| 3480 | IGF1R, CD221, IGFIR, IGFR, JTK13 | insulin-like growth factor 1 receptor (EC:2.7.10.1) |
C00034649
|
| 3481 | IGF2, C11orf43, IGF-II, PP9974 | insulin-like growth factor 2 (somatomedin A) |
C00034649
|
| 3484 | IGFBP1, AFBP, IBP1, IGF-BP25, PP12, hIGFBP-1 | insulin-like growth factor binding protein 1 |
C00034649
|
| 3486 | IGFBP3, BP-53, IBP3 | insulin-like growth factor binding protein 3 |
C00034649
|
| 3489 | IGFBP6, IBP6 | insulin-like growth factor binding protein 6 |
C00034649
|
| 3502 |
C00034649
|
||
| 3514 |
C00034649
|
||
| 28831 |
C00034649
|
||
| 3543 | IGLL1, 14.1, AGM2, CD179b, IGL1, IGL5, IGLJ14.1, IGLL, IGO, IGVPB, VPREB2 | immunoglobulin lambda-like polypeptide 1 |
C00034649
|
| 3549 | IHH, BDA1, HHG2 | indian hedgehog |
C00034649
|
| 3586 | IL10, CSIF, GVHDS, IL-10, IL10A, TGIF | interleukin 10 |
C00034649
|
| 3589 | IL11, AGIF, IL-11 | interleukin 11 |
C00034649
|
| 3592 | IL12A, CLMF, IL-12A, NFSK, NKSF1, P35 | interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
C00034649
|
| 7029 | TFDP2, DP2 | transcription factor Dp-2 (E2F dimerization partner 2) |
C00000094
|
| 3597 | IL13RA1, CD213A1, IL-13Ra, NR4 | interleukin 13 receptor, alpha 1 |
C00034649
|
| 3598 | IL13RA2, CD213A2, CT19, IL-13R, IL13BP | interleukin 13 receptor, alpha 2 |
C00034649
|
| 3600 | IL15, IL-15 | interleukin 15 |
C00034649
|
| 3601 | IL15RA, CD215 | interleukin 15 receptor, alpha |
C00034649
|
| 54756 | IL17RD, HH18, IL-17RD, IL17RLM, SEF | interleukin 17 receptor D |
C00034649
|
| 6016 | RIT1, NS8, RIBB, RIT, ROC1 | Ras-like without CAAX 1 |
C00000094
|
| 9173 | IL1RL1, DER4, FIT-1, IL33R, ST2, ST2L, ST2V, T1 | interleukin 1 receptor-like 1 |
C00034649
|
| 53832 | IL20RA, CRF2-8, IL-20R-alpha, IL-20R1, IL-20RA | interleukin 20 receptor, alpha |
C00034649
|
| 11009 | IL24, C49A, FISP, IL10B, MDA7, MOB5, ST16 | interleukin 24 |
C00034649
|
| 3560 | IL2RB, CD122, IL15RB, P70-75 | interleukin 2 receptor, beta |
C00034649
|
| 90865 | IL33, C9orf26, DVS27, IL1F11, NF-HEV, NFEHEV, RP11-575C20.2 | interleukin 33 |
C00034649
|
| 3565 | IL4, BCGF-1, BCGF1, BSF-1, BSF1, IL-4 | interleukin 4 |
C00034649
|
| 3566 | IL4R, CD124, IL-4RA, IL4RA | interleukin 4 receptor |
C00034649
|
| 3570 | IL6R, CD126, IL-6R-1, IL-6RA, IL6Q, IL6RA, IL6RQ, gp80 | interleukin 6 receptor |
C00034649
|
| 3575 | IL7R, CD127, CDW127, IL-7R-alpha, IL7RA, ILRA | interleukin 7 receptor |
C00034649
|
| 3623 | INHA | inhibin, alpha |
C00034649
|
| 3624 | INHBA, EDF, FRP | inhibin, beta A |
C00034649
|
| 3643 | INSR, CD220, HHF5 | insulin receptor (EC:2.7.10.1) |
C00034649
|
| 79711 | IPO4, Imp4 | importin 4 |
C00034649
|
| 10788 | IQGAP2 | IQ motif containing GTPase activating protein 2 |
C00034649
|
| 3654 | IRAK1, IRAK, pelle | interleukin-1 receptor-associated kinase 1 (EC:2.7.11.1) |
C00034649
|
| 3656 | IRAK2, IRAK-2 | interleukin-1 receptor-associated kinase 2 |
C00034649
|
| 11213 | IRAK3, ASRT5, IRAKM | interleukin-1 receptor-associated kinase 3 (EC:2.7.11.1) |
C00034649
|
| 51135 | IRAK4, IPD1, IRAK-4, NY-REN-64, REN64 | interleukin-1 receptor-associated kinase 4 (EC:2.7.11.1) |
C00034649
|
| 50805 | IRX4, IRXA3 | iroquois homeobox 4 |
C00034649
|
| 140862 | ISM1, C20orf82, ISM, Isthmin, bA149I18.1, dJ1077I2.1 | isthmin 1, angiogenesis inhibitor |
C00034649
|
| 51015 | ISOC1 | isochorismatase domain containing 1 |
C00034649
|
| 3672 | ITGA1, CD49a, VLA1 | integrin, alpha 1 |
C00034649
|
| 8515 | ITGA10, PRO827 | integrin, alpha 10 |
C00034649
|
| 3675 | ITGA3, CD49C, GAP-B3, GAPB3, ILNEB, MSK18, VCA-2, VL3A, VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
C00034649
|
| 3676 | ITGA4, CD49D, IA4 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
C00034649
|
| 3678 | ITGA5, CD49e, FNRA, VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
C00034649
|
| 3655 | ITGA6, CD49f, ITGA6B, VLA-6 | integrin, alpha 6 |
C00034649
|
| 3679 | ITGA7 | integrin, alpha 7 |
C00034649
|
| 8516 | ITGA8 | integrin, alpha 8 |
C00034649
|
| 3684 | ITGAM, CD11B, CR3A, MAC-1, MAC1A, MO1A, SLEB6 | integrin, alpha M (complement component 3 receptor 3 subunit) |
C00034649
|
| 9270 | ITGB1BP1, ICAP-1A, ICAP-1B, ICAP-1alpha, ICAP1, ICAP1A, ICAP1B | integrin beta 1 binding protein 1 |
C00034649
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00034649
|
| 6004 | RGS16, A28-RGS14, A28-RGS14P, RGS-R | regulator of G-protein signaling 16 |
C00000094
|
| 3691 | ITGB4, CD104 | integrin, beta 4 |
C00034649
|
| 3694 | ITGB6 | integrin, beta 6 |
C00034649
|
| 9358 | ITGBL1, OSCP, TIED | integrin, beta-like 1 (with EGF-like repeat domains) |
C00034649
|
| 9452 | ITM2A, BRICD2A, E25A | integral membrane protein 2A |
C00034649
|
| 81618 | ITM2C, BRI3, BRICD2C, E25, E25C, ITM3 | integral membrane protein 2C |
C00034649
|
| 79960 | JADE1, PHF17 | jade family PHD finger 1 |
C00034649
|
| 3717 | JAK2, JTK10, THCYT3 | Janus kinase 2 (EC:2.7.10.2) |
C00034649
|
| 122953 | JDP2, JUNDM2 | Jun dimerization protein 2 |
C00034649
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00034649
|
| 3730 | KAL1, ADMLX, HH1, HHA, KAL, KALIG-1, KMS, WFDC19 | Kallmann syndrome 1 sequence |
C00034649
|
| 151050 | KANSL1L, C2orf67, MSL1v2 | KAT8 regulatory NSL complex subunit 1-like |
C00034649
|
| 81621 | KAZALD1, BONO1, FKSG28, FKSG40, IGFBP-rP10 | Kazal-type serine peptidase inhibitor domain 1 |
C00034649
|
| 23254 | KAZN, KAZ | kazrin, periplakin interacting protein |
C00034649
|
| 9920 | KBTBD11, KLHDC7C | kelch repeat and BTB (POZ) domain containing 11 |
C00034649
|
| 3741 | KCNA5, ATFB7, HCK1, HK2, HPCN1, KV1.5, PCN1 | potassium voltage-gated channel, shaker-related subfamily, member 5 |
C00034649
|
| 3751 | KCND2, KV4.2, RK5 | potassium voltage-gated channel, Shal-related subfamily, member 2 |
C00034649
|
| 10008 | KCNE3, HOKPP, HYPP, MiRP2 | potassium voltage-gated channel, Isk-related family, member 3 |
C00034649
|
| 23704 | KCNE4, MIRP3 | potassium voltage-gated channel, Isk-related family, member 4 |
C00034649
|
| 3755 | KCNG1, K13, KCNG, KV6.1, kH2 | potassium voltage-gated channel, subfamily G, member 1 |
C00034649
|
| 3773 | KCNJ16, BIR9, KIR5.1 | potassium inwardly-rectifying channel, subfamily J, member 16 |
C00034649
|
| 3764 | KCNJ8, KIR6.1, uKATP-1 | potassium inwardly-rectifying channel, subfamily J, member 8 |
C00034649
|
| 3777 | KCNK3, K2p3.1, OAT1, PPH4, TASK, TASK-1, TBAK1 | potassium channel, subfamily K, member 3 |
C00034649
|
| 9424 | KCNK6, K2p6.1, KCNK8, TOSS, TWIK-2, TWIK2 | potassium channel, subfamily K, member 6 |
C00034649
|
| 3778 | KCNMA1, BKTM, KCa1.1, MaxiK, SAKCA, SLO, SLO-ALPHA, SLO1, bA205K10.1, mSLO1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
C00034649
|
| 10242 | KCNMB2 | potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
C00034649
|
| 3783 | KCNN4, IK1, IKCA1, KCA4, KCa3.1, SK4, hIKCa1, hKCa4, hSK4 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
C00034649
|
| 3790 | KCNS3, KV9.3 | potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
C00034649
|
| 343450 | KCNT2, KCa4.2, SLICK, SLO2.1 | potassium channel, subfamily T, member 2 |
C00034649
|
| 115207 | KCTD12, C13orf2, PFET1, PFETIN | potassium channel tetramerization domain containing 12 |
C00034649
|
| 9682 | KDM4A, JHDM3A, JMJD2, JMJD2A, TDRD14A | lysine (K)-specific demethylase 4A (EC:1.14.11.-) |
C00034649
|
| 3791 | KDR, CD309, FLK1, VEGFR, VEGFR2 | kinase insert domain receptor (a type III receptor tyrosine kinase) (EC:2.7.10.1) |
C00034649
|
| 2531 | KDSR, DHSR, FVT1, SDR35C1 | 3-ketodihydrosphingosine reductase (EC:1.1.1.102) |
C00034649
|
| 57242 |
C00034649
|
||
| 23366 | KIAA0895 |
C00034649
|
|
| 22832 | C6orf84, CEP162, QN1 | KIAA1009 |
C00034649
|
| 57214 | CCSP1, TMEM2L | KIAA1199 |
C00034649
|
| 56243 | SKT | KIAA1217 |
C00034649
|
| 23303 | KIF13B, GAKIN | kinesin family member 13B |
C00034649
|
| 24137 | KIF4A, KIF4, KIF4G1 | kinesin family member 4A |
C00034649
|
| 3806 | KIR2DS1, CD158H, CD158a, p50.1 | killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 1 |
C00034649
|
| 7071 | KLF10, EGR-alpha, EGRA, TIEG, TIEG1 | Kruppel-like factor 10 |
C00034649
|
| 688 | KLF5, BTEB2, CKLF, IKLF | Kruppel-like factor 5 (intestinal) |
C00034649
|
| 151230 | KLHL23, DITHP | kelch-like family member 23 |
C00034649
|
| 114818 | KLHL29, KBTBD9 | kelch-like family member 29 |
C00034649
|
| 401265 | KLHL31, BKLHD6, KBTBD1, KLHL, bA345L23.2 | kelch-like family member 31 |
C00034649
|
| 5655 | KLK10, NES1, PRSSL1 | kallikrein-related peptidase 10 (EC:3.4.21.35) |
C00034649
|
| 3820 | KLRB1, CD161, CLEC5B, NKR, NKR-P1, NKR-P1A, NKRP1A, hNKR-P1A | killer cell lectin-like receptor subfamily B, member 1 |
C00034649
|
| 3823 | KLRC3, NKG2-E, NKG2E | killer cell lectin-like receptor subfamily C, member 3 |
C00034649
|
| 3872 | KRT17, K17, PC, PC2, PCHC1 | keratin 17 |
C00034649
|
| 3875 | KRT18, CYK18, K18 | keratin 18 |
C00034649
|
| 25984 | KRT23, CK23, HAIK1, K23 | keratin 23 (histone deacetylase inducible) |
C00034649
|
| 3885 | KRT34, HA4, Ha-4, KRTHA4, hHa4 | keratin 34 |
C00034649
|
| 3853 | KRT6A, CK6A, CK6C, CK6D, K6A, K6C, K6D, KRT6C, KRT6D | keratin 6A |
C00034649
|
| 3854 | KRT6B, CK-6B, CK6B, K6B, KRTL1, PC2 | keratin 6B |
C00034649
|
| 3855 | KRT7, CK7, K2C7, K7, SCL | keratin 7 |
C00034649
|
| 3856 | KRT8, CARD2, CK-8, CK8, CYK8, K2C8, K8, KO | keratin 8 |
C00034649
|
| 3892 | KRT86, HB6, Hb1, KRTHB1, KRTHB6, MNX, hHb6 | keratin 86 |
C00034649
|
| 200634 | KRTCAP3, KCP3, MRV222, PRO9898 | keratinocyte associated protein 3 |
C00034649
|
| 8942 | KYNU | kynureninase (EC:3.7.1.3) |
C00034649
|
| 3909 | LAMA3, BM600, E170, LAMNA, LOCS, lama3a | laminin, alpha 3 |
C00034649
|
| 3910 | LAMA4, CMD1JJ, LAMA3, LAMA4*-1 | laminin, alpha 4 |
C00034649
|
| 3914 | LAMB3, BM600-125KDA, LAM5, LAMNB1 | laminin, beta 3 |
C00034649
|
| 3915 | LAMC1, LAMB2 | laminin, gamma 1 (formerly LAMB2) |
C00034649
|
| 3918 | LAMC2, B2T, BM600, CSF, EBR2, EBR2A, LAMB2T, LAMNB2 | laminin, gamma 2 |
C00034649
|
| 113251 | LARP4 | La ribonucleoprotein domain family, member 4 |
C00034649
|
| 3929 | LBP, BPIFD2 | lipopolysaccharide binding protein |
C00034649
|
| 3932 | LCK, LSK, YT16, p56lck, pp58lck | lymphocyte-specific protein tyrosine kinase (EC:2.7.10.2) |
C00034649
|
| 3936 | LCP1, CP64, L-PLASTIN, LC64P, LPL, PLS2 | lymphocyte cytosolic protein 1 (L-plastin) |
C00034649
|
| 3939 | LDHA, GSD11, LDH1, LDHM | lactate dehydrogenase A (EC:1.1.1.27) |
C00034649
|
| 753 | LDLRAD4, C18orf1 | low density lipoprotein receptor class A domain containing 4 |
C00034649
|
| 7044 | LEFTY2, EBAF, LEFTA, LEFTYA, TGFB4 | left-right determination factor 2 |
C00034649
|
| 93273 | LEMD1, CT50, LEMP-1 | LEM domain containing 1 |
C00034649
|
| 3952 | LEP, LEPD, OB, OBS | leptin |
C00034649
|
| 55214 | LEPREL1, MCVD, MLAT4, P3H2 | leprecan-like 1 (EC:1.14.11.7) |
C00034649
|
| 3955 | LFNG, SCDO3 | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (EC:2.4.1.222) |
C00034649
|
| 3958 | LGALS3, CBP35, GAL3, GALBP, GALIG, L31, LGALS2, MAC2 | lectin, galactoside-binding, soluble, 3 |
C00034649
|
| 29094 | LGALSL, GRP | lectin, galactoside-binding-like |
C00034649
|
| 3972 | LHB, CGB4, LSH-B, hLHB | luteinizing hormone beta polypeptide |
C00034649
|
| 10186 | LHFP | lipoma HMGIC fusion partner (EC:3.6.5.4) |
C00034649
|
| 3976 | LIF, CDF, DIA, HILDA, MLPLI | leukemia inhibitory factor |
C00034649
|
| 29931 | LINC00312, ERR-10, ERR10, LMCD1DN, LOH3CR2A, NAG-7, NCRNA00312 | long intergenic non-protein coding RNA 312 |
C00034649
|
| 9388 | LIPG, EDL, EL, PRO719 | lipase, endothelial (EC:3.1.1.3) |
C00034649
|
| 8543 | LMO4 | LIM domain only 4 |
C00034649
|
| 25802 | LMOD1, 1D, 64kD, D1, SM-LMOD | leiomodin 1 (smooth muscle) |
C00034649
|
| 4017 | LOXL2, LOR2, WS9-14 | lysyl oxidase-like 2 (EC:1.4.3.-) |
C00034649
|
| 84171 | LOXL4, LOXC | lysyl oxidase-like 4 |
C00034649
|
| 23566 | LPAR3, EDG7, Edg-7, GPCR, HOFNH30, LP-A3, LPA3, RP4-678I3 | lysophosphatidic acid receptor 3 |
C00034649
|
| 10162 | LPCAT3, C3F, LPCAT, LPLAT_5, LPSAT, MBOAT5, OACT5, nessy | lysophosphatidylcholine acyltransferase 3 (EC:2.3.1.23 2.3.1.n6) |
C00034649
|
| 23266 | LPHN2, CIRL2, CL2, LEC1, LPHH1 | latrophilin 2 |
C00034649
|
| 23175 | LPIN1, PAP1 | lipin 1 (EC:3.1.3.4) |
C00034649
|
| 4038 | LRP4, CLSS, LRP-4, LRP10, MEGF7, SOST2 | low density lipoprotein receptor-related protein 4 |
C00034649
|
| 55227 | LRRC1, LANO, dJ523E19.1 | leucine rich repeat containing 1 |
C00034649
|
| 10234 | LRRC17, P37NB | leucine rich repeat containing 17 |
C00034649
|
| 2615 | LRRC32, D11S833E, GARP | leucine rich repeat containing 32 |
C00034649
|
| 84230 | LRRC8C, AD158, FAD158 | leucine rich repeat containing 8 family, member C |
C00034649
|
| 55144 | LRRC8D, LRRC5 | leucine rich repeat containing 8 family, member D |
C00034649
|
| 57633 | LRRN1, FIGLER3, NLRR-1 | leucine rich repeat neuronal 1 |
C00034649
|
| 4047 | LSS, OSC | lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (EC:5.4.99.7) |
C00034649
|
| 4050 | LTB, TNFC, TNFSF3, p33 | lymphotoxin beta (TNF superfamily, member 3) |
C00034649
|
| 4052 | LTBP1 | latent transforming growth factor beta binding protein 1 |
C00034649
|
| 4060 | LUM, LDC, SLRR2D | lumican |
C00034649
|
| 56925 | LXN, ECI, TCI | latexin |
C00034649
|
| 116372 | LYPD1, LYPDC1, PHTS | LY6/PLAUR domain containing 1 |
C00034649
|
| 4069 | LYZ, LZM | lysozyme (EC:3.2.1.17) |
C00034649
|
| 54585 | LZTFL1, BBS17 | leucine zipper transcription factor-like 1 |
C00034649
|
| 4094 | MAF, CCA4, c-MAF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
C00034649
|
| 9935 | MAFB, KRML, MCTO | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
C00034649
|
| 728239 | MAGED4, MAGE-E1, MAGE1, MAGEE1 | melanoma antigen family D, 4 |
C00034649
|
| 256691 | MAMDC2 | MAM domain containing 2 |
C00034649
|
| 23324 | MAN2B2 | mannosidase, alpha, class 2B, member 2 (EC:3.2.1.24) |
C00034649
|
| 7873 | MANF, ARMET, ARP | mesencephalic astrocyte-derived neurotrophic factor |
C00034649
|
| 4129 | MAOB | monoamine oxidase B (EC:1.4.3.4) |
C00034649
|
| 4133 | MAP2, MAP2A, MAP2B, MAP2C | microtubule-associated protein 2 |
C00034649
|
| 5608 | MAP2K6, MAPKK6, MEK6, MKK6, PRKMK6, SAPKK-3, SAPKK3 | mitogen-activated protein kinase kinase 6 (EC:2.7.12.2) |
C00034649
|
| 4216 | MAP3K4, MAPKKK4, MEKK4, MTK1, PRO0412 | mitogen-activated protein kinase kinase kinase 4 (EC:2.7.11.25) |
C00034649
|
| 56911 | MAP3K7CL, C21orf7, HC21ORF7, TAK1L, TAKL, TAKL-1, TAKL-2, TAKL-4 | MAP3K7 C-terminal like |
C00034649
|
| 1326 | MAP3K8, COT, EST, ESTF, MEKK8, TPL2, Tpl-2, c-COT | mitogen-activated protein kinase kinase kinase 8 (EC:2.7.11.25) |
C00034649
|
| 9053 | MAP7, E-MAP-115, EMAP115 | microtubule-associated protein 7 |
C00034649
|
| 64757 | MARC1, MOSC1 | mitochondrial amidoxime reducing component 1 |
C00034649
|
| 65108 | MARCKSL1, F52, MACMARCKS, MLP, MLP1, MRP | MARCKS-like 1 |
C00034649
|
| 4141 | MARS, METRS, MRS, MTRNS | methionyl-tRNA synthetase (EC:6.1.1.10) |
C00034649
|
| 4144 | MAT2A, MATA2, MATII, SAMS2 | methionine adenosyltransferase II, alpha (EC:2.5.1.6) |
C00034649
|
| 4145 | MATK, CHK, CTK, HHYLTK, HYL, HYLTK, Lsk | megakaryocyte-associated tyrosine kinase (EC:2.7.10.2) |
C00034649
|
| 4147 | MATN2 | matrilin 2 |
C00034649
|
| 8785 | MATN4 | matrilin 4 |
C00034649
|
| 56890 | MDM1 | Mdm1 nuclear protein homolog (mouse) |
C00034649
|
| 84935 | MEDAG, AWMS3, C13orf33, MEDA-4, MEDA4, hAWMS3 | mesenteric estrogen-dependent adipogenesis |
C00034649
|
| 84466 | MEGF10, EMARDD | multiple EGF-like-domains 10 |
C00034649
|
| 92312 | MEX3A, MEX-3A, RKHD4 | mex-3 RNA binding family member A |
C00034649
|
| 4237 | MFAP2, MAGP, MAGP-1, MAGP1 | microfibrillar-associated protein 2 |
C00034649
|
| 4239 | MFAP4 | microfibrillar-associated protein 4 |
C00034649
|
| 8076 | MFAP5, MAGP2, MP25 | microfibrillar associated protein 5 |
C00034649
|
| 4242 | MFNG | MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (EC:2.4.1.222) |
C00034649
|
| 4257 | MGST1, GST12, MGST, MGST-I | microsomal glutathione S-transferase 1 (EC:2.5.1.18) |
C00034649
|
| 9645 | MICAL2, MICAL-2, MICAL2PV1, MICAL2PV2 | microtubule associated monooxygenase, calponin and LIM domain containing 2 |
C00034649
|
| 4281 | MID1, BBBG1, FXY, GBBB1, MIDIN, OGS1, OS, OSX, RNF59, TRIM18, XPRF, ZNFXY | midline 1 (Opitz/BBB syndrome) |
C00034649
|
| 406892 | MIR100, MIRN100, miR-100 | microRNA 100 |
C00034649
|
| 406900 | MIR106B, MIRN106B | microRNA 106b |
C00034649
|
| 406906 | MIR122, MIR122A, MIRN122, MIRN122A, hsa-mir-122, miRNA122, miRNA122A | microRNA 122 |
C00034649
|
| 100302289 | MIR1251, MIRN1251, hsa-mir-1251 | microRNA 1251 |
C00034649
|
| 406916 | MIR128-2, MIR128B, MIRN128-2, MIRN128B, mir-128b | microRNA 128-2 |
C00034649
|
| 387148 |
C00034649
|
||
| 100302150 | MIR1296, MIRN1296, hsa-mir-1296 | microRNA 1296 |
C00034649
|
| 406924 | MIR134, MIRN134 | microRNA 134 |
C00034649
|
| 442891 | MIR135B, MIRN135B | microRNA 135b |
C00034649
|
| 406934 | MIR142, MIRN142 | microRNA 142 |
C00034649
|
| 406936 | MIR144, MIRN144 | microRNA 144 |
C00034649
|
| 100302115 | MIR1468, MIRN1468, hsa-mir-1468 | microRNA 1468 |
C00034649
|
| 406938 | MIR146A, MIRN146, MIRN146A, miR-146a, miRNA146A | microRNA 146a |
C00034649
|
| 574447 | MIR146B, MIRN146B, miRNA146B | microRNA 146b |
C00034649
|
| 406947 | MIR155, MIRN155, miRNA155 | microRNA 155 |
C00034649
|
| 406948 | MIR15A, MIRN15A, hsa-mir-15a, miRNA15A | microRNA 15a |
C00034649
|
| 406949 | MIR15B, MIRN15B, hsa-mir-15b, miR-15b | microRNA 15b |
C00034649
|
| 406950 | MIR16-1, MIRN16-1, miRNA16-1 | microRNA 16-1 |
C00034649
|
| 406957 | MIR181C, MIRN181C, mir-181c | microRNA 181c |
C00034649
|
| 574457 | MIR181D, MIRN181D | microRNA 181d |
C00034649
|
| 406962 | MIR186, MIRN186, miR-186 | microRNA 186 |
C00034649
|
| 406966 | MIR191, MIRN191, miR-191 | microRNA 191 |
C00034649
|
| 406971 | MIR195, MIRN195, miRNA195 | microRNA 195 |
C00034649
|
| 442920 | MIR196B, MIRN196B, miR-196b, miRNA196B | microRNA 196b |
C00034649
|
| 406975 | MIR198, MIRN198, miR-198 | microRNA 198 |
C00034649
|
| 574448 | MIR202, MIRN202, hsa-mir-202 | microRNA 202 |
C00034649
|
| 406987 | MIR204, MIRN204, miRNA204 | microRNA 204 |
C00034649
|
| 406989 | MIR206, MIRN206, miRNA206 | microRNA 206 |
C00034649
|
| 574032 | MIR20B, MIRN20B, hsa-mir-20b | microRNA 20b |
C00034649
|
| 407006 | MIR221, MIRN221, miRNA221, mir-221 | microRNA 221 |
C00034649
|
| 407014 | MIR25, MIRN25, hsa-mir-25, miR-25 | microRNA 25 |
C00034649
|
| 407017 | MIR26B, MIRN26B, hsa-mir-26b, miR-26b | microRNA 26b |
C00034649
|
| 407022 | MIR296, MIRN296, miRNA296 | microRNA 296 |
C00034649
|
| 407023 | MIR299, MIRN299 | microRNA 299 |
C00034649
|
| 407028 | MIR302A, MIRN302, MIRN302A, hsa-mir-302 | microRNA 302a |
C00034649
|
| 442894 | MIR302B, MIRN302B | microRNA 302b |
C00034649
|
| 442896 | MIR302D, MIRN302D | microRNA 302d |
C00034649
|
| 407033 | MIR30D, MIRN30D | microRNA 30d |
C00034649
|
| 407034 | MIR30E, MIRN30E | microRNA 30e |
C00034649
|
| 442901 | MIR328, MIRN328, hsa-mir-328 | microRNA 328 |
C00034649
|
| 442904 | MIR335, MIRN335, hsa-mir-335, miRNA335 | microRNA 335 |
C00034649
|
| 442905 | MIR337, MIRN337, hsa-mir-337 | microRNA 337 |
C00034649
|
| 442906 | MIR338, MIRN338, hsa-mir-338 | microRNA 338 |
C00034649
|
| 693120 | MIR33B, MIRN33B, hsa-mir-33b | microRNA 33b |
C00034649
|
| 442910 | MIR345, MIRN345, hsa-mir-345 | microRNA 345 |
C00034649
|
| 442911 | MIR346, MIRN346, hsa-mir-346, miR-346, miRNA346 | microRNA 346 |
C00034649
|
| 407041 | MIR34B, MIRN34B, miRNA34B | microRNA 34b |
C00034649
|
| 574031 | MIR363, MIR-363, MIRN363, hsa-mir-363 | microRNA 363 |
C00034649
|
| 442913 | MIR376C, MIR368, MIRN368, MIRN376C, hsa-mir-368, miRNA368 | microRNA 376c |
C00034649
|
| 693121 | MIR411, MIRN411, hsa-mir-411 | microRNA 411 |
C00034649
|
| 574433 | MIR412, MIRN412, hsa-mir-412 | microRNA 412 |
C00034649
|
| 494337 | MIR425, MIRN425, hsa-mir-425 | microRNA 425 |
C00034649
|
| 554212 | MIR448, MIRN448, hsa-mir-448, miRNA448 | microRNA 448 |
C00034649
|
| 554213 | MIR449A, MIRN449, MIRN449A, hsa-mir-449 | microRNA 449a |
C00034649
|
| 619556 | MIR455, MIRN455, hsa-mir-455 | microRNA 455 |
C00034649
|
| 574436 | MIR485, MIRN485, hsa-mir-485 | microRNA 485 |
C00034649
|
| 619554 | MIR486, MIRN486, hsa-mir-486 | microRNA 486 |
C00034649
|
| 574444 | MIR491, MIRN491, hsa-mir-491 | microRNA 491 |
C00034649
|
| 574453 | MIR495, MIRN495, hsa-mir-495 | microRNA 495 |
C00034649
|
| 574503 | MIR501, MIRN501, hsa-mir-501 | microRNA 501 |
C00034649
|
| 574504 | MIR502, MIRN502, hsa-mir-502 | microRNA 502 |
C00034649
|
| 574506 | MIR503, MIRN503, hsa-mir-503 | microRNA 503 |
C00034649
|
| 574507 | MIR504, MIRN504, hsa-mir-504 | microRNA 504 |
C00034649
|
| 574511 | MIR506, MIRN506, hsa-mir-506 | microRNA 506 |
C00034649
|
| 574513 | MIR508, MIRN508, hsa-mir-508 | microRNA 508 |
C00034649
|
| 100126337 | MIR509-3, MIRN509-3 | microRNA 509-3 |
C00034649
|
| 100315541 |
C00034649
|
||
| 100315577 |
C00034649
|
||
| 574479 | MIR517A, MIRN517A | microRNA 517a |
C00034649
|
| 574483 | MIR517B, MIRN517B | microRNA 517b |
C00034649
|
| 574492 | MIR517C, MIRN517C | microRNA 517c |
C00034649
|
| 574474 | MIR518B, MIRN518B | microRNA 518b |
C00034649
|
| 574477 | MIR518C, MIRN518C | microRNA 518c |
C00034649
|
| 574489 | MIR518D, MIRN518D | microRNA 518d |
C00034649
|
| 574487 | MIR518E, MIRN518E | microRNA 518e |
C00034649
|
| 574469 | MIR519B, MIRN519B | microRNA 519b |
C00034649
|
| 574482 | MIR520D, MIRN520D | microRNA 520d |
C00034649
|
| 574484 | MIR520G, MIRN520G | microRNA 520g |
C00034649
|
| 574495 | MIR522, MIRN522, hsa-mir-522 | microRNA 522 |
C00034649
|
| 574471 | MIR523, MIRN523, hsa-mir-523 | microRNA 523 |
C00034649
|
| 574470 | MIR525, MIRN525, hsa-mir-525 | microRNA 525 |
C00034649
|
| 664617 | MIR542, MIRN542, hsa-mir-542 | microRNA 542 |
C00034649
|
| 693128 | MIR548B, MIRN548B | microRNA 548b |
C00034649
|
| 693144 | MIR559, MIRN559, hsa-mir-559 | microRNA 559 |
C00034649
|
| 693148 | MIR563, MIRN563, hsa-mir-563 | microRNA 563 |
C00034649
|
| 693149 | MIR564, MIRN564, hsa-mir-564 | microRNA 564 |
C00034649
|
| 693151 | MIR566, MIRN566, hsa-mir-566 | microRNA 566 |
C00034649
|
| 693154 | MIR569, MIRN569, hsa-mir-569 | microRNA 569 |
C00034649
|
| 693155 | MIR570, MIRN570, hsa-mir-570 | microRNA 570 |
C00034649
|
| 693156 | MIR571, MIRN571, hsa-mir-571 | microRNA 571 |
C00034649
|
| 693163 | MIR578, MIRN578, hsa-mir-578 | microRNA 578 |
C00034649
|
| 693169 | MIR584, MIRN584, hsa-mir-584 | microRNA 584 |
C00034649
|
| 693175 | MIR590, MIRN590, hsa-mir-590 | microRNA 590 |
C00034649
|
| 693176 | MIR591, MIRN591, hsa-mir-591 | microRNA 591 |
C00034649
|
| 693177 | MIR592, MIRN592, hsa-mir-592 | microRNA 592 |
C00034649
|
| 693180 | MIR595, MIRN595, hsa-mir-595 | microRNA 595 |
C00034649
|
| 693181 | MIR596, MIRN596, hsa-mir-596 | microRNA 596 |
C00034649
|
| 693182 | MIR597, MIRN597, hsa-mir-597 | microRNA 597 |
C00034649
|
| 693187 | MIR602, MIRN602, hsa-mir-602 | microRNA 602 |
C00034649
|
| 693188 | MIR603, MIRN603, hsa-mir-603 | microRNA 603 |
C00034649
|
| 693189 | MIR604, MIRN604, hsa-mir-604 | microRNA 604 |
C00034649
|
| 693192 | MIR607, MIRN607, hsa-mir-607 | microRNA 607 |
C00034649
|
| 693195 | MIR610, MIRN610, hsa-mir-610 | microRNA 610 |
C00034649
|
| 693198 | MIR613, MIRN613, hsa-mir-613 | microRNA 613 |
C00034649
|
| 693199 | MIR614, MIRN614, hsa-mir-614 | microRNA 614 |
C00034649
|
| 693203 | MIR618, MIRN618, hsa-mir-618 | microRNA 618 |
C00034649
|
| 693207 | MIR622, MIRN622, hsa-mir-622 | microRNA 622 |
C00034649
|
| 693216 | MIR631, MIRN631, hsa-mir-631 | microRNA 631 |
C00034649
|
| 693217 | MIR632, MIRN632, hsa-mir-632 | microRNA 632 |
C00034649
|
| 693223 | MIR638, MIRN638, hsa-mir-638 | microRNA 638 |
C00034649
|
| 693226 | MIR641, MIRN641, hsa-mir-641 | microRNA 641 |
C00034649
|
| 693230 | MIR645, MIRN645, hsa-mir-645 | microRNA 645 |
C00034649
|
| 693231 | MIR646, MIRN646, hsa-mir-646 | microRNA 646 |
C00034649
|
| 693232 | MIR647, MIRN647, hsa-mir-647 | microRNA 647 |
C00034649
|
| 724033 | MIR663A, MIR663, MIRN663, hsa-mir-663, hsa-mir-663a | microRNA 663a |
C00034649
|
| 768213 | MIR671, MIRN671, hsa-mir-671 | microRNA 671 |
C00034649
|
| 100033819 | MIR675, MIRN675, hsa-mir-675 | microRNA 675 |
C00034649
|
| 768220 | MIR765, MIRN765, hsa-mir-765 | microRNA 765 |
C00034649
|
| 100126310 | MIR876, MIRN876, hsa-mir-876 | microRNA 876 |
C00034649
|
| 100126341 | MIR891A, MIRN891A | microRNA 891a |
C00034649
|
| 693235 | MIR92B, MIRN92B, hsa-mir-92b | microRNA 92b |
C00034649
|
| 407050 | MIR93, MIRN9, MIRN93, hsa-mir-93, miR-93 | microRNA 93 |
C00034649
|
| 407052 | MIR95, MIRN95, hsa-mir-95, miR-95 | microRNA 95 |
C00034649
|
| 407055 | MIR99A, MIRN99A | microRNA 99a |
C00034649
|
| 4286 | MITF, CMM8, MI, WS2, WS2A, bHLHe32 | microphthalmia-associated transcription factor |
C00034649
|
| 10962 | MLLT11, AF1Q, RP11-316M1.10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 |
C00034649
|
| 79083 | MLPH, SLAC2-A | melanophilin |
C00034649
|
| 23531 | MMD, MMA, MMD1, PAQR11 | monocyte to macrophage differentiation-associated |
C00034649
|
| 4311 | MME, CALLA, CD10, NEP, SFE | membrane metallo-endopeptidase (EC:3.4.24.11) |
C00034649
|
| 4312 | MMP1, CLG, CLGN | matrix metallopeptidase 1 (interstitial collagenase) (EC:3.4.24.7) |
C00034649
|
| 4319 | MMP10, SL-2, STMY2 | matrix metallopeptidase 10 (stromelysin 2) (EC:3.4.24.22) |
C00034649
|
| 4320 | MMP11, SL-3, ST3, STMY3 | matrix metallopeptidase 11 (stromelysin 3) (EC:3.4.24.-) |
C00034649
|
| 4323 | MMP14, MMP-14, MMP-X1, MT-MMP, MT-MMP_1, MT1-MMP, MT1MMP, MTMMP1, WNCHRS | matrix metallopeptidase 14 (membrane-inserted) (EC:3.4.24.80) |
C00034649
|
| 4313 | MMP2, CLG4, CLG4A, MMP-II, MONA, TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) |
C00034649
|
| 56547 | MMP26 | matrix metallopeptidase 26 (EC:3.4.24.1) |
C00034649
|
| 64066 | MMP27, MMP-27 | matrix metallopeptidase 27 |
C00034649
|
| 4314 | MMP3, CHDS6, MMP-3, SL-1, STMY, STMY1, STR1 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) (EC:3.4.24.17) |
C00034649
|
| 4316 | MMP7, MMP-7, MPSL1, PUMP-1 | matrix metallopeptidase 7 (matrilysin, uterine) (EC:3.4.24.23) |
C00034649
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00034649
|
| 22915 | MMRN1, ECM, EMILIN4, GPIa*, MMRN | multimerin 1 |
C00034649
|
| 431645 |
C00034649
|
||
| 23041 | MON2 | MON2 homolog (S. cerevisiae) |
C00034649
|
| 5774 | PTPN3, PTP-H1, PTPH1 | protein tyrosine phosphatase, non-receptor type 3 (EC:3.1.3.48) |
C00000094
|
| 4354 | MPP1, AAG12, DXS552E, EMP55, MRG1, PEMP | membrane protein, palmitoylated 1, 55kDa |
C00034649
|
| 10205 | MPZL2, EVA, EVA1 | myelin protein zero-like 2 |
C00034649
|
| 22808 | MRAS, M-RAs, R-RAS3, RRAS3 | muscle RAS oncogene homolog |
C00034649
|
| 51264 | MRPL27, L27mt | mitochondrial ribosomal protein L27 |
C00034649
|
| 740 | MRPL49, C11orf4, L49mt, MRP-L49, NOF, NOF1 | mitochondrial ribosomal protein L49 |
C00034649
|
| 28957 | MRPS28, MRP-S28, MRP-S35, MRPS35 | mitochondrial ribosomal protein S28 |
C00034649
|
| 51154 | MRTO4, C1orf33, MRT4, dJ657E11.4 | mRNA turnover 4 homolog (S. cerevisiae) |
C00034649
|
| 10232 | MSLN, MPF, SMRP | mesothelin |
C00034649
|
| 6307 | MSMO1, DESP4, ERG25, SC4MOL | methylsterol monooxygenase 1 (EC:1.14.13.72) |
C00034649
|
| 4489 | MT1A, MT1, MT1S, MTC | metallothionein 1A |
C00034649
|
| 4490 | MT1B, MT1, MT1Q, MTP | metallothionein 1B |
C00034649
|
| 4493 | MT1E, MT1, MTD | metallothionein 1E |
C00034649
|
| 4494 | MT1F, MT1 | metallothionein 1F |
C00034649
|
| 4495 | MT1G, MT1, MT1K | metallothionein 1G |
C00034649
|
| 4496 | MT1H, MT-0, MT-1H, MT-IH, MT1 | metallothionein 1H |
C00034649
|
| 4500 | MT1L, MT1, MT1R, MTF | metallothionein 1L (gene/pseudogene) |
C00034649
|
| 4499 | MT1M, MT-1M, MT-IM, MT1, MT1K | metallothionein 1M |
C00034649
|
| 4501 | MT1X, MT-1l, MT1 | metallothionein 1X |
C00034649
|
| 4502 | MT2A, MT2 | metallothionein 2A |
C00034649
|
| 9112 | MTA1 | metastasis associated 1 |
C00034649
|
| 22823 | MTF2, M96, PCL2, RP5-976O13.1, TDRD19A, dJ976O13.2 | metal response element binding transcription factor 2 |
C00034649
|
| 10797 | MTHFD2, NMDMC | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase (EC:3.5.4.9 1.5.1.15) |
C00034649
|
| 2475 | MTOR, FRAP, FRAP1, FRAP2, RAFT1, RAPT1 | mechanistic target of rapamycin (serine/threonine kinase) (EC:2.7.11.1) |
C00034649
|
| 4548 | MTR, HMAG, MS, cblG | 5-methyltetrahydrofolate-homocysteine methyltransferase (EC:2.1.1.13) |
C00034649
|
| 4582 | MUC1, CA_15-3, CD227, EMA, H23AG, KL-6, MAM6, MCKD1, MUC-1, MUC-1/SEC, MUC-1/X, MUC1/ZD, PEM, PEMT, PUM | mucin 1, cell surface associated |
C00034649
|
| 4585 | MUC4, ASGP, HSA276359, MUC-4 | mucin 4, cell surface associated |
C00034649
|
| 727897 | MUC5B, MG1, MUC-5B, MUC5, MUC9 | mucin 5B, oligomeric mucus/gel-forming |
C00034649
|
| 139221 | MUM1L1 | melanoma associated antigen (mutated) 1-like 1 |
C00034649
|
| 4598 | MVK, LRBP, MK, MVLK, POROK3 | mevalonate kinase (EC:2.7.1.36) |
C00034649
|
| 25878 | MXRA5 | matrix-remodelling associated 5 |
C00034649
|
| 91663 | MYADM, SB135 | myeloid-associated differentiation marker |
C00034649
|
| 4613 | MYCN, MODED, MYCNOT, N-myc, NMYC, ODED, bHLHe37 | v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
C00034649
|
| 4619 | MYH1, MYHSA1, MYHa, MyHC-2X/D, MyHC-2x | myosin, heavy chain 1, skeletal muscle, adult |
C00034649
|
| 4628 | MYH10, NMMHC-IIB, NMMHCB | myosin, heavy chain 10, non-muscle |
C00034649
|
| 140465 | MYL6B, MLC1SA | myosin, light chain 6B, alkali, smooth muscle and non-muscle |
C00034649
|
| 10398 | MYL9, LC20, MLC-2C, MLC2, MRLC1, MYRL2 | myosin, light chain 9, regulatory |
C00034649
|
| 4638 | MYLK, AAT7, KRP, MLCK, MLCK1, MLCK108, MLCK210, MSTP083, MYLK1, smMLCK | myosin light chain kinase (EC:2.7.11.18) |
C00034649
|
| 23026 | MYO16, MYR8, Myo16b, NYAP3 | myosin XVI |
C00034649
|
| 93649 | MYOCD, MYCD | myocardin |
C00034649
|
| 25924 | MYRIP, SLAC2-C, SLAC2C | myosin VIIA and Rab interacting protein |
C00034649
|
| 27163 | NAAA, ASAHL, PLT | N-acylethanolamine acid amidase (EC:3.5.1.-) |
C00034649
|
| 259232 | NALCN, CanIon, INNFD, VGCNL1, bA430M15.1 | sodium leak channel, non-selective |
C00034649
|
| 10135 | NAMPT, 1110035O14Rik, PBEF, PBEF1, VF, VISFATIN | nicotinamide phosphoribosyltransferase (EC:2.4.2.12) |
C00034649
|
| 256236 | NAPSB, NAP1L, NAP2, NAPB, NAPSBP | napsin B aspartic peptidase, pseudogene |
C00034649
|
| 9 | NAT1, AAC1, MNAT, NAT-1, NATI | N-acetyltransferase 1 (arylamine N-acetyltransferase) (EC:2.3.1.5) |
C00034649
|
| 89795 | NAV3, POMFIL1, STEERIN3, unc53H3 | neuron navigator 3 |
C00034649
|
| 400746 | NCMAP, C1orf130, MP11 | noncompact myelin associated protein |
C00034649
|
| 8031 | NCOA4, ARA70, ELE1, PTC3, RFG | nuclear receptor coactivator 4 |
C00034649
|
| 9436 | NCR2, CD336, LY95, NK-p44, NKP44, dJ149M18.1 | natural cytotoxicity triggering receptor 2 |
C00034649
|
| 10403 | NDC80, HEC, HEC1, HsHec1, KNTC2, TID3, hsNDC80 | NDC80 kinetochore complex component |
C00034649
|
| 4693 | NDP, EVR2, FEVR, ND | Norrie disease (pseudoglioma) |
C00034649
|
| 10397 | NDRG1, CAP43, CMT4D, DRG-1, DRG1, GC4, HMSNL, NDR1, NMSL, PROXY1, RIT42, RTP, TARG1, TDD5 | N-myc downstream regulated 1 |
C00034649
|
| 57447 | NDRG2, SYLD | NDRG family member 2 |
C00034649
|
| 4744 | NEFH, NFH | neurofilament, heavy polypeptide |
C00034649
|
| 4741 | NEFM, NEF3, NF-M, NFM | neurofilament, medium polypeptide |
C00034649
|
| 257194 | NEGR1, DMML2433, IGLON4, KILON, Ntra | neuronal growth regulator 1 |
C00034649
|
| 4756 | NEO1, IGDCC2, NGN | neogenin 1 |
C00034649
|
| 10763 | NES | nestin |
C00034649
|
| 4781 | NFIB, CTF, HMGIC/NFIB, NF-I/B, NF1-B, NFI-B, NFI-RED, NFIB2, NFIB3 | nuclear factor I/B |
C00034649
|
| 64332 | NFKBIZ, IKBZ, INAP, MAIL | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
C00034649
|
| 152518 | NFXL1, CDZFP, HOZFP, OZFP, URCC5 | nuclear transcription factor, X-box binding-like 1 |
C00034649
|
| 55768 | NGLY1, CDG1V, PNG1, PNGase | N-glycanase 1 (EC:3.5.1.52) |
C00034649
|
| 4818 | NKG7, GIG1, GMP-17, p15-TIA-1 | natural killer cell group 7 sequence |
C00034649
|
| 57502 | NLGN4X, ASPGX2, AUTSX2, HLNX, HNL4X, NLGN4 | neuroligin 4, X-linked |
C00034649
|
| 126206 | NLRP5, CLR19.8, MATER, NALP5, PAN11, PYPAF8 | NLR family, pyrin domain containing 5 |
C00034649
|
| 4828 | NMB | neuromedin B |
C00034649
|
| 4831 | NME2, NDKB, NDPK-B, NDPKB, NM23-H2, NM23B, PUF | NME/NM23 nucleoside diphosphate kinase 2 (EC:2.7.4.6 2.7.13.3) |
C00034649
|
| 9241 | NOG, SYM1, SYNS1 | noggin |
C00034649
|
| 10528 | NOP56, NOL5A, SCA36 | NOP56 ribonucleoprotein |
C00034649
|
| 4843 | NOS2, HEP-NOS, INOS, NOS, NOS2A | nitric oxide synthase 2, inducible (EC:1.14.13.39) |
C00034649
|
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00034649
|
| 4853 | NOTCH2, AGS2, HJCYS, hN2 | notch 2 |
C00034649
|
| 4856 | NOV, CCN3, IBP-9, IGFBP-9, IGFBP9, NOVh | nephroblastoma overexpressed |
C00034649
|
| 50508 | NOX3, GP91-3, MOX-2 | NADPH oxidase 3 |
C00034649
|
| 4862 | NPAS2, MOP4, PASD4, bHLHe9 | neuronal PAS domain protein 2 |
C00034649
|
| 4864 | NPC1, NPC | Niemann-Pick disease, type C1 |
C00034649
|
| 4878 | NPPA, ANF, ANP, ATFB6, CDD-ANF, PND | natriuretic peptide A |
C00034649
|
| 4883 | NPR3, ANP-C, ANPR-C, ANPRC, C5orf23, GUCY2B, NPR-C, NPRC | natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
C00034649
|
| 4885 | NPTX2, NARP, NP-II, NP2 | neuronal pentraxin II |
C00034649
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00034649
|
| 9970 | NR1I3, CAR, CAR1, MB67 | nuclear receptor subfamily 1, group I, member 3 |
C00034649
|
| 4306 | NR3C2, MCR, MLR, MR, NR3C2VIT | nuclear receptor subfamily 3, group C, member 2 |
C00034649
|
| 3164 | NR4A1, GFRP1, HMR, N10, NAK-1, NGFIB, NP10, NUR77, TR3 | nuclear receptor subfamily 4, group A, member 1 |
C00034649
|
| 9360 | PPIG, CARS-Cyp, CYP, SCAF10, SRCyp | peptidylprolyl isomerase G (cyclophilin G) (EC:5.2.1.8) |
C00000094
|
| 8829 | NRP1, BDCA4, CD304, NP1, NRP, VEGF165R | neuropilin 1 |
C00034649
|
| 9369 | NRXN3, C14orf60 | neurexin 3 |
C00034649
|
| 50814 | NSDHL, H105E3, SDR31E1, XAP104 | NAD(P) dependent steroid dehydrogenase-like (EC:1.1.1.170) |
C00034649
|
| 27065 | NSG1, D4S234, D4S234E, NEEP21, P21 | neuron specific gene family member 1 |
C00034649
|
| 50863 | NTM, HNT, IGLON2, NTRI | neurotrimin |
C00034649
|
| 4916 | NTRK3, TRKC, gp145(trkC) | neurotrophic tyrosine kinase, receptor, type 3 (EC:2.7.10.1) |
C00034649
|
| 4922 | NTS, NMN-125, NN, NT, NT/N, NTS1 | neurotensin |
C00034649
|
| 11162 | NUDT6, ASFGF2, FGF-AS, FGF2AS, GFG-1, GFG1 | nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
C00034649
|
| 83540 | NUF2, CDCA1, CT106, NUF2R | NUF2, NDC80 kinetochore complex component |
C00034649
|
| 4926 | NUMA1, NUMA | nuclear mitotic apparatus protein 1 |
C00034649
|
| 23279 | NUP160 | nucleoporin 160kDa |
C00034649
|
| 4938 | OAS1, IFI-4, OIAS, OIASI | 2'-5'-oligoadenylate synthetase 1, 40/46kDa (EC:2.7.7.84) |
C00034649
|
| 132299 | OCIAD2 | OCIA domain containing 2 |
C00034649
|
| 4953 | ODC1, ODC | ornithine decarboxylase 1 (EC:4.1.1.17) |
C00034649
|
| 8473 | OGT, HRNT1, O-GLCNAC | O-linked N-acetylglucosamine (GlcNAc) transferase (EC:2.4.1.255) |
C00034649
|
| 283298 | OLFML1, UNQ564 | olfactomedin-like 1 |
C00034649
|
| 25903 | OLFML2B, RP11-227F8.1 | olfactomedin-like 2B |
C00034649
|
| 56944 | OLFML3, HNOEL-iso, OLF44 | olfactomedin-like 3 |
C00034649
|
| 4958 | OMD, OSAD, SLRR2C | osteomodulin |
C00034649
|
| 23596 | OPN3, ECPN | opsin 3 |
C00034649
|
| 4986 | OPRK1, K-OR-1, KOR, KOR-1, OPRK | opioid receptor, kappa 1 |
C00034649
|
| 80228 | ORAI2, C7orf19, CBCIP2, MEM142B, TMEM142B | ORAI calcium release-activated calcium modulator 2 |
C00034649
|
| 4999 | ORC2, ORC2L | origin recognition complex, subunit 2 |
C00034649
|
| 5000 | ORC4, ORC4L, ORC4P | origin recognition complex, subunit 4 |
C00034649
|
| 5004 | ORM1, AGP-A, AGP1, ORM | orosomucoid 1 |
C00034649
|
| 5005 | ORM2, AGP-B, AGP-B', AGP2 | orosomucoid 2 |
C00034649
|
| 114885 | OSBPL11, ORP-11, ORP11, OSBP12, TCCCIA00292 | oxysterol binding protein-like 11 |
C00034649
|
| 116039 | OSR2 | odd-skipped related transciption factor 2 |
C00034649
|
| 5016 | OVGP1, CHIT5, EGP, MUC9, OGP | oviductal glycoprotein 1, 120kDa |
C00034649
|
| 9934 | P2RY14, BPR105, GPR105, P2Y14 | purinergic receptor P2Y, G-protein coupled, 14 |
C00034649
|
| 5047 | PAEP, GD, GdA, GdF, GdS, PAEG, PEP, PP14 | progestagen-associated endometrial protein |
C00034649
|
| 55824 | PAG1, CBP, PAG | phosphoprotein associated with glycosphingolipid microdomains 1 |
C00034649
|
| 55003 | PAK1IP1, MAK11, PIP1, RP11-421M1.5, WDR84, bA421M1.5, hPIP1 | PAK1 interacting protein 1 |
C00034649
|
| 54873 | PALMD, C1orf11, PALML | palmdelphin |
C00034649
|
| 5069 | PAPPA, ASBABP2, DIPLA1, IGFBP-4ase, PAPA, PAPP-A, PAPPA1 | pregnancy-associated plasma protein A, pappalysin 1 (EC:3.4.24.79) |
C00034649
|
| 23598 | PATZ1, MAZR, PATZ, RIAZ, ZBTB19, ZNF278, ZSG, dJ400N23 | POZ (BTB) and AT hook containing zinc finger 1 |
C00034649
|
| 5079 | PAX5, BSAP | paired box 5 |
C00034649
|
| 54039 | PCBP3, ALPHA-CP3 | poly(rC) binding protein 3 |
C00034649
|
| 5095 | PCCA | propionyl CoA carboxylase, alpha polypeptide (EC:6.4.1.3) |
C00034649
|
| 54510 | PCDH18, PCDH68L | protocadherin 18 |
C00034649
|
| 5099 | PCDH7, BH-Pcdh, BHPCDH | protocadherin 7 |
C00034649
|
| 5101 | PCDH9 | protocadherin 9 |
C00034649
|
| 56114 | PCDHGA1, PCDH-GAMMA-A1 | protocadherin gamma subfamily A, 1 |
C00034649
|
| 56105 | PCDHGA11, PCDH-GAMMA-A11 | protocadherin gamma subfamily A, 11 |
C00034649
|
| 56112 | PCDHGA3, PCDH-GAMMA-A3 | protocadherin gamma subfamily A, 3 |
C00034649
|
| 5105 | PCK1, PEPCK-C, PEPCK1, PEPCKC | phosphoenolpyruvate carboxykinase 1 (soluble) (EC:4.1.1.32) |
C00034649
|
| 5125 | PCSK5, PC5, PC6, PC6A, SPC6, subtilase | proprotein convertase subtilisin/kexin type 5 (EC:3.4.21.-) |
C00034649
|
| 11333 | PDAP1, HASPP28, PAP, PAP1 | PDGFA associated protein 1 |
C00034649
|
| 79031 | PDCL3, HTPHLP, PHLP2A, PHLP3, VIAF, VIAF1 | phosducin-like 3 |
C00034649
|
| 5137 | PDE1C, Hcam3 | phosphodiesterase 1C, calmodulin-dependent 70kDa (EC:3.1.4.17) |
C00034649
|
| 5152 | PDE9A, HSPDE9A2 | phosphodiesterase 9A (EC:3.1.4.35) |
C00034649
|
| 5154 | PDGFA, PDGF-A, PDGF1 | platelet-derived growth factor alpha polypeptide |
C00034649
|
| 5155 | PDGFB, IBGC5, PDGF-2, PDGF2, SIS, SSV, c-sis | platelet-derived growth factor beta polypeptide |
C00034649
|
| 5156 | PDGFRA, CD140A, PDGFR-2, PDGFR2, RHEPDGFRA | platelet-derived growth factor receptor, alpha polypeptide (EC:2.7.10.1) |
C00034649
|
| 5159 | PDGFRB, CD140B, IBGC4, IMF1, JTK12, PDGFR, PDGFR-1, PDGFR1 | platelet-derived growth factor receptor, beta polypeptide (EC:2.7.10.1) |
C00034649
|
| 9260 | PDLIM7, LMP1, LMP3 | PDZ and LIM domain 7 (enigma) |
C00034649
|
| 5170 | PDPK1, PDK1, PDPK2, PRO0461 | 3-phosphoinositide dependent protein kinase-1 (EC:2.7.11.1) |
C00034649
|
| 10630 | PDPN, AGGRUS, GP36, GP40, Gp38, HT1A-1, OTS8, PA2.26, T1A, T1A-2 | podoplanin |
C00034649
|
| 23244 | PDS5A, SCC-112, SCC112 | PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
C00034649
|
| 23037 | PDZD2, AIPC, PAPIN, PDZK3, PIN1 | PDZ domain containing 2 |
C00034649
|
| 5175 | PECAM1, CD31, CD31/EndoCAM, GPIIA', PECA1, PECAM-1, endoCAM | platelet/endothelial cell adhesion molecule 1 |
C00034649
|
| 23089 | PEG10, EDR, HB-1, MEF3L, Mar2, Mart2, RGAG3 | paternally expressed 10 |
C00034649
|
| 53918 | PELO, PRO1770 | pelota homolog (Drosophila) |
C00034649
|
| 10400 | PEMT, PEAMT, PEMPT, PEMT2, PNMT | phosphatidylethanolamine N-methyltransferase (EC:2.1.1.17 2.1.1.71) |
C00034649
|
| 5187 | PER1, PER, RIGUI, hPER | period circadian clock 1 |
C00034649
|
| 8864 | PER2, FASPS, FASPS1 | period circadian clock 2 |
C00034649
|
| 8863 | PER3, GIG13 | period circadian clock 3 |
C00034649
|
| 5198 | PFAS, FGAMS, FGARAT, PURL | phosphoribosylformylglycinamidine synthase (EC:6.3.5.3) |
C00034649
|
| 80055 | PGAP1, Bst1, ISPD3024 | post-GPI attachment to proteins 1 |
C00034649
|
| 79605 | PGBD5 | piggyBac transposable element derived 5 |
C00034649
|
| 116154 | PHACTR3, C20orf101, H17739, SCAPIN1, SCAPININ | phosphatase and actin regulator 3 |
C00034649
|
| 5245 | PHB, PHB1 | prohibitin |
C00034649
|
| 22822 | PHLDA1, DT1P1B11, PHRIP, TDAG51 | pleckstrin homology-like domain, family A, member 1 |
C00034649
|
| 51050 | PI15, CRISP8, P24TI, P25TI | peptidase inhibitor 15 |
C00034649
|
| 10464 | PIBF1, C13orf24, CEP90, PIBF, RP11-505F3.1 | progesterone immunomodulatory binding factor 1 |
C00034649
|
| 55022 | PID1, NYGGF4, P-CLI1, PCLI1 | phosphotyrosine interaction domain containing 1 |
C00034649
|
| 8503 | PIK3R3, p55, p55-GAMMA | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
C00034649
|
| 5292 | PIM1, PIM | pim-1 oncogene (EC:2.7.11.1) |
C00034649
|
| 8395 | PIP5K1B, MSS4, STM7 | phosphatidylinositol-4-phosphate 5-kinase, type I, beta (EC:2.7.1.68) |
C00034649
|
| 93035 | PKHD1L1, PKHDL1 | polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 |
C00034649
|
| 5569 | PKIA, PRKACN1 | protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
C00034649
|
| 5321 | PLA2G4A, PLA2G4, cPLA2-alpha | phospholipase A2, group IVA (cytosolic, calcium-dependent) (EC:3.1.1.5 3.1.1.4) |
C00034649
|
| 5327 | PLAT, T-PA, TPA | plasminogen activator, tissue (EC:3.4.21.68) |
C00034649
|
| 5328 | PLAU, ATF, BDPLT5, QPD, UPA, URK, u-PA | plasminogen activator, urokinase (EC:3.4.21.73) |
C00034649
|
| 5329 | PLAUR, CD87, U-PAR, UPAR, URKR | plasminogen activator, urokinase receptor |
C00034649
|
| 23236 | PLCB1, EIEE12, PI-PLC, PLC-154, PLC-I, PLC154, PLCB1A, PLCB1B | phospholipase C, beta 1 (phosphoinositide-specific) (EC:3.1.4.11) |
C00034649
|
| 51196 | PLCE1, NPHS3, PLCE, PPLC | phospholipase C, epsilon 1 (EC:3.1.4.11) |
C00034649
|
| 23007 | PLCH1, PLCL3 | phospholipase C, eta 1 (EC:3.1.4.11) |
C00034649
|
| 5339 | PLEC, EBS1, EBSO, HD1, LGMD2Q, PCN, PLEC1, PLEC1b, PLTN | plectin |
C00034649
|
| 26499 | PLEK2 | pleckstrin 2 |
C00034649
|
| 144100 | PLEKHA7 | pleckstrin homology domain containing, family A member 7 |
C00034649
|
| 51177 | PLEKHO1, CKIP-1, OC120 | pleckstrin homology domain containing, family O member 1 |
C00034649
|
| 55857 | PLK1S1, C20orf19, Kiz, Kizuna, NCRNA00153 | polo-like kinase 1 substrate 1 |
C00034649
|
| 10769 | PLK2, SNK, hPlk2, hSNK | polo-like kinase 2 (EC:2.7.11.21) |
C00034649
|
| 57088 | PLSCR4, TRA1 | phospholipid scramblase 4 |
C00034649
|
| 57125 | PLXDC1, TEM3, TEM7 | plexin domain containing 1 |
C00034649
|
| 10154 | PLXNC1, CD232, PLXN-C1, VESPR | plexin C1 |
C00034649
|
| 55228 | PNMAL1 | paraneoplastic Ma antigen family-like 1 |
C00034649
|
| 55629 | PNRC2 | proline-rich nuclear receptor coactivator 2 |
C00034649
|
| 23509 | POFUT1, DDD2, FUT12, O-FUT, O-Fuc-T, O-FucT-1, OFUCT1 | protein O-fucosyltransferase 1 (EC:2.4.1.221) |
C00034649
|
| 56655 | POLE4, YHHQ1, p12 | polymerase (DNA-directed), epsilon 4, accessory subunit (EC:2.7.7.7) |
C00034649
|
| 5439 | POLR2J, POLR2J1, RPB11, RPB11A, RPB11m, hRPB14 | polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa (EC:2.7.7.6) |
C00034649
|
| 51728 | POLR3K, C11, C11-RNP3, RPC10, RPC11, RPC12.5, hRPC11 | polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
C00034649
|
| 1075 | CTSC, CPPI, DPP-I, DPP1, DPPI, HMS, JP, JPD, PALS, PDON1, PLS | cathepsin C (EC:3.4.14.1) |
C00002514
|
| 64840 | PORCN, DHOF, FODH, MG61, PORC, PPN | porcupine homolog (Drosophila) |
C00034649
|
| 5460 | POU5F1, OCT3, OCT4, OTF-3, OTF3, OTF4, Oct-3, Oct-4 | POU class 5 homeobox 1 |
C00034649
|
| 8613 | PPAP2B, Dri42, LPP3, PAP2B, VCIP | phosphatidic acid phosphatase type 2B (EC:3.1.3.4) |
C00034649
|
| 8544 | PIR | pirin (iron-binding nuclear protein) (EC:1.13.11.24) |
C00000094
|
| 728045 |
C00034649
|
||
| 8496 | PPFIBP1, L2, SGT2, hSGT2, hSgt2p | PTPRF interacting protein, binding protein 1 (liprin beta 1) |
C00034649
|
| 8495 | PPFIBP2, Cclp1 | PTPRF interacting protein, binding protein 2 (liprin beta 2) |
C00034649
|
| 94274 | PPP1R14A, CPI-17, CPI17, PPP1INL | protein phosphatase 1, regulatory (inhibitor) subunit 14A |
C00034649
|
| 81706 | PPP1R14C, CPI17-like, KEPI, NY-BR-81 | protein phosphatase 1, regulatory (inhibitor) subunit 14C |
C00034649
|
| 26051 | PPP1R16B, ANKRD4, TIMAP | protein phosphatase 1, regulatory subunit 16B |
C00034649
|
| 5502 | PPP1R1A, I1, IPP1 | protein phosphatase 1, regulatory (inhibitor) subunit 1A (EC:3.1.3.16) |
C00034649
|
| 55012 | PPP2R3C, C14orf10, G4-1, G5pr | protein phosphatase 2, regulatory subunit B'', gamma |
C00034649
|
| 306506 |
C00034649
|
||
| 23532 | PRAME, CT130, MAPE, OIP-4, OIP4 | preferentially expressed antigen in melanoma |
C00034649
|
| 9055 | PRC1, ASE1 | protein regulator of cytokinesis 1 |
C00034649
|
| 59335 | PRDM12, PFM9 | PR domain containing 12 |
C00034649
|
| 51422 | PRKAG2, AAKG, AAKG2, CMH6, H91620p, WPWS | protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
C00034649
|
| 5588 | PRKCQ, PRKCT, nPKC-theta | protein kinase C, theta (EC:2.7.11.13) |
C00034649
|
| 5613 | PRKX, PKX1 | protein kinase, X-linked (EC:2.7.11.1) |
C00034649
|
| 5617 | PRL | prolactin |
C00034649
|
| 5618 | PRLR, hPRLrI | prolactin receptor |
C00034649
|
| 54496 | PRMT7 | protein arginine methyltransferase 7 (EC:2.1.1.125 2.1.1.126) |
C00034649
|
| 5621 | PRNP, ASCR, AltPrP, CD230, CJD, GSS, KURU, PRIP, PrP, PrP27-30, PrP33-35C, PrPc, p27-30 | prion protein |
C00034649
|
| 84432 | PROK1, EGVEGF, PK1, PRK1 | prokineticin 1 |
C00034649
|
| 8842 | PROM1, AC133, CD133, CORD12, MCDR2, PROML1, RP41, STGD4 | prominin 1 |
C00034649
|
| 55660 | PRPF40A, FBP-11, FBP11, FLAF1, FNBP3, HIP-10, HIP10, HYPA, NY-REN-6, Prp40 | PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
C00034649
|
| 79170 | PRR15L, ATAD4 | proline rich 15-like |
C00034649
|
| 5396 | PRRX1, AGOTC, PHOX1, PMX1, PRX-1, PRX1 | paired related homeobox 1 |
C00034649
|
| 158471 | PRUNE2, A214N16.3, BMCC1, BNIPXL, C9orf65, KIAA0367, RP11-58J3.2, bA214N16.3 | prune homolog 2 (Drosophila) |
C00034649
|
| 29968 | PSAT1, EPIP, PSA, PSAT | phosphoserine aminotransferase 1 (EC:2.6.1.52) |
C00034649
|
| 55269 | PSPC1, PSP1 | paraspeckle component 1 |
C00034649
|
| 8781 |
C00034649
|
||
| 442213 | PTCHD4, C6orf138, dJ402H5.2 | patched domain containing 4 |
C00034649
|
| 5732 | PTGER2, EP2 | prostaglandin E receptor 2 (subtype EP2), 53kDa |
C00034649
|
| 5737 | PTGFR, FP | prostaglandin F receptor (FP) |
C00034649
|
| 5738 | PTGFRN, CD315, CD9P-1, EWI-F, FPRP, SMAP-6 | prostaglandin F2 receptor inhibitor |
C00034649
|
| 22949 | PTGR1, LTB4DH, PGR1, ZADH3 | prostaglandin reductase 1 (EC:1.3.1.48 1.3.1.74) |
C00034649
|
| 5742 | PTGS1, COX1, COX3, PCOX1, PES-1, PGG/HS, PGHS-1, PGHS1, PHS1, PTGHS | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00034649
|
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00034649
|
| 5746 | PTH2R, PTHR2 | parathyroid hormone 2 receptor |
C00034649
|
| 5744 | PTHLH, BDE2, HHM, PLP, PTHR, PTHRP | parathyroid hormone-like hormone |
C00034649
|
| 5764 | PTN, HARP, HBGF8, HBNF, NEGF1 | pleiotrophin |
C00034649
|
| 9200 | PTPLA, CAP, HACD1 | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A (EC:4.2.1.134) |
C00034649
|
| 401494 | PTPLAD2, HACD4 | protein tyrosine phosphatase-like A domain containing 2 (EC:4.2.1.134) |
C00034649
|
| 5795 | PTPRJ, CD148, DEP1, HPTPeta, R-PTP-ETA, SCC1 | protein tyrosine phosphatase, receptor type, J (EC:3.1.3.48) |
C00034649
|
| 5801 | PTPRR, EC-PTP, PCPTP1, PTP-SL, PTPBR7, PTPRQ | protein tyrosine phosphatase, receptor type, R (EC:3.1.3.48) |
C00034649
|
| 5806 | PTX3, TNFAIP5, TSG-14 | pentraxin 3, long |
C00034649
|
| 25797 | QPCT, GCT, QC, sQC | glutaminyl-peptide cyclotransferase (EC:2.3.2.5) |
C00034649
|
| 376267 | RAB15 | RAB15, member RAS oncogene family |
C00034649
|
| 5874 | RAB27B, C25KG | RAB27B, member RAS oncogene family |
C00034649
|
| 5865 | RAB3B | RAB3B, member RAS oncogene family |
C00034649
|
| 5880 | RAC2, EN-7, Gx, HSPC022, p21-Rac2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
C00034649
|
| 5894 | RAF1, CRAF, NS5, Raf-1, c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 (EC:2.7.11.1) |
C00034649
|
| 26064 | RAI14, NORPEG, RAI13 | retinoic acid induced 14 |
C00034649
|
| 5901 | RAN, ARA24, Gsp1, TC4 | RAN, member RAS oncogene family |
C00034649
|
| 5918 | RARRES1, LXNL, TIG1 | retinoic acid receptor responder (tazarotene induced) 1 |
C00034649
|
| 5919 | RARRES2, HP10433, TIG2 | retinoic acid receptor responder (tazarotene induced) 2 |
C00034649
|
| 10235 | RASGRP2, CALDAG-GEFI, CDC25L | RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
C00034649
|
| 387496 | RASL11A | RAS-like, family 11, member A |
C00034649
|
| 51285 | RASL12, RIS | RAS-like, family 12 |
C00034649
|
| 83937 | RASSF4, AD037 | Ras association (RalGDS/AF-6) domain family member 4 |
C00034649
|
| 9182 | RASSF9, P-CIP1, PAMCI, PCIP1 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
C00034649
|
| 5930 | RBBP6, MY038, P2P-R, PACT, RBQ-1, SNAMA | retinoblastoma binding protein 6 |
C00034649
|
| 27303 | RBMS3 | RNA binding motif, single stranded interacting protein 3 |
C00034649
|
| 5947 | RBP1, CRABP-I, CRBP, CRBP1, CRBPI, RBPC | retinol binding protein 1, cellular |
C00034649
|
| 5950 | RBP4, RDCCAS | retinol binding protein 4, plasma |
C00034649
|
| 1104 | RCC1, CHC1, RCC1-I, SNHG3-RCC1 | regulator of chromosome condensation 1 |
C00034649
|
| 8434 | RECK, ST15 | reversion-inducing-cysteine-rich protein with kazal motifs |
C00034649
|
| 5980 | REV3L, POLZ, REV3 | REV3-like, polymerase (DNA directed), zeta, catalytic subunit (EC:2.7.7.7) |
C00034649
|
| 28984 | RGCC, C13orf15, RGC-32, RGC32, bA157L14.2 | regulator of cell cycle |
C00034649
|
| 5996 | RGS1, 1R20, BL34, IER1, IR20 | regulator of G-protein signaling 1 |
C00034649
|
| 5998 | RGS3, C2PA, RGP3 | regulator of G-protein signaling 3 |
C00034649
|
| 5999 | RGS4, RGP4, SCZD9 | regulator of G-protein signaling 4 |
C00034649
|
| 8490 | RGS5, MST092, MST106, MST129, MSTP032, MSTP092, MSTP106, MSTP129 | regulator of G-protein signaling 5 |
C00034649
|
| 58480 | RHOU, ARHU, CDC42L1, DJ646B12.2, WRCH1, fJ646B12.2, hG28K | ras homolog family member U |
C00034649
|
| 55183 | RIF1 | RAP1 interacting factor homolog (yeast) |
C00034649
|
| 196383 | RILPL2, RLP2 | Rab interacting lysosomal protein-like 2 |
C00034649
|
| 23504 | RIMBP2, RBP2, RIM-BP2 | RIMS binding protein 2 |
C00034649
|
| 6038 | RNASE4, RNS4 | ribonuclease, RNase A family, 4 (EC:3.1.27.-) |
C00034649
|
| 8635 | RNASET2, RNASE6PL, bA514O12.3 | ribonuclease T2 (EC:3.1.27.1) |
C00034649
|
| 54941 | RNF125, TRAC-1, TRAC1 | ring finger protein 125, E3 ubiquitin protein ligase |
C00034649
|
| 11236 | RNF139, HRCA1, RCA1, TRC8 | ring finger protein 139 |
C00034649
|
| 57484 | RNF150 | ring finger protein 150 |
C00034649
|
| 6048 | RNF5, RING5, RMA1 | ring finger protein 5, E3 ubiquitin protein ligase |
C00034649
|
| 26824 | RNU11, RNU11-1, U11 | RNA, U11 small nuclear |
C00034649
|
| 6091 | ROBO1, DUTT1, SAX3 | roundabout, axon guidance receptor, homolog 1 (Drosophila) |
C00034649
|
| 79641 | ROGDI, KTZS | rogdi homolog (Drosophila) |
C00034649
|
| 6096 | RORB, NR1F2, ROR-BETA, RZR-BETA, RZRB, bA133M9.1 | RAR-related orphan receptor B |
C00034649
|
| 200916 | RPL22L1 | ribosomal protein L22-like 1 |
C00034649
|
| 6157 | RPL27A, L27A | ribosomal protein L27a |
C00034649
|
| 6224 | RPS20, S20 | ribosomal protein S20 |
C00034649
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00034649
|
| 9252 | RPS6KA5, MSK1, MSPK1, RLPK | ribosomal protein S6 kinase, 90kDa, polypeptide 5 (EC:2.7.11.1) |
C00034649
|
| 57521 | RPTOR, KOG1, Mip1 | regulatory associated protein of MTOR, complex 1 |
C00034649
|
| 9136 | RRP9, RNU3IP2, U3-55K | ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
C00034649
|
| 23212 | RRS1 | RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
C00034649
|
| 10313 | RTN3, ASYIP, HAP, NSPL2, NSPLII, RTN3-A1 | reticulon 3 |
C00034649
|
| 860 | RUNX2, AML3, CBF-alpha-1, CBFA1, CCD, CCD1, CLCD, OSF-2, OSF2, PEA2aA, PEBP2aA | runt-related transcription factor 2 |
C00034649
|
| 59350 | RXFP1, LGR7, RXFPR1 | relaxin/insulin-like family peptide receptor 1 |
C00034649
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00034649
|
| 6263 | RYR3 | ryanodine receptor 3 |
C00034649
|
| 6273 | S100A2, CAN19, S100L | S100 calcium binding protein A2 |
C00034649
|
| 6289 | SAA2 | serum amyloid A2 |
C00034649
|
| 148398 | SAMD11 | sterile alpha motif domain containing 11 |
C00034649
|
| 401474 | SAMD12 | sterile alpha motif domain containing 12 |
C00034649
|
| 54809 | SAMD9, C7orf5, DRIF1, NFTC, OEF1, OEF2 | sterile alpha motif domain containing 9 |
C00034649
|
| 6301 | SARS, SERRS, SERS | seryl-tRNA synthetase (EC:6.1.1.11) |
C00034649
|
| 388228 | SBK1, SBK | SH3 domain binding kinase 1 (EC:2.7.11.1) |
C00034649
|
| 6309 | SC5D, ERG3, S5DES, SC5DL | sterol-C5-desaturase (EC:1.14.21.6) |
C00034649
|
| 20249 |
C00034649
|
||
| 10648 | SCGB1D1, LIPA, LPHA, LPNA | secretoglobin, family 1D, member 1 |
C00034649
|
| 10647 | SCGB1D2, LIPB, LPHB, LPNB | secretoglobin, family 1D, member 2 |
C00034649
|
| 4246 | SCGB2A1, LPHC, LPNC, MGB2, UGB3 | secretoglobin, family 2A, member 1 |
C00034649
|
| 4250 | SCGB2A2, MGB1, UGB2 | secretoglobin, family 2A, member 2 |
C00034649
|
| 85477 | SCIN | scinderin |
C00034649
|
| 6337 | SCNN1A, BESC2, ENaCa, ENaCalpha, SCNEA, SCNN1 | sodium channel, non-voltage-gated 1 alpha subunit |
C00034649
|
| 6382 | SDC1, CD138, SDC, SYND1, syndecan | syndecan 1 |
C00034649
|
| 6385 | SDC4, SYND4 | syndecan 4 |
C00034649
|
| 23753 | SDF2L1 | stromal cell-derived factor 2-like 1 |
C00034649
|
| 6397 | SEC14L1, PRELID4A, SEC14L | SEC14-like 1 (S. cerevisiae) |
C00034649
|
| 23541 | SEC14L2, C22orf6, SPF, TAP, TAP1 | SEC14-like 2 (S. cerevisiae) |
C00034649
|
| 6398 | SECTM1, K12 | secreted and transmembrane 1 |
C00034649
|
| 81929 | SEH1L, SEC13L, SEH1A, SEH1B, Seh1 | SEH1-like (S. cerevisiae) |
C00034649
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00034649
|
| 7869 | SEMA3B, LUCA-1, SEMA5, SEMAA, SemA, semaV | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
C00034649
|
| 6405 | SEMA3F, SEMA-IV, SEMA4, SEMAK | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
C00034649
|
| 10507 | SEMA4D, C9orf164, CD100, M-sema-G, SEMAJ, coll-4 | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
C00034649
|
| 12 | SERPINA3, AACT, ACT, GIG25 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
C00034649
|
| 866 | SERPINA6, CBG | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
C00034649
|
| 5055 | SERPINB2, HsT1201, PAI, PAI-2, PAI2, PLANH2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
C00034649
|
| 5268 | SERPINB5, PI5, maspin | serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
C00034649
|
| 5272 | SERPINB9, CAP-3, CAP3, PI-9, PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
C00034649
|
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00034649
|
| 5176 | SERPINF1, EPC-1, OI12, OI6, PEDF | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
C00034649
|
| 710 | SERPING1, C1IN, C1INH, C1NH, HAE1, HAE2 | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
C00034649
|
| 56256 | SERTAD4, DJ667H12.2 | SERTA domain containing 4 |
C00034649
|
| 143686 | SESN3, SEST3 | sestrin 3 |
C00034649
|
| 91404 | SESTD1, SOLO | SEC14 and spectrin domains 1 |
C00034649
|
| 6418 | SET, 2PP2A, I2PP2A, IGAAD, IPP2A2, PHAPII, TAF-I, TAF-IBETA | SET nuclear oncogene |
C00034649
|
| 6421 | SFPQ, POMP100, PSF | splicing factor proline/glutamine-rich |
C00034649
|
| 389376 | SFTA2, GSGL541, SFTPG, UNQ541 | surfactant associated 2 |
C00034649
|
| 6439 | SFTPB, PSP-B, SFTB3, SFTP3, SMDP1, SP-B | surfactant protein B |
C00034649
|
| 6444 | SGCD, 35DAG, CMD1L, DAGD, SG-delta, SGCDP, SGD | sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
C00034649
|
| 84251 | SGIP1 | SH3-domain GRB2-like (endophilin) interacting protein 1 |
C00034649
|
| 6446 | SGK1, SGK | serum/glucocorticoid regulated kinase 1 (EC:2.7.11.1) |
C00034649
|
| 117157 | SH2D1B, EAT2 | SH2 domain containing 1B |
C00034649
|
| 153769 | SH3RF2, HEPP1, POSHER, PPP1R39, RNF158 | SH3 domain containing ring finger 2 |
C00034649
|
| 399694 | SHC4, RaLP, SHCD | SHC (Src homology 2 domain containing) family, member 4 |
C00034649
|
| 6469 | SHH, HHG1, HLP3, HPE3, MCOPCB5, SMMCI, TPT, TPTPS | sonic hedgehog |
C00034649
|
| 284266 | SIGLEC15, CD33L3, HsT1361, SIGLEC-15 | sialic acid binding Ig-like lectin 15 |
C00034649
|
| 26037 | SIPA1L1, E6TP1 | signal-induced proliferation-associated 1 like 1 |
C00034649
|
| 57568 | SIPA1L2, SPAL2 | signal-induced proliferation-associated 1 like 2 |
C00034649
|
| 140885 | SIRPA, BIT, CD172A, MFR, MYD-1, P84, PTPNS1, SHPS1, SIRP | signal-regulatory protein alpha (EC:3.1.3.48) |
C00034649
|
| 122060 | SLAIN1, C13orf32 | SLAIN motif family, member 1 |
C00034649
|
| 6563 | SLC14A1, HUT11, HsT1341, JK, RACH1, RACH2, UT-B1, UT1, UTE | solute carrier family 14 (urea transporter), member 1 |
C00034649
|
| 6564 | SLC15A1, HPECT1, HPEPT1, PEPT1 | solute carrier family 15 (oligopeptide transporter), member 1 |
C00034649
|
| 6565 | SLC15A2, PEPT2 | solute carrier family 15 (oligopeptide transporter), member 2 |
C00034649
|
| 9120 | SLC16A6, MCT6, MCT7 | solute carrier family 16, member 6 |
C00034649
|
| 9194 | SLC16A7, MCT2 | solute carrier family 16 (monocarboxylate transporter), member 7 |
C00034649
|
| 10560 | SLC19A2, TC1, THMD1, THT1, THTR1, TRMA | solute carrier family 19 (thiamine transporter), member 2 |
C00034649
|
| 6505 | SLC1A1, EAAC1, EAAT3, SCZD18 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
C00034649
|
| 6507 | SLC1A3, EA6, EAAT1, GLAST, GLAST1 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
C00034649
|
| 6580 | SLC22A1, HOCT1, OCT1, oct1_cds | solute carrier family 22 (organic cation transporter), member 1 |
C00034649
|
| 6581 | SLC22A3, EMT, EMTH, OCT3 | solute carrier family 22 (organic cation transporter), member 3 |
C00034649
|
| 6584 | SLC22A5, CDSP, OCTN2, OCTN2VT | solute carrier family 22 (organic cation/carnitine transporter), member 5 |
C00034649
|
| 114571 | SLC22A9, HOAT4, OAT4, OAT7, UST3H, ust3 | solute carrier family 22 (organic anion transporter), member 9 |
C00034649
|
| 9962 | SLC23A2, NBTL1, SLC23A1, SVCT2, YSPL2 | solute carrier family 23 (ascorbic acid transporter), member 2 |
C00034649
|
| 8034 | SLC25A16, D10S105E, GDA, GDC, HGT.1, ML7, hML7 | solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
C00034649
|
| 6513 | SLC2A1, DYT17, DYT18, DYT9, EIG12, GLUT, GLUT1, GLUT1DS, HTLVR, PED | solute carrier family 2 (facilitated glucose transporter), member 1 |
C00034649
|
| 7780 | SLC30A2, PP12488, TNZD, ZNT2, ZnT-2 | solute carrier family 30 (zinc transporter), member 2 |
C00034649
|
| 219855 | SLC37A2, pp11662 | solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
C00034649
|
| 54407 | SLC38A2, ATA2, PRO1068, SAT2, SNAT2 | solute carrier family 38, member 2 |
C00034649
|
| 55089 | SLC38A4, ATA3, NAT3, PAAT | solute carrier family 38, member 4 |
C00034649
|
| 23516 | SLC39A14, LZT-Hs4, NET34, ZIP14, cig19 | solute carrier family 39 (zinc transporter), member 14 |
C00034649
|
| 6519 | SLC3A1, ATR1, CSNU1, D2H, NBAT, RBAT | solute carrier family 3 (amino acid transporter heavy chain), member 1 |
C00034649
|
| 30061 | SLC40A1, FPN1, HFE4, IREG1, MST079, MTP1, SLC11A3 | solute carrier family 40 (iron-regulated transporter), member 1 |
C00034649
|
| 84102 | SLC41A2, SLC41A1-L1 | solute carrier family 41 (magnesium transporter), member 2 |
C00034649
|
| 57153 | SLC44A2, CTL2, PP1292 | solute carrier family 44 (choline transporter), member 2 |
C00034649
|
| 126969 | SLC44A3, CTL3 | solute carrier family 44, member 3 |
C00034649
|
| 283537 | SLC46A3, FKSG16 | solute carrier family 46, member 3 |
C00034649
|
| 55244 | SLC47A1, MATE1 | solute carrier family 47 (multidrug and toxin extrusion), member 1 |
C00034649
|
| 83959 | SLC4A11, BTR1, CDPD1, CHED2, NABC1, dJ794I6.2 | solute carrier family 4, sodium borate transporter, member 11 |
C00034649
|
| 9497 | SLC4A7, NBC2, NBC3, NBCN1, SBC2, SLC4A6 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
C00034649
|
| 200010 | SLC5A9, SGLT4 | solute carrier family 5 (sodium/sugar cotransporter), member 9 |
C00034649
|
| 6538 | SLC6A11, GAT-3, GAT3, GAT4 | solute carrier family 6 (neurotransmitter transporter), member 11 |
C00034649
|
| 6542 | SLC7A2, ATRC2, CAT2, HCAT2 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
C00034649
|
| 6545 | SLC7A4, CAT-4, CAT4, HCAT3, VH | solute carrier family 7, member 4 |
C00034649
|
| 23428 | SLC7A8, LAT2, LPI-PC1 | solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
C00034649
|
| 6546 | SLC8A1, NCX1 | solute carrier family 8 (sodium/calcium exchanger), member 1 |
C00034649
|
| 6578 | SLCO2A1, MATR1, OATP2A1, PGT, PHOAR2, SLC21A2 | solute carrier organic anion transporter family, member 2A1 |
C00034649
|
| 28232 | SLCO3A1, OATP-D, OATP3A1, OATPD, SLC21A11 | solute carrier organic anion transporter family, member 3A1 |
C00034649
|
| 6585 | SLIT1, MEGF4, SLIL1, SLIT-1, SLIT3 | slit homolog 1 (Drosophila) |
C00034649
|
| 139065 | SLITRK4 | SLIT and NTRK-like family, member 4 |
C00034649
|
| 4091 | SMAD6, AOVD2, HsT17432, MADH6, MADH7 | SMAD family member 6 |
C00034649
|
| 4093 | SMAD9, MADH6, MADH9, PPH2, SMAD8, SMAD8A, SMAD8B | SMAD family member 9 |
C00034649
|
| 85027 | SMIM3, C5orf62, MST150, NID67 | small integral membrane protein 3 |
C00034649
|
| 64094 | SMOC2, DTDP1, MST117, MSTP117, MSTP140, SMAP2, bA270C4A.1, bA37D8.1, dJ421D16.1 | SPARC related modular calcium binding 2 |
C00034649
|
| 10924 | SMPDL3A, ASM3A, ASML3a, yR36GH4.1 | sphingomyelin phosphodiesterase, acid-like 3A |
C00034649
|
| 6525 | SMTN | smoothelin |
C00034649
|
| 64754 | SMYD3, KMT3E, ZMYND1, ZNFN3A1, bA74P14.1 | SET and MYND domain containing 3 (EC:2.1.1.43) |
C00034649
|
| 9627 | SNCAIP, SYPH1, Sph1 | synuclein, alpha interacting protein |
C00034649
|
| 54861 | SNRK, HSNFRK | SNF related kinase (EC:2.7.11.1) |
C00034649
|
| 8651 | SOCS1, CIS1, CISH1, JAB, SOCS-1, SSI-1, SSI1, TIP3 | suppressor of cytokine signaling 1 |
C00034649
|
| 9021 | SOCS3, ATOD4, CIS3, Cish3, SOCS-3, SSI-3, SSI3 | suppressor of cytokine signaling 3 |
C00034649
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00034649
|
| 10580 | SORBS1, CAP, FLAF2, R85FL, SH3D5, SH3P12, SORB1 | sorbin and SH3 domain containing 1 |
C00034649
|
| 8470 | SORBS2, ARGBP2, PRO0618 | sorbin and SH3 domain containing 2 |
C00034649
|
| 6272 | SORT1, Gp95, LDLCQ6, NT3 | sortilin 1 |
C00034649
|
| 6664 | SOX11 | SRY (sex determining region Y)-box 11 |
C00034649
|
| 83595 | SOX7 | SRY (sex determining region Y)-box 7 |
C00034649
|
| 8404 | SPARCL1, PIG33, SC1 | SPARC-like 1 (hevin) |
C00034649
|
| 153218 | SPINK13, HBVDNAPTP1, HESPINTOR, LiESP6, SPINK5L3 | serine peptidase inhibitor, Kazal type 13 (putative) |
C00034649
|
| 11005 | SPINK5, LEKTI, LETKI, NETS, NS, VAKTI | serine peptidase inhibitor, Kazal type 5 |
C00034649
|
| 6693 | SPN, CD43, GALGP, GPL115, LSN | sialophorin |
C00034649
|
| 10418 | SPON1, VSGP/F-spondin, f-spondin | spondin 1, extracellular matrix protein |
C00034649
|
| 10417 | SPON2, DIL-1, DIL1, M-SPONDIN, MINDIN | spondin 2, extracellular matrix protein |
C00034649
|
| 6696 | SPP1, BNSP, BSPI, ETA-1, OPN | secreted phosphoprotein 1 |
C00034649
|
| 6701 | SPRR2B | small proline-rich protein 2B |
C00034649
|
| 10252 | SPRY1, hSPRY1 | sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
C00034649
|
| 81848 | SPRY4, HH17 | sprouty homolog 4 (Drosophila) |
C00034649
|
| 80176 | SPSB1, SSB-1, SSB1 | splA/ryanodine receptor domain and SOCS box containing 1 |
C00034649
|
| 6711 | SPTBN1, ELF, SPTB2, betaSpII | spectrin, beta, non-erythrocytic 1 |
C00034649
|
| 171546 | SPTSSA, C14orf147, SSSPTA | serine palmitoyltransferase, small subunit A |
C00034649
|
| 6716 | SRD5A2 | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) (EC:1.3.1.22) |
C00034649
|
| 6720 | SREBF1, SREBP-1c, SREBP1, bHLHd1 | sterol regulatory element binding transcription factor 1 |
C00034649
|
| 5552 | SRGN, PPG, PRG, PRG1 | serglycin |
C00034649
|
| 8406 | SRPX, DRS, ETX1, SRPX1 | sushi-repeat containing protein, X-linked |
C00034649
|
| 6431 | SRSF6, B52, SFRS6, SRP55 | serine/arginine-rich splicing factor 6 |
C00034649
|
| 6736 | SRY, SRXX1, SRXY1, TDF, TDY | sex determining region Y |
C00034649
|
| 8082 | SSPN, DAGA5, KRAG, NSPN, SPN1, SPN2 | sarcospan |
C00034649
|
| 6750 | SST, SMST | somatostatin |
C00034649
|
| 6751 | SSTR1, SRIF-2, SS-1-R, SS1-R, SS1R | somatostatin receptor 1 |
C00034649
|
| 6768 | ST14, HAI, MT-SP1, MTSP1, PRSS14, SNC19, TADG15, TMPRSS14 | suppression of tumorigenicity 14 (colon carcinoma) (EC:3.4.21.109) |
C00034649
|
| 8869 | ST3GAL5, SATI, SIAT9, SIATGM3S, ST3GalV | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (EC:2.4.99.9) |
C00034649
|
| 10402 | ST3GAL6, SIAT10, ST3GALVI | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 (EC:2.4.99.-) |
C00034649
|
| 84620 | ST6GAL2, SIAT2, ST6GalII | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 (EC:2.4.99.1) |
C00034649
|
| 55808 | ST6GALNAC1, HSY11339, SIAT7A, ST6GalNAcI, STYI | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC:2.4.99.3) |
C00034649
|
| 7982 | ST7, ETS7q, FAM4A, FAM4A1, HELG, RAY1, SEN4, TSG7 | suppression of tumorigenicity 7 |
C00034649
|
| 6489 | ST8SIA1, GD3S, SIAT8, SIAT8-A, SIAT8A, ST8SiaI | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 (EC:2.4.99.8) |
C00034649
|
| 57559 | STAMBPL1, ALMalpha, AMSH-FP, AMSH-LP, bA399O19.2 | STAM binding protein-like 1 (EC:3.1.2.15) |
C00034649
|
| 26228 | STAP1, BRDG1, STAP-1 | signal transducing adaptor family member 1 |
C00034649
|
| 6770 | STAR, STARD1 | steroidogenic acute regulatory protein |
C00034649
|
| 100135887 |
C00034649
|
||
| 6776 | STAT5A, MGF, STAT5 | signal transducer and activator of transcription 5A |
C00034649
|
| 6777 | STAT5B, STAT5 | signal transducer and activator of transcription 5B |
C00034649
|
| 79689 | STEAP4, STAMP2, TIARP, TNFAIP9 | STEAP family member 4 |
C00034649
|
| 11075 | STMN2, SCG10, SCGN10 | stathmin-like 2 |
C00034649
|
| 55342 | STRBP, ILF3L, SPNR, p74 | spermatid perinuclear RNA binding protein |
C00034649
|
| 29091 | STXBP6, amisyn | syntaxin binding protein 6 (amisyn) |
C00034649
|
| 6783 | SULT1E1, EST, EST-1, ST1E1, STE | sulfotransferase family 1E, estrogen-preferring, member 1 (EC:2.8.2.4) |
C00034649
|
| 7341 | SUMO1, DAP1, GMP1, OFC10, PIC1, SENP2, SMT3, SMT3C, SMT3H3, UBL1 | small ubiquitin-like modifier 1 |
C00034649
|
| 6840 | SVIL | supervillin |
C00034649
|
| 23345 | SYNE1, 8B, ARCA1, C6orf98, CPG2, EDMD4, MYNE1, Nesp1, SCAR8, dJ45H2.2 | spectrin repeat containing, nuclear envelope 1 |
C00034649
|
| 8871 | SYNJ2, INPP5H | synaptojanin 2 (EC:3.1.3.36) |
C00034649
|
| 171024 | SYNPO2 | synaptopodin 2 |
C00034649
|
| 79933 | SYNPO2L | synaptopodin 2-like |
C00034649
|
| 83851 | SYT16, CHR14SYT, SYT14L, Strep14, syt14r, yt14r | synaptotagmin XVI |
C00034649
|
| 84447 | SYVN1, DER3, HRD1 | synovial apoptosis inhibitor 1, synoviolin (EC:6.3.2.-) |
C00034649
|
| 10579 | TACC2, AZU-1, ECTACC | transforming, acidic coiled-coil containing protein 2 |
C00034649
|
| 6876 | TAGLN, SM22, SMCC, TAGLN1, WS3-10 | transgelin |
C00034649
|
| 6891 | TAP2, ABC18, ABCB3, APT2, D6S217E, PSF-2, PSF2, RING11 | transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
C00034649
|
| 23435 | TARDBP, ALS10, TDP-43 | TAR DNA binding protein |
C00034649
|
| 30851 | TAX1BP3, TIP-1 | Tax1 (human T-cell leukemia virus type I) binding protein 3 |
C00034649
|
| 23216 | TBC1D1, TBC, TBC1 | TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
C00034649
|
| 55296 | TBC1D19 | TBC1 domain family, member 19 |
C00034649
|
| 55357 | TBC1D2, PARIS-1, PARIS1, TBC1D2A | TBC1 domain family, member 2 |
C00034649
|
| 4943 | TBC1D25, MG81, OATL1 | TBC1 domain family, member 25 |
C00034649
|
| 55171 | TBCCD1 | TBCC domain containing 1 |
C00034649
|
| 6920 | TCEA3, TFIIS, TFIIS.H | transcription elongation factor A (SII), 3 |
C00034649
|
| 6925 | TCF4, E2-2, ITF-2, ITF2, PTHS, SEF-2, SEF2, SEF2-1, SEF2-1A, SEF2-1B, TCF-4, bHLHb19 | transcription factor 4 |
C00034649
|
| 6999 | TDO2, TDO, TO, TPH2, TRPO | tryptophan 2,3-dioxygenase (EC:1.13.11.11) |
C00034649
|
| 7010 | TEK, CD202B, TIE-2, TIE2, VMCM, VMCM1 | TEK tyrosine kinase, endothelial (EC:2.7.10.1) |
C00034649
|
| 26011 | TENM4, Doc4, ODZ4, TNM4, Ten-M4 | teneurin transmembrane protein 4 |
C00034649
|
| 29842 | TFCP2L1, CRTR1, LBP-9, LBP9 | transcription factor CP2-like 1 |
C00034649
|
| 7942 | TFEB, ALPHATFEB, BHLHE35, TCFEB | transcription factor EB |
C00034649
|
| 7980 | TFPI2, PP5, REF1, TFPI-2 | tissue factor pathway inhibitor 2 |
C00034649
|
| 23483 | TGDS, SDR2E1, TDPGD | TDP-glucose 4,6-dehydratase (EC:4.2.1.46) |
C00034649
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00034649
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00034649
|
| 7043 | TGFB3, ARVD, ARVD1, TGF-beta3 | transforming growth factor, beta 3 |
C00034649
|
| 7048 | TGFBR2, AAT3, FAA3, LDS1B, LDS2B, MFS2, RIIC, TAAD2, TGFR-2, TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) (EC:2.7.11.30) |
C00034649
|
| 7052 | TGM2, G-ALPHA-h, GNAH, TG2, TGC | transglutaminase 2 (EC:2.3.2.13) |
C00034649
|
| 7056 | THBD, AHUS6, BDCA3, CD141, THPH12, THRM, TM | thrombomodulin |
C00034649
|
| 7058 | THBS2, TSP2 | thrombospondin 2 |
C00034649
|
| 117145 | THEM4, CTMP | thioesterase superfamily member 4 (EC:3.1.2.2) |
C00034649
|
| 221981 | THSD7A | thrombospondin, type I, domain containing 7A |
C00034649
|
| 55623 | THUMPD1 | THUMP domain containing 1 |
C00034649
|
| 7070 | THY1, CD90 | Thy-1 cell surface antigen |
C00034649
|
| 166815 | TIGD2 | tigger transposable element derived 2 |
C00034649
|
| 7076 | TIMP1, CLGI, EPA, EPO, HCI, TIMP | TIMP metallopeptidase inhibitor 1 |
C00034649
|
| 25976 | TIPARP, ARTD14, PARP7, pART14 | TCDD-inducible poly(ADP-ribose) polymerase (EC:2.4.2.30) |
C00034649
|
| 9414 | TJP2, C9DUPq21.11, DFNA51, DUP9q21.11, X104, ZO2 | tight junction protein 2 |
C00034649
|
| 57707 | TLDC1, KIAA1609 | TBC/LysM-associated domain containing 1 |
C00034649
|
| 7089 | TLE2, ESG, ESG2, GRG2 | transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
C00034649
|
| 7093 | TLL2 | tolloid-like 2 |
C00034649
|
| 7097 | TLR2, CD282, TIL4 | toll-like receptor 2 |
C00034649
|
| 7099 | TLR4, ARMD10, CD284, TLR-4, TOLL | toll-like receptor 4 |
C00034649
|
| 7100 | TLR5, SLEB1, TIL3 | toll-like receptor 5 |
C00034649
|
| 51284 | TLR7, TLR7-like | toll-like receptor 7 |
C00034649
|
| 144404 | TMEM120B | transmembrane protein 120B |
C00034649
|
| 54972 | TMEM132A, GBP, HSPA5BP1 | transmembrane protein 132A |
C00034649
|
| 92293 | TMEM132C | transmembrane protein 132C |
C00034649
|
| 135932 | TMEM139 | transmembrane protein 139 |
C00034649
|
| 25907 | TMEM158, BBP, RIS1, p40BBP | transmembrane protein 158 (gene/pseudogene) |
C00034649
|
| 81615 | TMEM163, DC29, SV31 | transmembrane protein 163 |
C00034649
|
| 114801 | TMEM200A, KIAA1913, TTMA, TTMC | transmembrane protein 200A |
C00034649
|
| 79652 | TMEM204, C16orf30, CLP24 | transmembrane protein 204 |
C00034649
|
| 26175 | TMEM251, C14orf109 | transmembrane protein 251 |
C00034649
|
| 140738 | TMEM37, PR, PR1 | transmembrane protein 37 |
C00034649
|
| 55076 | TMEM45A, DERP7 | transmembrane protein 45A |
C00034649
|
| 83604 | TMEM47, BCMP1, TM4SF10 | transmembrane protein 47 |
C00034649
|
| 199964 | TMEM61 | transmembrane protein 61 |
C00034649
|
| 7111 | TMOD1, D9S57E, ETMOD, TMOD | tropomodulin 1 |
C00034649
|
| 11013 | TMSB15A, TMSB15, TMSB15B, TMSL8, TMSNB, Tb15, TbNB | thymosin beta 15a |
C00034649
|
| 286527 | TMSB15B, Tbeta15b | thymosin beta 15B |
C00034649
|
| 83857 | TMTC1, OLF, TMTC1A | transmembrane and tetratricopeptide repeat containing 1 |
C00034649
|
| 3371 | TNC, 150-225, DFNA56, GMEM, GP, HXB, JI, TN, TN-C | tenascin C |
C00034649
|
| 9818 | NUPL1, PRO2463 | nucleoporin like 1 |
C00000094
|
| 7130 | TNFAIP6, TSG-6, TSG6 | tumor necrosis factor, alpha-induced protein 6 |
C00034649
|
| 25816 | TNFAIP8, GG2-1, MDC-3.13, NDED, SCC-S2, SCCS2 | tumor necrosis factor, alpha-induced protein 8 |
C00034649
|
| 8792 | TNFRSF11A, CD265, FEO, LOH18CR1, ODFR, OFE, OPTB7, OSTS, PDB2, RANK, TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
C00034649
|
| 4982 | TNFRSF11B, OCIF, OPG, TR1 | tumor necrosis factor receptor superfamily, member 11b |
C00034649
|
| 55504 | TNFRSF19, TAJ, TAJ-alpha, TRADE, TROY | tumor necrosis factor receptor superfamily, member 19 |
C00034649
|
| 7133 | TNFRSF1B, CD120b, TBPII, TNF-R-II, TNF-R75, TNFBR, TNFR1B, TNFR2, TNFR80, p75, p75TNFR | tumor necrosis factor receptor superfamily, member 1B |
C00034649
|
| 7293 | TNFRSF4, ACT35, CD134, OX40, TXGP1L | tumor necrosis factor receptor superfamily, member 4 |
C00034649
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00034649
|
| 10673 | TNFSF13B, BAFF, BLYS, CD257, DTL, TALL-1, TALL1, THANK, TNFSF20, ZTNF4 | tumor necrosis factor (ligand) superfamily, member 13b |
C00034649
|
| 7134 | TNNC1, CMD1Z, CMH13, TN-C, TNC, TNNC | troponin C type 1 (slow) |
C00034649
|
| 27324 | TOX3, CAGF9, TNRC9 | TOX high mobility group box family member 3 |
C00034649
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00034649
|
| 9540 | TP53I3, PIG3 | tumor protein p53 inducible protein 3 |
C00034649
|
| 8626 | TP63, AIS, B(p51A), B(p51B), EEC3, KET, LMS, NBP, OFC8, RHS, SHFM4, TP53CP, TP53L, TP73L, p40, p51, p53CP, p63, p73H, p73L | tumor protein p63 |
C00034649
|
| 7162 | TPBG, 5T4, 5T4AG, M6P1 | trophoblast glycoprotein |
C00034649
|
| 29896 | TRA2A, AWMS1, HSU53209 | transformer 2 alpha homolog (Drosophila) |
C00034649
|
| 28639 |
C00034649
|
||
| 28951 | TRIB2, C5FW, GS3955, TRB2 | tribbles pseudokinase 2 |
C00034649
|
| 57761 | TRIB3, C20orf97, NIPK, SINK, SKIP3, TRB3 | tribbles pseudokinase 3 |
C00034649
|
| 10626 | TRIM16, EBBP | tripartite motif containing 16 |
C00034649
|
| 388610 | TRNP1, C1orf225, TNRP | TMF1-regulated nuclear protein 1 |
C00034649
|
| 7223 | TRPC4, HTRP-4, HTRP4, TRP4 | transient receptor potential cation channel, subfamily C, member 4 |
C00034649
|
| 7225 | TRPC6, FSGS2, TRP6 | transient receptor potential cation channel, subfamily C, member 6 |
C00034649
|
| 54795 | TRPM4, PFHB1B, TRPM4B | transient receptor potential cation channel, subfamily M, member 4 |
C00034649
|
| 85480 | TSLP | thymic stromal lymphopoietin |
C00034649
|
| 441631 | TSPAN11, VSSW1971 | tetraspanin 11 |
C00034649
|
| 23554 | TSPAN12, EVR5, NET-2, NET2, TM4SF12 | tetraspanin 12 |
C00034649
|
| 92104 | TTC30A | tetratricopeptide repeat domain 30A |
C00034649
|
| 23170 | TTLL12, dJ526I14.2 | tubulin tyrosine ligase-like family, member 12 |
C00034649
|
| 347733 | TUBB2B, PMGYSA, bA506K6.1 | tubulin, beta 2B class IIb |
C00034649
|
| 10628 | TXNIP, EST01027, HHCPA78, THIF, VDUP1 | thioredoxin interacting protein |
C00034649
|
| 84959 | UBASH3B, STS-1, STS1, TULA-2, p70 | ubiquitin associated and SH3 domain containing B (EC:3.1.3.48) |
C00034649
|
| 10537 | UBD, FAT10, GABBR1, UBD-3 | ubiquitin D |
C00034649
|
| 8266 | UBL4A, DX254E, DXS254E, G6PD, GDX, GET5, MDY2, TMA24, UBL4 | ubiquitin-like 4A |
C00034649
|
| 652995 | UCA1, CUDR, LINC00178, NCRNA00178, UCAT1 | urothelial cancer associated 1 (non-protein coding) |
C00034649
|
| 7357 | UGCG, GCS, GLCT1 | UDP-glucose ceramide glucosyltransferase (EC:2.4.1.80) |
C00034649
|
| 54986 | ULK4, FAM7C1, REC01035 | unc-51 like kinase 4 (EC:2.7.11.1) |
C00034649
|
| 7378 | UPP1, UDRPASE, UP, UPASE, UPP | uridine phosphorylase 1 (EC:2.4.2.3) |
C00034649
|
| 8615 | USO1, P115, TAP, VDP | USO1 vesicle transport factor |
C00034649
|
| 54532 | USP53 | ubiquitin specific peptidase 53 |
C00034649
|
| 22846 | VASH1, KIAA1036 | vasohibin 1 |
C00034649
|
| 7408 | VASP | vasodilator-stimulated phosphoprotein |
C00034649
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00034649
|
| 1462 | VCAN, CSPG2, ERVR, GHAP, PG-M, WGN, WGN1 | versican |
C00034649
|
| 7432 | VIP, PHM27 | vasoactive intestinal peptide |
C00034649
|
| 8876 | VNN1, HDLCQ8, Tiff66 | vanin 1 (EC:3.5.1.92) |
C00034649
|
| 196740 | VSTM4, C10orf72 | V-set and transmembrane domain containing 4 |
C00034649
|
| 79679 | VTCN1, B7-H4, B7H4, B7S1, B7X, B7h.5, PRO1291, VCTN1 | V-set domain containing T cell activation inhibitor 1 |
C00034649
|
| 7448 | VTN, V75, VN, VNT | vitronectin |
C00034649
|
| 7450 | VWF, F8VWF, VWD | von Willebrand factor |
C00034649
|
| 7454 | WAS, IMD2, SCNX, THC, THC1, WASP | Wiskott-Aldrich syndrome |
C00034649
|
| 10885 | WDR3, DIP2, UTP12 | WD repeat domain 3 |
C00034649
|
| 256764 | WDR72, AI2A3 | WD repeat domain 72 |
C00034649
|
| 79084 | WDR77, MEP-50, MEP50, Nbla10071, RP11-552M11.3, p44, p44/Mep50 | WD repeat domain 77 |
C00034649
|
| 10406 | WFDC2, EDDM4, HE4, WAP5, dJ461P17.6 | WAP four-disulfide core domain 2 |
C00034649
|
| 79971 | WLS, C1orf139, EVI, GPR177, MRP, mig-14 | wntless Wnt ligand secretion mediator |
C00034649
|
| 51384 | WNT16 | wingless-type MMTV integration site family, member 16 |
C00034649
|
| 54361 | WNT4, SERKAL, WNT-4 | wingless-type MMTV integration site family, member 4 |
C00034649
|
| 7474 | WNT5A, hWNT5A | wingless-type MMTV integration site family, member 5A |
C00034649
|
| 81029 | WNT5B | wingless-type MMTV integration site family, member 5B |
C00034649
|
| 7490 | WT1, AWT1, EWS-WT1, GUD, NPHS4, WAGR, WIT-2, WT33 | Wilms tumor 1 |
C00034649
|
| 7494 | XBP1, TREB5, XBP-1, XBP2 | X-box binding protein 1 |
C00034649
|
| 6375 | XCL1, ATAC, LPTN, LTN, SCM-1, SCM-1a, SCM1, SCM1A, SCYC1 | chemokine (C motif) ligand 1 |
C00034649
|
| 6846 | XCL2, SCM-1b, SCM1B, SCYC2 | chemokine (C motif) ligand 2 |
C00034649
|
| 10681 | GNB5, GB5 | guanine nucleotide binding protein (G protein), beta 5 |
C00000094
|
| 7512 | XPNPEP2, APP2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (EC:3.4.11.9) |
C00034649
|
| 7518 | XRCC4 | X-ray repair complementing defective repair in Chinese hamster cells 4 |
C00034649
|
| 64848 | YTHDC2, CAHL | YTH domain containing 2 (EC:3.6.4.13) |
C00034649
|
| 100301855 |
C00034649
|
||
| 79413 | ZBED2 | zinc finger, BED-type containing 2 |
C00034649
|
| 7704 | ZBTB16, PLZF, ZNF145 | zinc finger and BTB domain containing 16 |
C00034649
|
| 23174 | ZCCHC14, BDG-29, BDG29 | zinc finger, CCHC domain containing 14 |
C00034649
|
| 644353 | ZCCHC18, PNMA7B, SIZN2 | zinc finger, CCHC domain containing 18 |
C00034649
|
| 79670 | ZCCHC6, PAPD6 | zinc finger, CCHC domain containing 6 (EC:2.7.7.52) |
C00034649
|
| 79844 | ZDHHC11, ZNF399 | zinc finger, DHHC-type containing 11 (EC:2.3.1.225) |
C00034649
|
| 6935 | ZEB1, AREB6, BZP, DELTAEF1, FECD6, NIL2A, PPCD3, TCF8, ZFHEP, ZFHX1A | zinc finger E-box binding homeobox 1 |
C00034649
|
| 7538 | ZFP36, G0S24, GOS24, NUP475, RNF162A, TIS11, TTP, zfp-36 | ZFP36 ring finger protein |
C00034649
|
| 677 | ZFP36L1, BRF1, Berg36, ERF-1, ERF1, RNF162B, TIS11B, cMG1 | ZFP36 ring finger protein-like 1 |
C00034649
|
| 51663 | ZFR, ZFR1 | zinc finger RNA binding protein |
C00034649
|
| 83637 | ZMIZ2, NET27, TRAFIP20, ZIMP7, hZIMP7 | zinc finger, MIZ-type containing 2 |
C00034649
|
| 10795 | ZNF268, HZF3 | zinc finger protein 268 |
C00034649
|
| 91975 | ZNF300 | zinc finger protein 300 |
C00034649
|
| 167465 | ZNF366, DCSCRIPT | zinc finger protein 366 |
C00034649
|
| 79088 | ZNF426 | zinc finger protein 426 |
C00034649
|
| 126299 | ZNF428, C19orf37, Zfp428 | zinc finger protein 428 |
C00034649
|
| 90649 | ZNF486, KRBO2 | zinc finger protein 486 |
C00034649
|
| 25925 | ZNF521, EHZF, Evi3 | zinc finger protein 521 |
C00034649
|
| 55205 | ZNF532 | zinc finger protein 532 |
C00034649
|
| 51385 | ZNF589, SZF1 | zinc finger protein 589 |
C00034649
|
| 22834 | ZNF652 | zinc finger protein 652 |
C00034649
|
| 7552 | ZNF711, CMPX1, MRX97, ZNF4, ZNF5, ZNF6, Zfp711, dJ75N13.1 | zinc finger protein 711 |
C00034649
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00001429
|
| 2597 | GAPDH, G3PD, GAPD | glyceraldehyde-3-phosphate dehydrogenase (EC:1.2.1.12) |
C00001429
|
| 3352 | HTR1D, 5-HT1D, HT1DA, HTR1DA, HTRL, RDC4 | 5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
C00001429
|
| 3359 | HTR3A, 5-HT-3, 5-HT3A, 5-HT3R, 5HT3R, HTR3 | 5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
C00001429
|
| 9177 | HTR3B, 5-HT3B | 5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
C00001429
|
| 3363 | HTR7, 5-HT7 | 5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
C00001429
|
| 85413 | SLC22A16, CT2, FLIPT2, OAT6, OCT6, OKB1, dJ261K5.1 | solute carrier family 22 (organic cation/carnitine transporter), member 16 |
C00001429
|
| 6814 | STXBP3, MUNC18-3, MUNC18C, PSP, UNC-18C | syntaxin binding protein 3 |
C00001429
|
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00001429
|
| 7166 | TPH1, TPRH, TRPH, TPH | tryptophan hydroxylase 1 (EC:1.14.16.4) |
C00001429
|
| 8989 | TRPA1, ANKTM1, FEPS | transient receptor potential cation channel, subfamily A, member 1 |
C00001429
|
| 54578 | UGT1A6, GNT1, HLUGP, HLUGP1, UDPGT, UDPGT_1-6, UGT1, UGT1A6S, UGT1F | UDP glucuronosyltransferase 1 family, polypeptide A6 (EC:2.4.1.17) |
C00001429
|
| 31 | ACACA, ACAC, ACACAD, ACC, ACC1, ACCA | acetyl-CoA carboxylase alpha (EC:6.4.1.2 6.3.4.14) |
C00001200
|
| 1488 | CTBP2 | C-terminal binding protein 2 |
C00001200
|
| 14103 |
C00001200
|
||
| 6566 | SLC16A1, HHF7, MCT, MCT1 | solute carrier family 16 (monocarboxylate transporter), member 1 |
C00001200
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00001200
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (196)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #614279 | 46,xy sex reversal 8; srxy8 |
P17516
P52895 |
| #100100 | Abdominal muscles, absence of, with urinary tract abnormality and cryptorchidism |
P20309
|
| #219080 | Acth-independent macronodular adrenal hyperplasia; aimah |
P63092
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #103780 | Alcohol dependence |
P08172
P14416 P31645 P47869 |
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #614373 | Amyotrophic lateral sclerosis 16, juvenile; als16 |
Q99720
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #207410 | Antley-bixler syndrome without genital anomalies or disordered steroidogenesis; abs2 |
P21802
|
| #101200 | Apert syndrome |
P21802
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #600807 | Asthma, susceptibility to |
Q13093
|
| #608584 | Asthma-related traits, susceptibility to, 2 |
Q6W5P4
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #123790 | Beare-stevenson cutis gyrata syndrome; bstvs |
P21802
|
| #614592 | Bent bone dysplasia syndrome; bbds |
P21802
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #614490 | Blood group, junior system; jr |
Q9UNQ0
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #602025 | Body mass index quantitative trait locus 9; bmiq9 |
P41968
|
| %606641 | Body mass index; bmi |
P37231
|
| #612560 | Bone mineral density quantitative trait locus 12; bmnd12 |
O75795
|
| #114480 | Breast cancer |
P38398
|
| #604370 | Breast-ovarian cancer, familial, susceptibility to, 1; brovca1 |
P38398
|
| #300615 | Brunner syndrome |
P21397
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #605479 | Cholestasis, benign recurrent intrahepatic, 2; bric2 |
O95342
|
| #601847 | Cholestasis, progressive familial intrahepatic, 2; pfic2 |
O95342
|
| #302960 | Chondrodysplasia punctata 2, x-linked dominant; cdpx2 |
Q15125
|
| #114500 | Colorectal cancer; crc |
P84022
Q14191 |
| #611489 | Corticosteroid-binding globulin deficiency |
P08185
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #123500 | Crouzon syndrome |
P21802
|
| #162800 | Cyclic neutropenia |
P08246
|
| #612522 | Diabetes mellitus, insulin-dependent, 22; iddm22 |
P51681
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #607208 | Dravet syndrome |
P18507
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #237500 | Dubin-johnson syndrome; djs |
Q92887
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #607681 | Epilepsy, childhood absence, susceptibility to, 2; eca2 |
P18507
|
| #612269 | Epilepsy, childhood absence, susceptibility to, 5; eca5 |
P28472
|
| #613060 | Epilepsy, idiopathic generalized, susceptibility to, 10; eig10 |
O14764
|
| #611136 | Epilepsy, juvenile myoclonic, susceptibility to, 5; ejm5 |
P14867
|
| #612656 | Episodic ataxia, type 6; ea6 |
P43003
|
| #609820 | Erythrocytosis, familial, 3; ecyt3 |
Q9GZT9
|
| #133239 | Esophageal cancer |
P04637
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #612219 | Ewing sarcoma; es |
P11308
|
| #301500 | Fabry disease |
P06280
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613659 | Gastric cancer |
P04626
|
| #137215 | Gastric cancer, hereditary diffuse; hdgc |
P04626
|
| #606764 | Gastrointestinal stromal tumor; gist |
P10721
|
| #604233 | Generalized epilepsy with febrile seizures plus, type 1; gefsp1 |
P18507
|
| #611277 | Generalized epilepsy with febrile seizures plus, type 3; gefsp3 |
P18507
|
| #231095 | Ghosal hematodiaphyseal dysplasia; ghdd |
P24557
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #137800 | Glioma susceptibility 1; glm1 |
O75874
P04626 P37231 |
| #606824 | Glucose/galactose malabsorption; ggm |
P13866
|
| #232300 | Glycogen storage disease ii |
P10253
|
| #612938 | Growth retardation, developmental delay, coarse facies, and early death; gdfd |
Q9C0B1
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #602089 | Hemangioma, capillary infantile |
P35968
|
| #114550 | Hepatocellular carcinoma |
P08581
|
| #609423 | Human immunodeficiency virus type 1, susceptibility to |
P41597
P51681 |
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237450 | Hyperbilirubinemia, rotor type; hblrr |
Q9NPD5
Q9Y6L6 |
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #149400 | Hyperekplexia, hereditary 1; hkpx1 |
P23415
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #605115 | Hypertension, early-onset, autosomal dominant, with severe exacerbation in pregnancy |
P08235
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #614441 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 2; phoar2 |
Q92959
|
| #147950 | Hypogonadotropic hypogonadism 2 with or without anosmia; hh2 |
P11362
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #147050 | Ige responsiveness, atopic; iger |
Q13093
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #123150 | Jackson-weiss syndrome; jws |
P21802
|
| #607785 | Juvenile myelomonocytic leukemia; jmml |
Q06124
|
| #149730 | Lacrimoauriculodentodigital syndrome; ladd |
P21802
|
| #151100 | Leopard syndrome 1 |
Q06124
|
| #601626 | Leukemia, acute myeloid; aml |
P10721
|
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #211980 | Lung cancer |
P00533
P04626 P04637 |
| #608516 | Major depressive disorder; mdd |
P08172
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #174800 | Mccune-albright syndrome; mas |
P63092
|
| #613839 | Megaloblastic anemia due to dihydrofolate reductase deficiency |
P00374
|
| #611092 | Mental retardation, autosomal recessive 6; mrt6 |
Q13002
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| %300852 | Mental retardation, x-linked 88; mrx88 |
P50052
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #300699 | Mental retardation, x-linked, syndromic, wu type; mrxsw |
P42263
|
| #156250 | Metachondromatosis; metcds |
Q06124
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #612633 | Microvascular complications of diabetes, susceptibility to, 5; mvcd5 |
P27169
|
| #601492 | Mucopolysaccharidosis, type ix; mps9 |
Q12794
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #126200 | Multiple sclerosis, susceptibility to; ms |
P08575
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #254600 | Myeloperoxidase deficiency; mpod |
P05164
|
| #159900 | Myoclonic dystonia |
P14416
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #202700 | Neutropenia, severe congenital, 1, autosomal dominant; scn1 |
P08246
|
| #257200 | Niemann-pick disease, type a |
P17405
|
| #607616 | Niemann-pick disease, type b |
P17405
|
| #257270 | Night blindness, congenital stationary, type 1b; csnb1b |
O15303
|
| #163950 | Noonan syndrome 1; ns1 |
Q06124
|
| #601665 | Obesity |
P32245
P37231 |
| #164230 | Obsessive-compulsive disorder; ocd |
P31645
|
| #613705 | Orofacial cleft 10; ofc10 |
P63165
|
| #604715 | Orthostatic intolerance |
P23975
|
| #166350 | Osseous heteroplasia, progressive; poh |
P63092
|
| #166250 | Osteoglophonic dysplasia; ogd |
P11362
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #167000 | Ovarian cancer |
P04626
P38398 |
| #614320 | Pancreatic cancer, susceptibility to, 4; pnca4 |
P38398
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #613135 | Parkinsonism-dystonia, infantile; pkdys |
Q01959
|
| #614674 | Periodic fever, menstrual cycle-dependent |
P08908
|
| #101600 | Pfeiffer syndrome |
P11362
P21802 |
| #172700 | Pick disease of brain |
P10636
|
| #172800 | Piebald trait; pbt |
P10721
|
| #102200 | Pituitary adenoma, growth hormone-secreting |
P63092
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #614278 | Platelet-activating factor acetylhydrolase deficiency; pafad |
Q13093
|
| #177735 | Pseudohypoaldosteronism, type i, autosomal dominant; pha1a |
P08235
|
| #103580 | Pseudohypoparathyroidism, type ia; php1a |
P63092
|
| #603233 | Pseudohypoparathyroidism, type ib; php1b |
P63092
|
| #612462 | Pseudohypoparathyroidism, type ic; php1c |
P63092
|
| #605074 | Renal cell carcinoma, papillary, 1; rccp1 |
P08581
|
| #233100 | Renal glucosuria; glys1 |
P31639
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #609579 | Scaphocephaly, maxillary retrusion, and mental retardation |
P21802
|
| #615232 | Schizophrenia 18; sczd18 |
P43005
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #608971 | Severe combined immunodeficiency, autosomal recessive, t cell-negative, b cell-positive, nk cell-positive |
P08575
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #614831 | Spinocerebellar ataxia, autosomal recessive 13; scar13 |
Q13255
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
P04637
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #273300 | Testicular germ cell tumor; tgct |
P10721
|
| #187950 | Thrombocythemia 1; thcyt1 |
P40225
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #190300 | Tremor, hereditary essential, 1; etm1 |
P35462
|
| #190440 | Trigonocephaly 1; trigno1 |
P11362
|
| #138900 | Uric acid concentration, serum, quantitative trait locus 1; uaqtl1 |
Q9UNQ0
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #277700 | Werner syndrome; wrn |
Q14191
|
| #610379 | West nile virus, susceptibility to |
P51681
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (143)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00783 | Febrile seizures |
O14764
(related)
P18507 (related) |
| H00787 | Congenital stationary night blindness (CSNB) |
O15303
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
P04637 (marker) P08581 (related) Q16790 (marker) |
| H00624 | Familial cholestasis |
O95342
(related)
|
| H01197 | Dihydrofolate reductase (DHFR) deficiency |
P00374
(related)
|
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P04637 (related) P04637 (marker) P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
P04626 (related) P04637 (related) P08581 (related) P21802 (related) |
| H00022 | Bladder cancer |
P00533
(related)
P04626 (related) P04637 (related) |
| H00028 | Choriocarcinoma |
P00533
(related)
P03956 (related) P04626 (related) P04637 (related) P08253 (related) |
| H00030 | Cervical cancer |
P00533
(related)
P04626 (related) |
| H00042 | Glioma |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) |
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
Q01453 (related) |
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
P37231 (related) |
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04626 (related) P04637 (related) P06401 (marker) Q92731 (marker) |
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P04150
(related)
P11511 (related) |
| H00019 | Pancreatic cancer |
P04626
(related)
P04637 (related) P04637 (marker) |
| H00027 | Ovarian cancer |
P04626
(related)
P04637 (related) P38398 (related) |
| H00031 | Breast cancer |
P04626
(related)
P04626 (marker) P04637 (related) P38398 (related) |
| H00046 | Cholangiocarcinoma |
P04626
(related)
P04637 (related) P08581 (related) P35354 (related) |
| H00004 | Chronic myeloid leukemia (CML) |
P04637
(related)
Q01196 (related) |
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
P28907 (marker) |
| H00006 | Hairy-cell leukemia |
P04637
(related)
|
| H00008 | Burkitt lymphoma |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00010 | Multiple myeloma |
P04637
(related)
|
| H00013 | Small cell lung cancer |
P04637
(related)
|
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) |
| H00025 | Penile cancer |
P04637
(related)
P04637 (marker) P08253 (related) P14780 (related) P35354 (related) |
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00032 | Thyroid cancer |
P04637
(related)
P37231 (related) |
| H00036 | Osteosarcoma |
P04637
(related)
P08684 (marker) |
| H00038 | Malignant melanoma |
P04637
(related)
P14679 (marker) |
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00041 | Kaposi's sarcoma |
P04637
(related)
|
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00047 | Gallbladder cancer |
P04637
(related)
|
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00101 | Other phagocyte defects |
P05164
(related)
|
| H00003 | Acute myeloid leukemia (AML) |
P05164
(marker)
P10721 (related) P10721 (marker) Q01196 (related) Q01196 (marker) Q13951 (marker) |
| H00093 | Combined immunodeficiencies (CIDs) |
P06239
(related)
|
| H00125 | Fabry disease |
P06280
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H01163 | Corticosteroid-binding globulin (CBG) deficiency |
P08185
(related)
|
| H00243 | Hyperkalemic distal renal tubular acidosis (RTA type 4) |
P08235
(related)
|
| H00603 | Hypertension exacerbated in pregnancy |
P08235
(related)
|
| H00100 | Neutropenic disorders |
P08246
(related)
|
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00091 | T-B+Severe combined immunodeficiencies (SCIDs) |
P08575
(related)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00024 | Prostate cancer |
P10275
(related)
P11308 (related) |
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
Q13148 (related) |
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00170 | Piebaldism |
P10721
(related)
|
| H00023 | Testicular cancer |
P10721
(marker)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00035 | Ewing's sarcoma |
P11308
(related)
|
| H00255 | Hypogonadotropic hypogonadism |
P11362
(related)
|
| H00443 | Osteoglophonic dysplasia (OD) |
P11362
(related)
|
| H00458 | Craniosynostosis |
P11362
(related)
P21802 (related) |
| H00516 | Isolated orofacial clefts |
P11362
(related)
P63165 (related) |
| H01207 | Trigonocephaly |
P11362
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H01261 | Congenital glucose-galactose malabsorption (GGM) |
P13866
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
|
| H00808 | Idiopathic generalized epilepsies (IGEs) |
P14867
(related)
P18507 (related) |
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00137 | Niemann-Pick disease (NPD) typeA and B |
P17405
(related)
|
| H00424 | Defects in the degradation of sphingomyelin |
P17405
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H00642 | Lacrimo-auriculo-dento-digital syndrome (LADD) |
P21802
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
Q92887 (related) |
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H00769 | Hyperekplexia |
P23415
(related)
|
| H01031 | Orthostatic intolerance (OI) |
P23975
(related)
|
| H00490 | Diaphyseal dysplasia with anemia (Ghosal) |
P24557
(related)
|
| H01126 | Familial renal glucosuria (FRG) |
P31639
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
P49286 (related) Q9C0B1 (related) |
| H00227 | Congenital amegakaryocytic thrombocytopenia (CAMT) |
P40225
(marker)
|
| H00577 | Syndromic X-linked mental retardation with epilepsy or seizures |
P42263
(related)
|
| H00749 | Episodic ataxias |
P43003
(related)
|
| H00911 | Dicarboxylic aminoaciduria |
P43005
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
P50052
(related)
Q99714 (related) |
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
Q14191 (related) |
| H00244 | Pseudohypoparathyroidism |
P63092
(related)
|
| H00441 | Progressive osseous heteroplasia (POH) |
P63092
(related)
|
| H00501 | Fibrous dysplasia, polyostotic |
P63092
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
| H00523 | Noonan syndrome and related disorders |
Q06124
(related)
|
| H01018 | Metachondromatosis |
Q06124
(related)
|
| H00133 | Mucopolysaccharidosis type IX (MPS9) |
Q12794
(related)
|
| H00421 | Mucopolysaccharidosis (MPS) |
Q12794
(related)
|
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|
| H00768 | Nonsyndromic autosomal recessive mental retardation (NS-ARMR) |
Q13002
(related)
|
| H01194 | X-linked chondrodysplasia punctata |
Q15125
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
Q99700
(related)
Q9NUW8 (related) |
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00926 | Growth retardation, developmental delay, coarse facies, and early death |
Q9C0B1
(related)
|
| H00236 | Congenital polycythemia |
Q9GZT9
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
203 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D006973 | Hypertension |
C00001424
C00034649 C00001429 C00001435 |
| D009336 | Necrosis |
C00001424
C00001429 C00001200 |
| D002543 | Cerebral Hemorrhage |
C00001424
C00034649 C00001435 |
| D002375 | Catalepsy |
C00001424
C00034649 C00001429 |
| D007022 | Hypotension |
C00001424
C00001429 C00001435 |
| D013610 | Tachycardia |
C00001424
C00001429 C00001435 |
| D010146 | Pain |
C00001424
C00034649 C00001429 |
| D001008 | Anxiety Disorders |
C00001424
C00034649 C00001429 |
| D012640 | Seizures |
C00001424
C00034649 C00001429 |
| D013226 | Status Epilepticus |
C00001424
C00034649 C00001429 |
| D001321 | Autistic Disorder |
C00001424
C00001429 |
| D056486 | Drug-Induced Liver Injury |
C00000100
C00034649 |
| D000647 | Amnesia |
C00001424
C00000091 |
| D001145 | Arrhythmias, Cardiac |
C00001424
C00034649 |
| D004195 | Disease Models, Animal |
C00034649
C00001429 |
| D013375 | Substance Withdrawal Syndrome |
C00001424
C00034649 |
| D009410 | Nerve Degeneration |
C00001424
C00034649 |
| D001919 | Bradycardia |
C00001424
C00001429 |
| D007249 | Inflammation |
C00034649
C00001429 |
| D007674 | Kidney Diseases |
C00001424
C00034649 |
| D028361 | Mitochondrial Diseases |
C00034649
C00001200 |
| D017379 | Hypertrophy, Left Ventricular |
C00001424
C00001429 |
| D006984 | Hypertrophy |
C00001424
C00034649 |
| D009202 | Cardiomyopathies |
C00001424
C00001435 |
| D009422 | Nervous System Diseases |
C00034649
C00001429 |
| D020521 | Stroke |
C00034649
C00001429 |
| D019970 | Cocaine-Related Disorders |
C00001424
C00034649 |
| D006976 | Hypertension, Pulmonary |
C00001424
C00001429 |
| D004409 | Dyskinesia, Drug-Induced |
C00001424
C00034649 |
| D009135 | Muscular Diseases |
C00001429
C00001200 |
| D006930 | Hyperalgesia |
C00001424
C00001429 |
| D000544 | Alzheimer Disease |
C00001429
|
| D005921 | Glomerulonephritis |
C00001424
|
| D005923 | Glomerulosclerosis, Focal Segmental |
C00001424
|
| D006261 | Headache |
C00001424
|
| D006323 | Heart Arrest |
C00001424
|
| D006331 | Heart Diseases |
C00001424
|
| D006333 | Heart Failure |
C00001424
|
| D006470 | Hemorrhage |
C00001424
|
| D004827 | Epilepsy |
C00001424
|
| D006935 | Hypercapnia |
C00001424
|
| D006940 | Hyperemia |
C00001424
|
| D004421 | Dystonia |
C00001424
|
| D006974 | Hypertension, Malignant |
C00001424
|
| D003922 | Diabetes Mellitus, Type 1 |
C00001424
|
| D006332 | Cardiomegaly |
C00001424
|
| D002303 | Cardiac Output, Low |
C00001424
|
| D007008 | Hypokalemia |
C00001424
|
| D002100 | Cachexia |
C00001424
|
| D007024 | Hypotension, Orthostatic |
C00001424
|
| D007035 | Hypothermia |
C00001424
|
| D007511 | Ischemia |
C00001424
|
| D002037 | Bundle-Branch Block |
C00001424
|
| D008113 | Liver Neoplasms |
C00001424
|
| D008114 | Liver Neoplasms, Experimental |
C00001424
|
| D009203 | Myocardial Infarction |
C00001424
|
| D017202 | Myocardial Ischemia |
C00001424
|
| D015428 | Myocardial Reperfusion Injury |
C00001424
|
| D002545 | Brain Ischemia |
C00001424
|
| D001766 | Blindness |
C00001424
|
| D009798 | Ocular Hypertension |
C00001424
|
| D009846 | Oliguria |
C00001424
|
| D009901 | Optic Nerve Diseases |
C00001424
|
| D001237 | Asphyxia |
C00001424
|
| D010243 | Paralysis |
C00001424
|
| D010554 | Personality Disorders |
C00001424
|
| D011183 | Postoperative Complications |
C00001424
|
| D011249 | Pregnancy Complications, Cardiovascular |
C00001424
|
| D011297 | Prenatal Exposure Delayed Effects |
C00001424
|
| D011507 | Proteinuria |
C00001424
|
| D011605 | Psychoses, Substance-Induced |
C00001424
|
| D029424 | Pulmonary Disease, Chronic Obstructive |
C00001424
|
| D051437 | Renal Insufficiency |
C00001424
|
| D012120 | Respiration Disorders |
C00001424
|
| D012131 | Respiratory Insufficiency |
C00001424
|
| D012164 | Retinal Diseases |
C00001424
|
| D012206 | Rhabdomyolysis |
C00001424
|
| D001002 | Anuria |
C00001424
|
| D018805 | Sepsis |
C00001424
|
| D012769 | Shock |
C00001424
|
| D012770 | Shock, Cardiogenic |
C00001424
|
| D012771 | Shock, Hemorrhagic |
C00001424
|
| D012772 | Shock, Septic |
C00001424
|
| D013035 | Spasm |
C00001424
|
| D000860 | Anoxia |
C00001424
|
| D013274 | Stomach Neoplasms |
C00001424
|
| D013345 | Subarachnoid Hemorrhage |
C00001424
|
| D017542 | Aneurysm, Ruptured |
C00001424
|
| D019969 | Amphetamine-Related Disorders |
C00001424
|
| D013616 | Tachycardia, Sinus |
C00001424
|
| D017180 | Tachycardia, Ventricular |
C00001424
|
| D014511 | Uremia |
C00001424
|
| D056987 | Vasoplegia |
C00001424
|
| D018754 | Ventricular Dysfunction |
C00001424
|
| D018487 | Ventricular Dysfunction, Left |
C00001424
|
| D014693 | Ventricular Fibrillation |
C00001424
|
| D000034 | Abortion, Veterinary |
C00034649
|
| D000230 | Adenocarcinoma |
C00034649
|
| D000756 | Anemia, Sideroblastic |
C00034649
|
| D000799 | Angioedema |
C00034649
|
| D054179 | Angioedemas, Hereditary |
C00034649
|
| D000855 | Anorexia |
C00034649
|
| D001024 | Aortic Valve Stenosis |
C00034649
|
| D020201 | Brain Hemorrhage, Traumatic |
C00034649
|
| D001930 | Brain Injuries |
C00034649
|
| D001943 | Breast Neoplasms |
C00034649
|
| D018275 | Carcinoma, Lobular |
C00034649
|
| D002318 | Cardiovascular Diseases |
C00034649
|
| D002341 | Carotid Artery Thrombosis |
C00034649
|
| D002493 | Central Nervous System Diseases |
C00034649
|
| D002544 | Cerebral Infarction |
C00034649
|
| D002561 | Cerebrovascular Disorders |
C00034649
|
| D002769 | Cholelithiasis |
C00034649
|
| D002779 | Cholestasis |
C00034649
|
| D003704 | Dementia |
C00034649
|
| D003711 | Demyelinating Diseases |
C00034649
|
| D003866 | Depressive Disorder |
C00034649
|
| D003929 | Diabetic Neuropathies |
C00034649
|
| D058186 | Acute Kidney Injury |
C00001424
|
| D012734 | Disorders of Sex Development |
C00034649
|
| D004211 | Disseminated Intravascular Coagulation |
C00034649
|
| D004244 | Dizziness |
C00034649
|
| D004342 | Drug Hypersensitivity |
C00034649
|
| D020964 | Embryo Loss |
C00034649
|
| D004681 | Encephalomyelitis, Autoimmune, Experimental |
C00034649
|
| D016889 | Endometrial Neoplasms |
C00034649
|
| D004715 | Endometriosis |
C00034649
|
| D004832 | Epilepsy, Absence |
C00034649
|
| D005313 | Fetal Death |
C00034649
|
| D005348 | Fibrocystic Breast Disease |
C00034649
|
| D005705 | Gallbladder Diseases |
C00034649
|
| D005911 | Gliosis |
C00034649
|
| D018149 | Glucose Intolerance |
C00034649
|
| D006059 | Gonadal Dysgenesis |
C00034649
|
| D006330 | Heart Defects, Congenital |
C00034649
|
| D006345 | Heart Septal Defects, Ventricular |
C00034649
|
| D006606 | Hiccup |
C00034649
|
| D019584 | Hot Flashes |
C00034649
|
| D006937 | Hypercholesterolemia |
C00034649
|
| D006966 | Hyperprolactinemia |
C00034649
|
| D006968 | Hypersensitivity, Delayed |
C00034649
|
| D007018 | Hypopituitarism |
C00034649
|
| D020244 | Infarction, Middle Cerebral Artery |
C00034649
|
| D007247 | Infertility, Female |
C00034649
|
| D011471 | Prostatic Neoplasms |
C00002514
|
| D007889 | Leiomyoma |
C00034649
|
| D008105 | Liver Cirrhosis, Biliary |
C00034649
|
| D008106 | Liver Cirrhosis, Experimental |
C00034649
|
| D008325 | Mammary Neoplasms, Experimental |
C00034649
|
| D008413 | Mastitis |
C00034649
|
| D059373 | Mastodynia |
C00034649
|
| D001523 | Mental Disorders |
C00034649
|
| D008796 | Metrorrhagia |
C00034649
|
| D007248 | Infertility, Male |
C00002514
|
| D016472 | Motor Neuron Disease |
C00034649
|
| D009220 | Myositis |
C00034649
|
| D009120 | Muscle Cramp |
C00000977
|
| D009440 | Neurasthenia |
C00034649
|
| D020258 | Neurotoxicity Syndromes |
C00034649
|
| D007752 | Obstetric Labor, Premature |
C00034649
|
| D010048 | Ovarian Cysts |
C00034649
|
| D010051 | Ovarian Neoplasms |
C00034649
|
| D016584 | Panic Disorder |
C00034649
|
| D010523 | Peripheral Nervous System Diseases |
C00034649
|
| D011085 | Polycystic Ovary Syndrome |
C00034649
|
| D011248 | Pregnancy Complications |
C00034649
|
| D047928 | Premature Birth |
C00034649
|
| D012468 | Salivary Gland Neoplasms |
C00034649
|
| D012851 | Sinus Thrombosis, Intracranial |
C00034649
|
| D013119 | Spinal Cord Injuries |
C00034649
|
| D008104 | Liver Cirrhosis, Alcoholic |
C00001055
|
| D013927 | Thrombosis |
C00034649
|
| D014591 | Uterine Diseases |
C00034649
|
| D014625 | Vaginal Neoplasms |
C00034649
|
| D054556 | Venous Thromboembolism |
C00034649
|
| D020246 | VENOUS THROMBOSIS |
C00034649
|
| D005901 | Glaucoma |
C00001424
|
| D000648 | Amnesia, Retrograde |
C00001429
|
| D001022 | Aortic Valve Insufficiency |
C00001429
|
| D001049 | Apnea |
C00001429
|
| D001281 | Atrial Fibrillation |
C00001429
|
| D002389 | Catatonia |
C00001429
|
| D002659 | Child Development Disorders, Pervasive |
C00001429
|
| D021081 | Chronobiology Disorders |
C00001429
|
| D015140 | Dementia, Vascular |
C00001429
|
| D003967 | Diarrhea |
C00001429
|
| D004487 | Edema |
C00001429
|
| D005355 | Fibrosis |
C00001429
|
| D005483 | Flushing |
C00001429
|
| D006349 | Heart Valve Diseases |
C00001429
|
| D043183 | Irritable Bowel Syndrome |
C00001429
|
| D008944 | Mitral Valve Insufficiency |
C00001429
|
| D002471 | Cell Transformation, Neoplastic |
C00001183
|
| D011537 | Pruritus |
C00001429
|
| D020230 | Serotonin Syndrome |
C00001429
|
| D013276 | Stomach Ulcer |
C00001429
|
| D059246 | Tachypnea |
C00001429
|
| D014202 | Tremor |
C00001429
|
| D002637 | Chest Pain |
C00001435
|
| D008881 | Migraine Disorders |
C00001435
|
| D015878 | Mydriasis |
C00001435
|
| D011041 | Poisoning |
C00001435
|
| D006943 | Hyperglycemia |
C00001200
|