Vitis vinifera
Species
KNApSAcK Entry
| Organism name | Vitis vinifera |
|---|---|
| Genus | Vitis |
| Family | Vitaceae |
| Kingdom | Plantae |
NCBI taxonomy
Entry
| Linked NCBI taxonomy name | Vitis vinifera |
|---|---|
| Linked NCBI taxonomy ID | 29760 |
| Linked level | species |
Family
| Family in NCBI taxonomy | Vitaceae |
|---|---|
| ID | 3602 |
Kingdom (Superkingdom)
| Kingdom (Superkingdom) in NCBI taxonomy | Viridiplantae |
|---|---|
| ID | 33090 |
Plant class
| Plant class | rosids |
|---|---|
| ID | 71275 |
Metabolite list (82)
| KNApSAcK ID | name | ChEMBL link | CTD link |
# of proteins in
ChEMBL interaction / related OMIM / related KEGG DISEASE |
# of genes in
CTD interaction / related diseases |
KCF-S
cluster |
phytochemical
cluster |
figure |
|---|---|---|---|---|---|---|---|---|
|
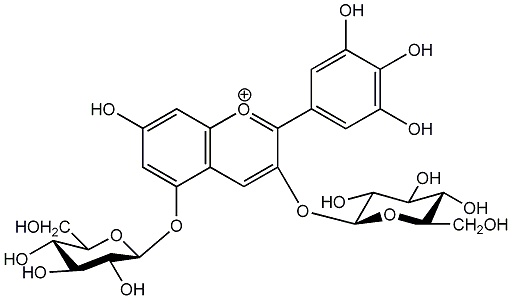
C00006709
|
Delphin
/ Delphinidin 3,5-diglucoside |
No. 1 | No. 15 |
|
||||
|
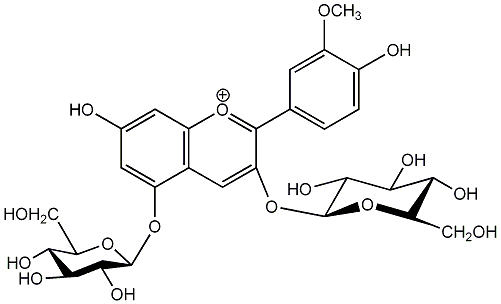
C00006688
|
Peonin
/ Peonidin 3,5-diglucoside / Peonidin 3-glucoside-5-glucoside |
C047592
|
No. 1 | No. 15 |
|
|||
|
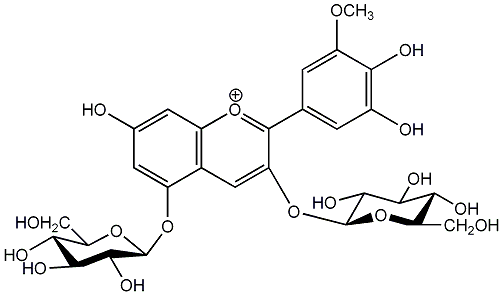
C00006728
|
Petunin
/ Petunidin 3,5-di-O-beta-D-glucoside |
No. 1 | No. 15 |
|
||||
|
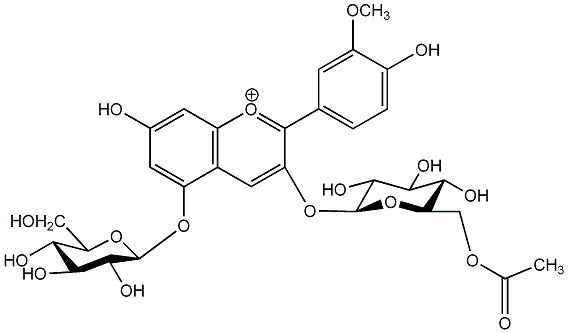
C00006865
|
Peonidin acetyl 3,5-diglucoside
|
No. 1 | No. 15 |
|
||||
|
C00006859
|
Peonidin 3-(6''-acetylglucoside)
|
No. 2 | No. 15 |

|
||||
|
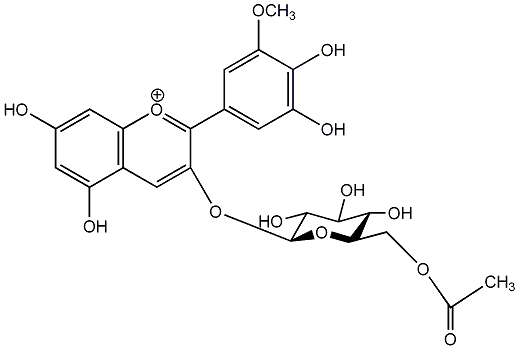
C00006894
|
Petunidin 3-(6''-acetylglucoside)
|
No. 2 | No. 15 |
|
||||
|
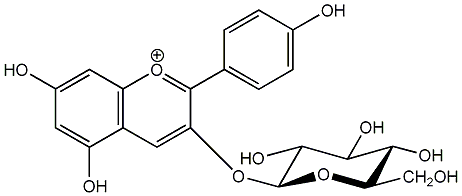
C00006630
|
Callistephin
/ Pelargonidin 3-O-beta-D-glucopyranoside |
C078485
|
No. 2 | No. 15 |
|
|||
|
C00006876
|
Delphinidin 3-(6''-acetylglucoside)
|
No. 2 | No. 15 |

|
||||
|
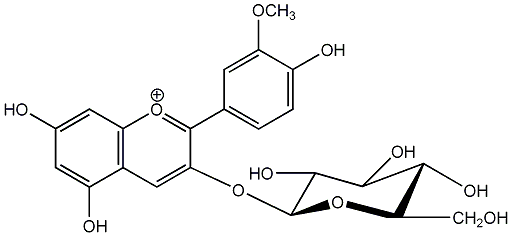
C00006681
|
Oxycoccicyanin
/ Peonidin 3-O-beta-D-glucopyranoside |
CHEMBL1784263
|
No. 2 | No. 15 |
|
|||
|
C00006698
|
Myrtillin
/ Delphinidin 3-O-beta-D-glucopyranoside |
CHEMBL518846
|
No. 2 | No. 15 |
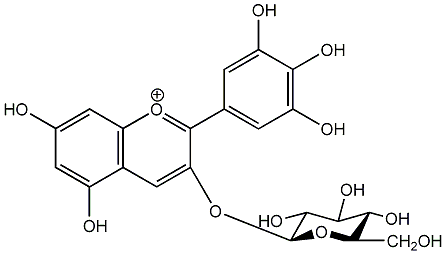
|
|||
|
C00006901
|
Malvidin 3-(6''-acetylglucoside)
|
No. 2 | No. 15 |

|
||||
|
C00006793
|
Cyanidin 3-(6''-acetylglucoside)
|
No. 2 | No. 15 |

|
||||
|
C00006722
|
Petunidin 3-glucoside
/ Petunidin 3-O-beta-D-glucopyranoside |
CHEMBL1784264
|
C455107
|
No. 2 | No. 15 |
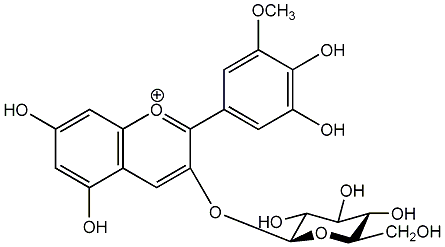
|
||
|
C00006735
|
Oenin
/ Malvidin 3-O-beta-D-glucopyranoside |
CHEMBL403236
|
C458419
|
1 / 0 | No. 2 | No. 15 |
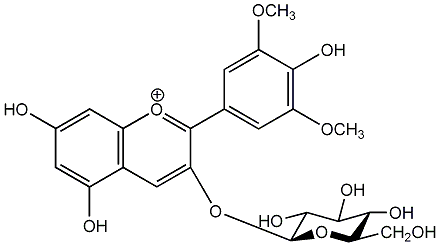
|
|
|
C00002374
|
Chrysanthemin
/ Cyanidin 3-O-glucoside |
CHEMBL257839
CHEMBL1197952 |
C114438
|
1 / 0 / 1 | No. 2 | No. 15 |
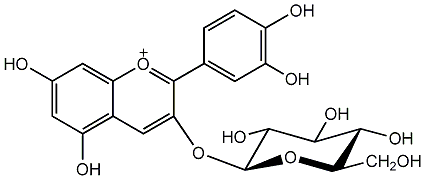
|
|
|
C00006884
|
Awobanin
/ Delphinidin-3-(6-O-p-coumarylglucoside)-5-glucoside |
No. 7 | No. 15 |
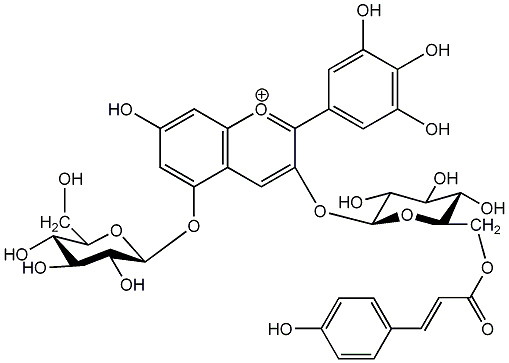
|
||||
|
C00006910
|
Tibouchinin
/ Malvidin-3-(6-O-p-coumarylglucoside)-5-glucoside |
No. 7 | No. 15 |

|
||||
|
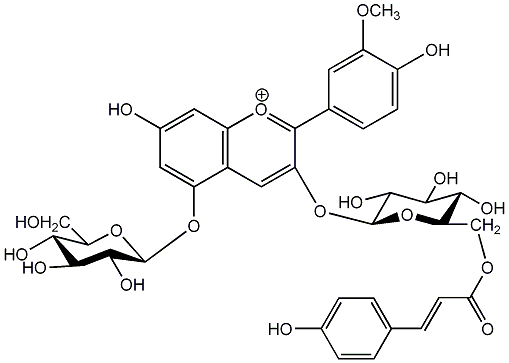
C00006867
|
Peonidin 3-(6''-p-coumarylglucoside)-5-glucoside
|
No. 7 | No. 15 |
|
||||
|
C00008703
|
Astilbin
|
CHEMBL486017
CHEMBL1159406 CHEMBL1159407 CHEMBL1314522 |
C099069
|
8 / 1 / 2 | No. 12 | No. 14 |

|
|
|
C00008672
|
Engeletin
|
No. 12 | No. 14 |

|
||||
|
C00002935
|
Procyanidin B4
/ Catechin-(4alpha->8)-epicatechin |
CHEMBL38714
CHEMBL81753 CHEMBL504937 CHEMBL501490 CHEMBL447373 CHEMBL1253314 CHEMBL1590914 |
20 / 7 / 8 | No. 16 | No. 19 |
|
||
|
C00009073
|
Procyanidin B8
/ Catechin-(4alpha->6)-epicatechin |
CHEMBL506487
CHEMBL502984 CHEMBL451115 |
No. 16 | No. 19 |

|
|||
|
C00009074
|
Procyanidin B7
/ Catechin-(4beta->6)-catechin |
CHEMBL506487
CHEMBL502984 CHEMBL451115 |
No. 16 | No. 19 |

|
|||
|
C00009075
|
Procyanidin B1
/ Epicatechin-(4beta->8)-catechin |
CHEMBL38714
CHEMBL81753 CHEMBL504937 CHEMBL501490 CHEMBL447373 CHEMBL1253314 CHEMBL1590914 |
C479579
|
20 / 7 / 8 | No. 16 | No. 19 |

|
|
|
C00009077
|
Procyanidin B2
/ Epicathechin-(4beta->8)-epicathechin |
CHEMBL38714
CHEMBL81753 CHEMBL504937 CHEMBL501490 CHEMBL447373 CHEMBL1253314 CHEMBL1590914 |
C479580
|
20 / 7 / 8 | 10 / 0 | No. 16 | No. 19 |
|
|
C00009071
|
Procyanidin B3
/ Catechin-(4alpha->8)-catechin |
CHEMBL38714
CHEMBL81753 CHEMBL504937 CHEMBL501490 CHEMBL447373 CHEMBL1253314 CHEMBL1590914 |
20 / 7 / 8 | No. 16 | No. 19 |

|
||
|
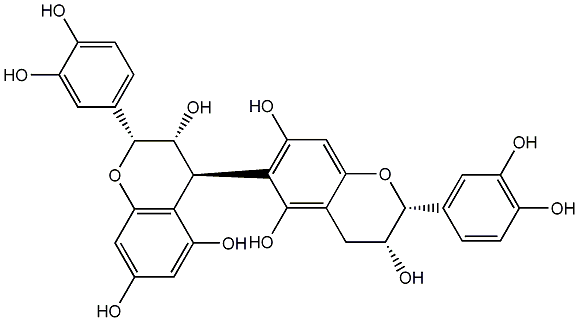
C00009076
|
Procyanidin B5
/ Epicatechin-(4beta-6)-epicatechin |
CHEMBL506487
CHEMBL502984 CHEMBL451115 |
No. 16 | No. 19 |
|
|||
|
C00009070
|
Procyanidin B6
/ Catechin-(4alpha->6)-catechin |
CHEMBL506487
CHEMBL502984 CHEMBL451115 |
No. 16 | No. 19 |
|
|||
|
C00009086
|
Epicatechin-(4beta->6)-epicatechin-(4beta->8)-catechin
|
CHEMBL592228
|
2 / 3 / 4 | No. 29 | No. 19 |

|
||
|
C00009213
|
3-O-Galloylepicatechin-(4beta->8)-epicatechin-3-O-gallate
|
CHEMBL39504
|
11 / 2 / 2 | No. 29 | No. 19 |

|
||
|
C00009087
|
Epicatechin-(4beta->6)-epicatechin-(4beta->8)-epicatechin
|
CHEMBL592228
|
2 / 3 / 4 | No. 29 | No. 19 |

|
||
|
C00009212
|
Epicatechin-(4beta->8)-epicatechin-3-O-gallate
|
No. 29 | No. 19 |

|
||||
|
C00009089
|
Arecatannin B1
|
CHEMBL592329
|
2 / 3 / 4 | No. 29 | No. 19 |
|
||
|
C00009211
|
3-O-Galloylepicatechin-(4beta->8)-catechin
|
No. 29 | No. 19 |

|
||||
|
C00009210
|
Catechin-(4alpha->8)-epicatechin-3-O-gallate
|
No. 29 | No. 19 |

|
||||
|
C00009208
|
3-O-Galloylepicatechin-(4beta->8)-epicatechin
|
No. 29 | No. 19 |

|
||||
|
C00009097
|
Arecatamnin A1
/ [Epicatechin-(4beta->8)]2-catechin |
CHEMBL290632
|
13 / 5 / 6 | No. 29 | No. 19 |
|
||
|
C00009098
|
Procyanidin C1
/ [Epicatechin-(4beta->8)]2-epicatechin |
CHEMBL290632
|
13 / 5 / 6 | No. 29 | No. 19 |
|
||
|
C00009206
|
3-O-Galloylepicatechin-(4beta->6)-catechin
|
No. 29 | No. 19 |

|
||||
|
C00009090
|
Epicatechin-(4beta->8)-epicatechin-(4beta->6)-epicatechin
|
CHEMBL592329
|
2 / 3 / 4 | No. 29 | No. 19 |

|
||
|
C00006878
|
Delphinidin 3-(6''-p-coumarylglucoside)
|
No. 30 | No. 15 |

|
||||
|
C00006879
|
Delphinidin 3-caffeylglucoside
|
No. 30 | No. 15 |

|
||||
|
C00006896
|
Petunidin 3-(6''-p-coumarylglucoside)
|
No. 30 | No. 15 |

|
||||
|
C00006862
|
Peonidin 3-(6''-caffeylgucoside)
|
No. 30 | No. 15 |

|
||||
|
C00006903
|
Malvidin 3-(6''-p-coumarylglucoside)
|
No. 30 | No. 15 |

|
||||
|
C00006756
|
Pelargonidin 3-p-coumarylglucoside
|
No. 30 | No. 15 |

|
||||
|
C00006861
|
Peonidin 3-(6''-p-coumarylglucoside)
|
No. 30 | No. 15 |

|
||||
|
C00006800
|
Hyacinthin
/ Cyanidin 3-(6-O-p-coumarylglucoside) |
No. 30 | No. 15 |

|
||||
|
C00006904
|
Malvidin 3-(6''-p-caffeyglucoside)
|
No. 30 | No. 15 |

|
||||
|
C00002896
|
Piceid
/ Resveratrol 3-O-beta-glucopyranoside / 3,3',4,5'-Tetrahydroxystilbene 3-O-beta-D-glucopyranoside |
CHEMBL142652
CHEMBL509386 CHEMBL597338 CHEMBL1990895 |
7 / 6 / 4 | No. 36 | No. 13 |
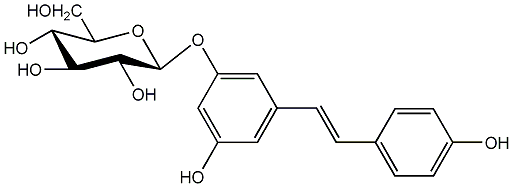
|
||
|
C00000001
|
GA1
/ Gibberellin A1 |
C422660
|
No. 40 | No. 41 |
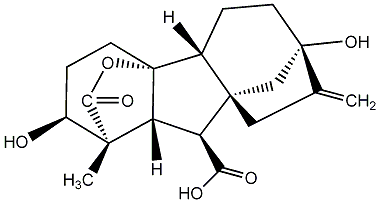
|
|||
|
C00000003
|
GA3
/ Gibberellin A3 |
CHEMBL513241
CHEMBL566653 CHEMBL1232952 CHEMBL1476967 CHEMBL1487394 |
C007842
|
10 / 18 / 17 | No. 40 | No. 41 |
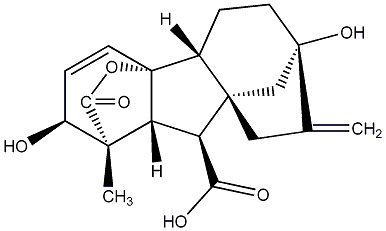
|
|
|
C00000956
|
(-)-Epicatechin
|
CHEMBL80941
CHEMBL311498 CHEMBL129482 CHEMBL200715 CHEMBL206452 CHEMBL583912 |
85 / 58 / 54 | No. 52 | No. 14 |
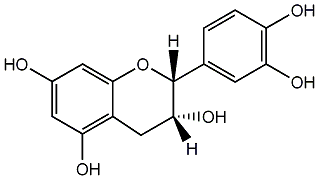
|
||
|
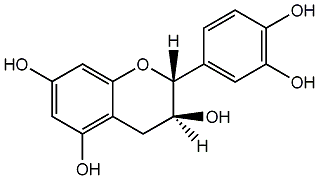
C00000947
|
D-Catechol
/ D-Catechin / Teafuran 30A / (+)-Catechin / Dexcyanidanol / (+)-3',4',5,7-Tetrahydroxy-2,3-trans-flavan-3-ol |
CHEMBL80941
CHEMBL311498 CHEMBL129482 CHEMBL200715 CHEMBL206452 CHEMBL583912 |
D002392
|
85 / 58 / 54 | 91 / 13 | No. 52 | No. 14 |
|
|
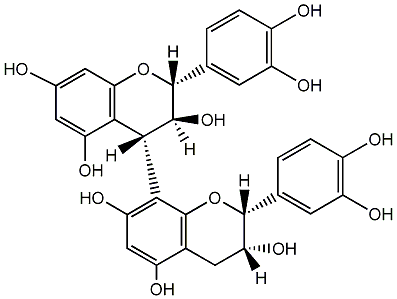
C00015837
|
Pallidol
/ (-)-Pallidol |
CHEMBL480462
|
2 / 0 / 3 | No. 108 | No. 30 |

|
||
|
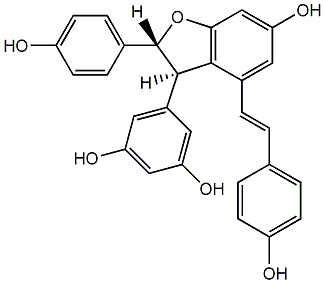
C00002905
|
epsilon-Viniferin
/ (-)-epsilon-Viniferin |
CHEMBL498439
CHEMBL1224875 CHEMBL1224889 |
C091891
|
9 / 0 | No. 108 | No. 30 |
|
|
|
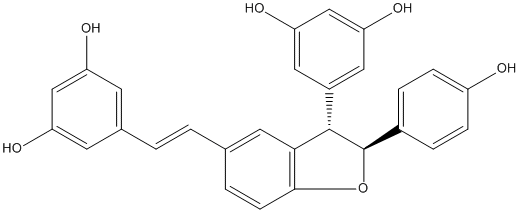
C00046353
|
Resveratrol (E)-dehydrodimer
|
CHEMBL518935
|
2 / 0 / 3 | No. 108 | No. 30 |
|
||
|
C00009228
|
Epicatechin-(4beta->8)-3-O-galloylepicatechin-(4beta->8)-epicatechin-3-O-gallate
|
No. 113 | No. 19 |

|
||||
|
C00009224
|
[Epicatechin-(4beta-8)]2-epicatechin-3-O-gallate
|
No. 113 | No. 19 |

|
||||
|
C00009225
|
Epicatechin-(4beta->8)-3-O-galloylepicatechin-(4beta->8)-catechin
|
No. 113 | No. 19 |

|
||||
|
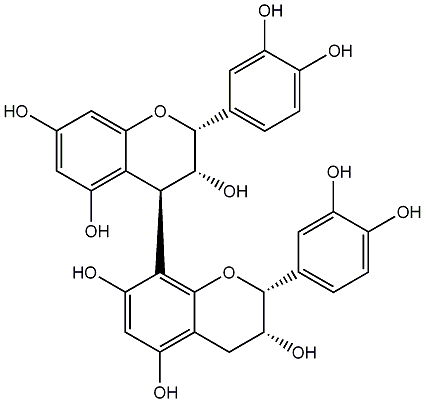
C00015693
|
lpha-Viniferin
/ (+)-alpha-Viniferin |
CHEMBL443463
CHEMBL1997745 |
C064176
|
1 / 0 / 3 | 1 / 2 | No. 163 | No. 30 |

|
|
C00011164
|
cis-Resveratrol-3,4'-O-beta-diglucoside
|
CHEMBL1812996
|
No. 169 | No. 13 |
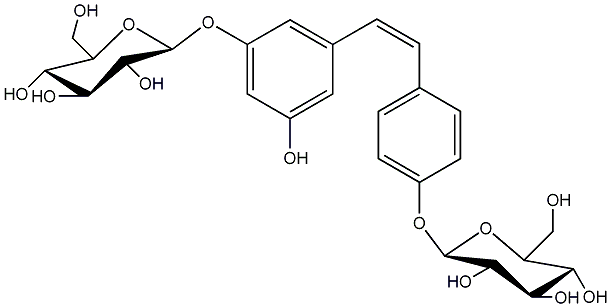
|
|||
|
C00000019
|
GA19
/ Gibberellin A19 |
C120175
|
No. 187 | No. 41 |
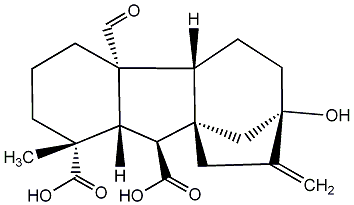
|
|||
|
C00008882
|
Gallocatechin 3-O-gallate
|
CHEMBL297453
CHEMBL311663 CHEMBL126079 CHEMBL338988 CHEMBL264938 |
49 / 48 / 54 | No. 219 | No. 14 |
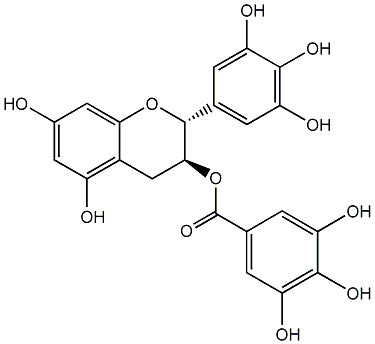
|
||
|
C00000958
|
(-)-Epigallocatechin gallate
/ epi-Gallocatechin 3-O-gallate / (2R-cis)-3,4-Dihydro-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-2H-1-benzopyran-3-yl ester 3,4,5-trihydroxybenzoic acid |
CHEMBL297453
CHEMBL311663 CHEMBL126079 CHEMBL338988 CHEMBL264938 |
C045651
|
49 / 48 / 54 | 2031 / 30 | No. 219 | No. 14 |
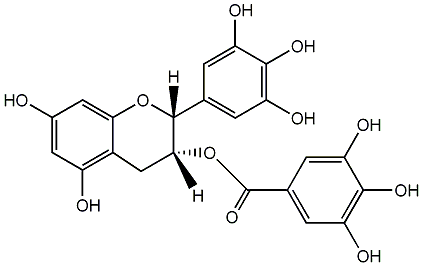
|
|
C00008866
|
(-)-Epicatechin gallate
/ (-)-Epicatechin 3-O-gallate |
CHEMBL36327
CHEMBL78573 CHEMBL328085 CHEMBL126142 CHEMBL129451 CHEMBL483083 CHEMBL1514905 |
41 / 20 / 30 | No. 219 | No. 14 |

|
||
|
C00002903
|
Resveratrol
/ 3,4',5-Trihydroxystilbene |
CHEMBL165
CHEMBL87333 CHEMBL2019155 |
C059514
|
179 / 114 / 101 | 1572 / 186 | No. 295 | No. 13 |
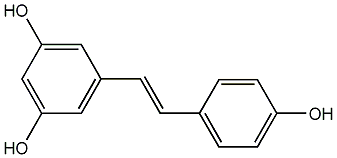
|
|
C00002902
|
Pterostilbene
/ 3,5-Dimethoxy-4'-hydroxystilbene |
CHEMBL87712
CHEMBL83527 |
C107773
|
19 / 19 / 20 | 54 / 15 | No. 344 | No. 13 |
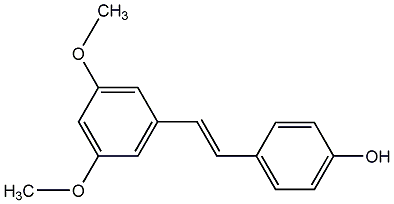
|
|
C00000856
|
4-Hydroxybenzoic acid
/ p-Hydroxybenzoic acid |
CHEMBL441343
|
C038193
|
21 / 7 / 16 | 2 / 1 | No. 817 | No. 81 |
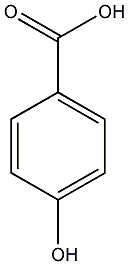
|
|
C00002648
|
Gentisic acid
/ 2,5-Dihydroxybenzoic acid |
CHEMBL1461
|
C010925
|
15 / 6 / 8 | 2 / 1 | No. 817 | No. 81 |
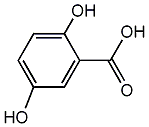
|
|
C00002647
|
Gallic acid
|
CHEMBL288114
|
D005707
|
42 / 53 / 68 | 52 / 16 | No. 817 | No. 81 |
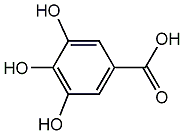
|
|
C00011188
|
Malvidin 3-glucoside-4-vinylcatechol
|
No. 1049 |

|
|||||
|
C00011189
|
Malvidin 3-glucoside-4-vinylguaiacol
|
No. 1049 |

|
|||||
|
C00011187
|
Malvidin 3-glucoside-4-vinylphenol
|
No. 1049 |

|
|||||
|
C00011186
|
Malvidin 3-glucoside-pyruvate
|
No. 1049 |

|
|||||
|
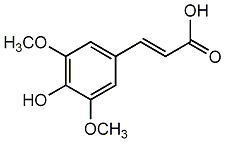
C00002776
|
Sinapic acid
|
CHEMBL109341
|
C073734
|
5 / 5 / 5 | 0 / 5 | No. 1366 | No. 6 |
|
|
C00000100
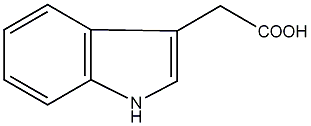
|
IAA
/ Indole-3-acetic acid |
CHEMBL82411
|
C030737
|
3 / 0 / 0 | 2 / 1 | No. 1432 | No. 4 |
|
|
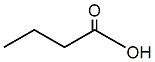
C00001180
|
Butyric acid
|
CHEMBL14227
|
D020148
|
12 / 3 / 2 | 40 / 5 | No. 3692 | No. 68 |
|
|
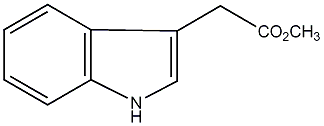
C00000101
|
IAA methyl ester
/ Methyl indole-3-acetate |
No. 4225 |
|
|||||
|
C00010310
|
2,6-Dimethyl-7-octene-2,3,6-triol
|
No. 5364 |

|
|||||
|
C00001206
|
L-(+)-Tartaric acid
|
CHEMBL225983
CHEMBL1200861 CHEMBL1236315 |
11 / 5 / 5 | No. 5453 |
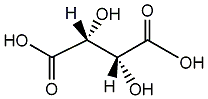
|
|||
|
C00001282
|
Eutypine
|
C102200
|
No. 6464 |

|
Human Protein / Gene in interactions
275 ChEMBL Protein in interactions
| accession | description | class description | KNApSAcK metabolite in interactions |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | C00000003 C00000947 C00000956 C00000958 C00001206 C00002903 C00002935 C00008866 C00008882 C00009071 C00009075 C00009077 | 0 / 1 |
| P17252 | Protein kinase C alpha type | Alpha | C00000947 C00000956 C00002903 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | C00000947 C00000956 C00000958 C00002647 C00002776 C00002902 C00002903 C00008882 C00015693 C00015837 C00046353 | 0 / 3 |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002903 C00002935 C00008866 C00008882 C00009071 C00009075 C00009077 | 3 / 3 |
| O94806 | Serine/threonine-protein kinase D3 | Pkd | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| P16050 | Arachidonate 15-lipoxygenase | Enzyme | C00000947 C00000956 C00000958 C00002903 C00002935 C00008866 C00008882 C00009071 C00009075 C00009077 | 0 / 0 |
| Q02156 | Protein kinase C epsilon type | Eta | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| Q05513 | Protein kinase C zeta type | Iota | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| Q05655 | Protein kinase C delta type | Delta | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| P05129 | Protein kinase C gamma type | Alpha | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 1 / 1 |
| P05771 | Protein kinase C beta type | Alpha | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| Q04759 | Protein kinase C theta type | Delta | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 1 |
| P24723 | Protein kinase C eta type | Eta | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 1 / 0 |
| Q15139 | Serine/threonine-protein kinase D1 | Pkd | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| P41743 | Protein kinase C iota type | Iota | C00000947 C00000956 C00002935 C00008866 C00009071 C00009075 C00009077 C00009097 C00009098 C00009213 | 0 / 0 |
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | C00000003 C00000947 C00000956 C00001206 C00002903 C00002935 C00009071 C00009075 C00009077 | 1 / 0 |
| P08253 | 72 kDa type IV collagenase | M10A | C00000958 C00002648 C00008882 C00009086 C00009087 C00009089 C00009090 C00009097 C00009098 | 1 / 3 |
| B2RXH2 | Lysine-specific demethylase 4E | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002896 C00002902 C00002903 C00008866 C00008882 | 0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | C00000003 C00000947 C00000956 C00001206 C00002903 C00002935 C00009071 C00009075 C00009077 | 0 / 0 |
| P10636 | Microtubule-associated protein tau | Unclassified protein | C00000947 C00000956 C00000958 C00002647 C00002896 C00002902 C00002903 C00008866 C00008882 | 4 / 3 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | C00000003 C00000947 C00000956 C00001206 C00002903 C00002935 C00009071 C00009075 C00009077 | 0 / 1 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | C00000003 C00000947 C00000956 C00001206 C00002903 C00002935 C00009071 C00009075 C00009077 | 1 / 1 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | C00000947 C00000956 C00000958 C00002776 C00002902 C00002903 C00008882 C00015837 C00046353 | 0 / 0 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002896 C00002903 C00008866 C00008882 | 0 / 0 |
| P28482 | Mitogen-activated protein kinase 1 | Erk | C00000947 C00000956 C00000958 C00002647 C00002896 C00002902 C00002903 C00008882 | 0 / 0 |
| P14780 | Matrix metalloproteinase-9 | M10A | C00002648 C00002903 C00009086 C00009087 C00009089 C00009090 C00009097 C00009098 | 2 / 2 |
| O00255 | Menin | Unclassified protein | C00000947 C00000956 C00000958 C00002902 C00002903 C00008866 C00008882 | 2 / 5 |
| Q9NUW8 | Tyrosyl-DNA phosphodiesterase 1 | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002903 C00008866 C00008882 | 1 / 1 |
| P02768 | Serum albumin | Secreted protein | C00000100 C00000947 C00000956 C00000958 C00002903 C00008866 C00008882 | 0 / 0 |
| P10253 | Lysosomal alpha-glucosidase | Hydrolase | C00000947 C00000956 C00000958 C00002896 C00002902 C00002903 C00008882 | 1 / 1 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | C00000947 C00000956 C00000958 C00002902 C00002903 C00008866 C00008882 | 1 / 2 |
| Q9UBT6 | DNA polymerase kappa | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002903 C00008866 C00008882 | 0 / 0 |
| P00918 | Carbonic anhydrase 2 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002648 C00002903 | 1 / 2 |
| Q9ULX7 | Carbonic anhydrase 14 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002648 C00002903 | 0 / 0 |
| P00352 | Retinal dehydrogenase 1 | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002903 C00008882 | 0 / 0 |
| P00915 | Carbonic anhydrase 1 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002648 C00002903 | 0 / 0 |
| P43166 | Carbonic anhydrase 7 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002648 C00002903 | 0 / 0 |
| P06746 | DNA polymerase beta | Enzyme | C00000947 C00000956 C00000958 C00002896 C00002903 C00008882 | 0 / 0 |
| Q16790 | Carbonic anhydrase 9 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002648 C00002903 | 0 / 1 |
| P15428 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002903 C00008882 | 2 / 2 |
| O43570 | Carbonic anhydrase 12 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002648 C00002903 | 1 / 2 |
| Q96KQ7 | Histone-lysine N-methyltransferase EHMT2 | Enzyme | C00000947 C00000956 C00000958 C00002647 C00002903 C00008882 | 0 / 0 |
| P11473 | Vitamin D3 receptor | NR1I1 | C00000947 C00000956 C00000958 C00002647 C00008866 C00008882 | 2 / 3 |
| P08581 | Hepatocyte growth factor receptor | TK tyrosine-protein kinase MET subfamily | C00000947 C00000956 C00000958 C00002647 C00008866 C00008882 | 2 / 3 |
| P52209 | 6-phosphogluconate dehydrogenase, decarboxylating | Enzyme | C00000947 C00000956 C00000958 C00008866 C00008882 | 0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | C00000947 C00000956 C00000958 C00008866 C00008882 | 0 / 0 |
| P54132 | Bloom syndrome protein | Enzyme | C00000947 C00000956 C00000958 C00002903 C00008882 | 1 / 2 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | C00000947 C00000956 C00000958 C00008866 C00008882 | 0 / 0 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | C00000947 C00000956 C00000958 C00002903 C00008882 | 0 / 0 |
| P23280 | Carbonic anhydrase 6 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002903 | 0 / 0 |
| P56817 | Beta-secretase 1 | A1A | C00000947 C00000956 C00000958 C00008866 C00008882 | 0 / 0 |
| P00374 | Dihydrofolate reductase | Oxidoreductase | C00000947 C00000956 C00000958 C00008866 C00008882 | 1 / 1 |
| P10415 | Apoptosis regulator Bcl-2 | Other cytosolic protein | C00000947 C00000956 C00000958 C00008866 C00008882 | 0 / 7 |
| P22748 | Carbonic anhydrase 4 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002903 | 1 / 1 |
| Q9Y2D0 | Carbonic anhydrase 5B, mitochondrial | Lyase | C00000856 C00000947 C00000956 C00002647 C00002903 | 0 / 0 |
| P35218 | Carbonic anhydrase 5A, mitochondrial | Lyase | C00000856 C00000947 C00000956 C00002647 C00002903 | 0 / 0 |
| P07451 | Carbonic anhydrase 3 | Lyase | C00000856 C00000947 C00000956 C00002647 C00002903 | 0 / 0 |
| P35372 | Mu-type opioid receptor | Opioid receptor | C00000947 C00000956 C00002903 C00008866 | 0 / 0 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | Unclassified protein | C00000958 C00002647 C00002903 C00008882 | 0 / 0 |
| Q9UIF8 | Bromodomain adjacent to zinc finger domain protein 2B | Unclassified protein | C00000947 C00000956 C00000958 C00008882 | 0 / 0 |
| Q16637 | Survival motor neuron protein | Unclassified protein | C00000947 C00000956 C00002647 C00002903 | 4 / 1 |
| P18433 | Receptor-type tyrosine-protein phosphatase alpha | Receptor tyrosine-protein phosphatase | C00002935 C00009071 C00009075 C00009077 | 0 / 0 |
| O75496 | Geminin | Unclassified protein | C00000003 C00001206 C00002902 C00002903 | 0 / 0 |
| P41143 | Delta-type opioid receptor | Opioid receptor | C00000947 C00000956 C00002903 C00008866 | 0 / 0 |
| P49327 | Fatty acid synthase | Transferase | C00000947 C00000956 C00000958 C00008882 | 0 / 0 |
| P23467 | Receptor-type tyrosine-protein phosphatase beta | Receptor tyrosine-protein phosphatase | C00002935 C00009071 C00009075 C00009077 | 0 / 0 |
| Q9UNA4 | DNA polymerase iota | Enzyme | C00000947 C00000956 C00002648 C00002903 | 0 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | C00000947 C00000956 C00002648 C00002903 | 0 / 0 |
| Q16539 | Mitogen-activated protein kinase 14 | p38 | C00000958 C00002903 C00008866 C00008882 | 0 / 0 |
| Q13093 | Platelet-activating factor acetylhydrolase | Enzyme | C00000947 C00000956 C00002647 C00002776 | 3 / 0 |
| P41145 | Kappa-type opioid receptor | Opioid receptor | C00000947 C00000956 C00002903 C00008866 | 0 / 0 |
| P28907 | ADP-ribosyl cyclase 1 | Enzyme | C00000947 C00000956 C00002374 C00002903 | 0 / 1 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | C00000958 C00002903 C00008866 C00008882 | 0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | C00000958 C00002647 C00002903 C00008882 | 0 / 0 |
| P07711 | Cathepsin L1 | C1A | C00000947 C00000956 C00002903 | 0 / 0 |
| P84022 | Mothers against decapentaplegic homolog 3 | Unclassified protein | C00000003 C00000947 C00000956 | 2 / 0 |
| P22309 | UDP-glucuronosyltransferase 1-1 | Enzyme | C00000947 C00000956 C00002647 | 5 / 1 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | C00000958 C00002903 C00008882 | 3 / 1 |
| P04626 | Receptor tyrosine-protein kinase erbB-2 | TK tyrosine-protein kinase EGFR subfamily | C00000958 C00002903 C00008882 | 5 / 9 |
| P16473 | Thyrotropin receptor | Glycohormone receptor | C00000947 C00000956 C00001206 | 3 / 2 |
| P37231 | Peroxisome proliferator-activated receptor gamma | NR1C3 | C00000958 C00002903 C00008882 | 5 / 3 |
| P02545 | Prelamin-A/C | Unclassified protein | C00000003 C00002647 C00002903 | 11 / 10 |
| Q8N1Q1 | Carbonic anhydrase 13 | Lyase | C00000947 C00000956 C00002903 | 0 / 0 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | C00002647 C00002776 C00008703 | 0 / 0 |
| P14618 | Pyruvate kinase PKM | Enzyme | C00000958 C00002903 C00008882 | 0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | C00000003 C00002648 C00002903 | 0 / 0 |
| P08246 | Neutrophil elastase | S1A | C00000958 C00002903 C00008882 | 2 / 1 |
| Q12809 | Potassium voltage-gated channel subfamily H member 2 | KCNH, Kv10-12.x (Ether-a-go-go) | C00000958 C00002903 C00008882 | 2 / 2 |
| Q01453 | Peripheral myelin protein 22 | Unclassified protein | C00000947 C00000956 C00002903 | 5 / 2 |
| P09237 | Matrilysin | M10A | C00000958 C00008882 | 0 / 0 |
| O15296 | Arachidonate 15-lipoxygenase B | Enzyme | C00002903 C00008866 | 0 / 0 |
| O15118 | Niemann-Pick C1 protein | Unclassified protein | C00002902 C00002903 | 1 / 1 |
| Q9HAW9 | UDP-glucuronosyltransferase 1-8 | Enzyme | C00000856 C00001180 | 0 / 0 |
| P03372 | Estrogen receptor | NR3A1 | C00002647 C00002903 | 1 / 1 |
| P22303 | Acetylcholinesterase | Hydrolase | C00002903 C00008866 | 1 / 0 |
| P50281 | Matrix metalloproteinase-14 | M10A | C00000958 C00008882 | 0 / 0 |
| Q92887 | Canalicular multispecific organic anion transporter 1 | Unclassified protein | C00002903 C00008866 | 1 / 1 |
| Q9H0H5 | Rac GTPase-activating protein 1 | Unclassified protein | C00000947 C00000956 | 0 / 0 |
| P26358 | DNA (cytosine-5)-methyltransferase 1 | Transferase | C00000958 C00008882 | 2 / 0 |
| P11388 | DNA topoisomerase 2-alpha | Isomerase | C00000947 C00000956 | 0 / 0 |
| P00533 | Epidermal growth factor receptor | TK tyrosine-protein kinase EGFR subfamily | C00000856 C00002903 | 1 / 8 |
| Q9Y3R4 | Sialidase-2 | Enzyme | C00000947 C00000956 | 0 / 0 |
| Q16665 | Hypoxia-inducible factor 1-alpha | Transcription Factor | C00002903 C00008866 | 0 / 0 |
| Q9NR56 | Muscleblind-like protein 1 | Unclassified protein | C00002896 C00002903 | 1 / 0 |
| P39748 | Flap endonuclease 1 | Enzyme | C00000947 C00000956 | 0 / 0 |
| P51570 | Galactokinase | Enzyme | C00000958 C00008882 | 1 / 1 |
| P22310 | UDP-glucuronosyltransferase 1-4 | Enzyme | C00000947 C00000956 | 3 / 0 |
| P07998 | Ribonuclease pancreatic | Enzyme | C00000947 C00000956 | 0 / 0 |
| P10696 | Alkaline phosphatase, placental-like | Enzyme | C00000947 C00000956 | 0 / 1 |
| Q6W5P4 | Neuropeptide S receptor | Neuropeptide receptor | C00000947 C00000956 | 1 / 0 |
| P08183 | Multidrug resistance protein 1 | drug | C00002902 C00002903 | 1 / 0 |
| P51151 | Ras-related protein Rab-9A | Unclassified protein | C00002902 C00002903 | 0 / 0 |
| P10275 | Androgen receptor | NR3C4 | C00000003 C00002903 | 3 / 4 |
| P38398 | Breast cancer type 1 susceptibility protein | Enzyme | C00002902 C00002903 | 4 / 2 |
| Q13627 | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | CMGC dual-specificity kinase DYRK1 | C00000958 C00008882 | 1 / 0 |
| P43220 | Glucagon-like peptide 1 receptor | Glucagon-like peptide receptor | C00002902 C00002903 | 0 / 0 |
| P04745 | Alpha-amylase 1 | Enzyme | C00000947 C00000956 | 0 / 0 |
| P47989 | Xanthine dehydrogenase/oxidase | Oxidoreductase | C00000947 C00000956 | 1 / 1 |
| P55789 | FAD-linked sulfhydryl oxidase ALR | Enzyme | C00002902 C00002903 | 1 / 0 |
| P04637 | Cellular tumor antigen p53 | Transcription Factor | C00002647 C00002903 | 7 / 37 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | C00002902 C00002903 | 2 / 2 |
| P04150 | Glucocorticoid receptor | NR3C1 | C00002647 C00002903 | 0 / 1 |
| P00734 | Prothrombin | S1A | C00000947 C00000956 | 4 / 2 |
| Q9HAW8 | UDP-glucuronosyltransferase 1-10 | Enzyme | C00000856 C00001180 | 0 / 0 |
| P11274 | Breakpoint cluster region protein | Bcr | C00002902 C00002903 | 1 / 2 |
| P05186 | Alkaline phosphatase, tissue-nonspecific isozyme | Enzyme | C00000947 C00000956 | 3 / 1 |
| P09923 | Intestinal-type alkaline phosphatase | Enzyme | C00000947 C00000956 | 0 / 0 |
| P00519 | Tyrosine-protein kinase ABL1 | Abl | C00002902 C00002903 | 1 / 2 |
| Q9Y253 | DNA polymerase eta | Enzyme | C00000947 C00000956 | 1 / 1 |
| P03956 | Interstitial collagenase | M10A | C00002648 C00002903 | 0 / 1 |
| O95342 | Bile salt export pump | drug | C00000958 C00008882 | 2 / 1 |
| O14746 | Telomerase reverse transcriptase | Enzyme | C00000958 C00008882 | 5 / 5 |
| Q99720 | Sigma non-opioid intracellular receptor 1 | Membrane receptor | C00002903 | 1 / 0 |
| P08311 | Cathepsin G | S1A | C00002903 | 0 / 0 |
| P32241 | Vasoactive intestinal polypeptide receptor 1 | Vasoactive intestinal peptide receptor | C00002903 | 0 / 0 |
| Q9GZT4 | Serine racemase | Enzyme | C00001206 | 0 / 0 |
| P32238 | Cholecystokinin receptor type A | Cholecystokinin receptor | C00002903 | 0 / 0 |
| P32245 | Melanocortin receptor 4 | Melanocortin receptor | C00002903 | 1 / 0 |
| P10145 | Interleukin-8 | Secreted protein | C00002647 | 0 / 0 |
| P11308 | Transcriptional regulator ERG | Unclassified protein | C00008703 | 1 / 2 |
| P30411 | B2 bradykinin receptor | Bradykinin receptor | C00002903 | 0 / 0 |
| Q9HC16 | DNA dC->dU-editing enzyme APOBEC-3G | Enzyme | C00008703 | 0 / 0 |
| P11021 | 78 kDa glucose-regulated protein | Unclassified protein | C00002903 | 0 / 0 |
| P25101 | Endothelin-1 receptor | Endothelin receptor | C00002903 | 0 / 0 |
| Q9NZ45 | CDGSH iron-sulfur domain-containing protein 1 | Unclassified protein | C00002903 | 0 / 0 |
| P01106 | Myc proto-oncogene protein | Unclassified protein | C00002903 | 1 / 13 |
| P15121 | Aldose reductase | Enzyme | C00002647 | 0 / 0 |
| P41595 | 5-hydroxytryptamine receptor 2B | Serotonin receptor | C00002903 | 0 / 0 |
| P08172 | Muscarinic acetylcholine receptor M2 | Acetylcholine receptor | C00002903 | 2 / 0 |
| P11229 | Muscarinic acetylcholine receptor M1 | Acetylcholine receptor | C00002903 | 0 / 0 |
| Q92731 | Estrogen receptor beta | NR3A2 | C00002903 | 0 / 1 |
| P14679 | Tyrosinase | Oxidoreductase | C00002903 | 4 / 2 |
| P21554 | Cannabinoid receptor 1 | Cannabinoid receptor | C00002903 | 0 / 0 |
| P31645 | Sodium-dependent serotonin transporter | Serotonin | C00002903 | 2 / 0 |
| P35462 | D(3) dopamine receptor | Dopamine receptor | C00002903 | 1 / 0 |
| P80404 | 4-aminobutyrate aminotransferase, mitochondrial | Transferase | C00000856 | 1 / 1 |
| P30988 | Calcitonin receptor | Calcitonin receptor | C00002903 | 0 / 0 |
| P20309 | Muscarinic acetylcholine receptor M3 | Acetylcholine receptor | C00002903 | 1 / 0 |
| Q9HBH1 | Peptide deformylase, mitochondrial | Enzyme | C00000100 | 0 / 0 |
| P16083 | Ribosyldihydronicotinamide dehydrogenase [quinone] | Enzyme | C00002903 | 0 / 0 |
| P21452 | Substance-K receptor | Neurokinin receptor | C00002903 | 0 / 0 |
| P51679 | C-C chemokine receptor type 4 | CC chemokine receptor | C00002903 | 0 / 0 |
| P51681 | C-C chemokine receptor type 5 | CC chemokine receptor | C00002903 | 3 / 0 |
| P50406 | 5-hydroxytryptamine receptor 6 | Serotonin receptor | C00002903 | 0 / 0 |
| P21917 | D(4) dopamine receptor | Dopamine receptor | C00002903 | 0 / 0 |
| O15379 | Histone deacetylase 3 | Hydrolase | C00001180 | 0 / 0 |
| P08913 | Alpha-2A adrenergic receptor | Adrenergic receptor | C00002903 | 0 / 0 |
| P04035 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Oxidoreductase | C00002903 | 0 / 0 |
| P41597 | C-C chemokine receptor type 2 | CC chemokine receptor | C00002903 | 1 / 0 |
| O60656 | UDP-glucuronosyltransferase 1-9 | Enzyme | C00000856 | 0 / 0 |
| P42858 | Huntingtin | Unclassified protein | C00002903 | 1 / 1 |
| P08575 | Receptor-type tyrosine-protein phosphatase C | Enzyme | C00002903 | 2 / 1 |
| Q12778 | Forkhead box protein O1 | Unclassified protein | C00002903 | 1 / 1 |
| Q8IXJ6 | NAD-dependent protein deacetylase sirtuin-2 | Enzyme | C00002903 | 0 / 0 |
| Q7Z4W1 | L-xylulose reductase | Enzyme | C00001180 | 0 / 1 |
| P11509 | Cytochrome P450 2A6 | Cytochrome P450 2A6 | C00002903 | 0 / 0 |
| Q9UQL6 | Histone deacetylase 5 | Hydrolase | C00001180 | 0 / 0 |
| P17405 | Sphingomyelin phosphodiesterase | Enzyme | C00002776 | 2 / 2 |
| Q9NTG7 | NAD-dependent protein deacetylase sirtuin-3, mitochondrial | Enzyme | C00002903 | 0 / 0 |
| P16581 | E-selectin | Adhesion | C00002647 | 0 / 0 |
| Q92769 | Histone deacetylase 2 | Hydrolase | C00001180 | 0 / 0 |
| O43524 | Forkhead box protein O3 | Unclassified protein | C00002903 | 0 / 0 |
| P27540 | Aryl hydrocarbon receptor nuclear translocator | Unclassified protein | C00008866 | 0 / 0 |
| P41968 | Melanocortin receptor 3 | Melanocortin receptor | C00002903 | 1 / 0 |
| P17948 | Vascular endothelial growth factor receptor 1 | Vegfr | C00002903 | 0 / 0 |
| P50052 | Type-2 angiotensin II receptor | Angiotensin receptor | C00002903 | 1 / 1 |
| P51449 | Nuclear receptor ROR-gamma | Nuclear hormone receptor subfamily 1 group F member 3 | C00002903 | 0 / 0 |
| Q96QE3 | ATPase family AAA domain-containing protein 5 | Unclassified protein | C00002903 | 0 / 0 |
| P25929 | Neuropeptide Y receptor type 1 | Neuropeptide Y receptor | C00002903 | 0 / 0 |
| O76074 | cGMP-specific 3',5'-cyclic phosphodiesterase | PDE_5A | C00002903 | 0 / 0 |
| P11926 | Ornithine decarboxylase | Lyase | C00002903 | 0 / 0 |
| P29274 | Adenosine receptor A2a | Adenosine receptor | C00002903 | 0 / 0 |
| P08588 | Beta-1 adrenergic receptor | Adrenergic receptor | C00002903 | 1 / 0 |
| P12821 | Angiotensin-converting enzyme | M2 | C00002647 | 4 / 2 |
| Q9Y271 | Cysteinyl leukotriene receptor 1 | Leukotriene receptor | C00002903 | 0 / 0 |
| P28223 | 5-hydroxytryptamine receptor 2A | Serotonin receptor | C00002903 | 0 / 0 |
| P28335 | 5-hydroxytryptamine receptor 2C | Serotonin receptor | C00002903 | 0 / 0 |
| P37288 | Vasopressin V1a receptor | Vasopressin and oxytocin receptor | C00002903 | 0 / 0 |
| P08173 | Muscarinic acetylcholine receptor M4 | Acetylcholine receptor | C00002903 | 0 / 0 |
| P25103 | Substance-P receptor | Neurokinin receptor | C00002903 | 0 / 0 |
| P25105 | Platelet-activating factor receptor | PAF receptor | C00002903 | 0 / 0 |
| P14416 | D(2) dopamine receptor | Dopamine receptor | C00002903 | 2 / 0 |
| P33032 | Melanocortin receptor 5 | Melanocortin receptor | C00002903 | 0 / 0 |
| P56524 | Histone deacetylase 4 | Hydrolase | C00001180 | 1 / 1 |
| Q9HAW7 | UDP-glucuronosyltransferase 1-7 | Enzyme | C00000856 | 0 / 0 |
| P14920 | D-amino-acid oxidase | Enzyme | C00001206 | 0 / 0 |
| P07237 | Protein disulfide-isomerase | Enzyme | C00008703 | 0 / 0 |
| Q8WUI4 | Histone deacetylase 7 | Hydrolase | C00001180 | 0 / 0 |
| P31749 | RAC-alpha serine/threonine-protein kinase | Akt | C00002903 | 4 / 1 |
| P51580 | Thiopurine S-methyltransferase | Enzyme | C00000856 | 1 / 1 |
| P14151 | L-selectin | Adhesion | C00002647 | 0 / 0 |
| P16109 | P-selectin | Adhesion | C00002647 | 1 / 0 |
| Q08209 | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Ser_Thr | C00002903 | 0 / 0 |
| P06241 | Tyrosine-protein kinase Fyn | Src | C00002903 | 0 / 0 |
| P05181 | Cytochrome P450 2E1 | Cytochrome P450 2E1 | C00002903 | 0 / 0 |
| Q96EB6 | NAD-dependent protein deacetylase sirtuin-1 | Enzyme | C00002903 | 0 / 0 |
| Q9UQ49 | Sialidase-3 | Enzyme | C00000856 | 0 / 0 |
| P25024 | C-X-C chemokine receptor type 1 | CXC chemokine receptor | C00002903 | 0 / 0 |
| O75164 | Lysine-specific demethylase 4A | Enzyme | C00001206 | 0 / 0 |
| P78536 | Disintegrin and metalloproteinase domain-containing protein 17 | M12B | C00002647 | 1 / 0 |
| P04798 | Cytochrome P450 1A1 | Cytochrome P450 1A1 | C00002903 | 0 / 0 |
| Q9BY41 | Histone deacetylase 8 | Hydrolase | C00001180 | 2 / 0 |
| Q99549 | M-phase phosphoprotein 8 | Unclassified protein | C00002903 | 0 / 0 |
| P19224 | UDP-glucuronosyltransferase 1-6 | Enzyme | C00000856 | 0 / 0 |
| P13945 | Beta-3 adrenergic receptor | Adrenergic receptor | C00002903 | 0 / 0 |
| P18825 | Alpha-2C adrenergic receptor | Adrenergic receptor | C00002903 | 0 / 0 |
| P23975 | Sodium-dependent noradrenaline transporter | Norepinephrine | C00002903 | 1 / 1 |
| P25100 | Alpha-1D adrenergic receptor | Adrenergic receptor | C00002903 | 0 / 0 |
| P30542 | Adenosine receptor A1 | Adenosine receptor | C00002903 | 0 / 0 |
| P18089 | Alpha-2B adrenergic receptor | Adrenergic receptor | C00002903 | 0 / 0 |
| P24557 | Thromboxane-A synthase | Cytochrome P450 5A1 | C00002903 | 1 / 1 |
| P08912 | Muscarinic acetylcholine receptor M5 | Acetylcholine receptor | C00002903 | 0 / 0 |
| P06239 | Tyrosine-protein kinase Lck | Src | C00002903 | 0 / 1 |
| P35869 | Aryl hydrocarbon receptor | Transcription Factor | C00002903 | 0 / 0 |
| P25025 | C-X-C chemokine receptor type 2 | CXC chemokine receptor | C00002903 | 0 / 0 |
| P08254 | Stromelysin-1 | M10A | C00002648 | 1 / 0 |
| Q9UBN7 | Histone deacetylase 6 | Hydrolase | C00001180 | 0 / 0 |
| Q9UKV0 | Histone deacetylase 9 | Hydrolase | C00001180 | 0 / 0 |
| Q01959 | Sodium-dependent dopamine transporter | Dopamine | C00002903 | 1 / 0 |
| P35367 | Histamine H1 receptor | Histamine receptor | C00002903 | 0 / 0 |
| P25021 | Histamine H2 receptor | Histamine receptor | C00002903 | 0 / 0 |
| Q07817 | Bcl-2-like protein 1 | Other cytosolic protein | C00002903 | 0 / 0 |
| P21397 | Amine oxidase [flavin-containing] A | Oxidoreductase | C00002903 | 1 / 1 |
| P07550 | Beta-2 adrenergic receptor | Adrenergic receptor | C00002903 | 0 / 1 |
| P11169 | Solute carrier family 2, facilitated glucose transporter member 3 | Unclassified protein | C00002903 | 0 / 0 |
| P51649 | Succinate-semialdehyde dehydrogenase, mitochondrial | Oxidoreductase | C00000856 | 1 / 1 |
| P09960 | Leukotriene A-4 hydrolase | M1 | C00002903 | 0 / 0 |
| P40225 | Thrombopoietin | Unclassified protein | C00002903 | 1 / 1 |
| Q99489 | D-aspartate oxidase | Enzyme | C00001206 | 0 / 0 |
| Q13526 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | Enzyme | C00008703 | 0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | C00002903 | 1 / 0 |
| Q13547 | Histone deacetylase 1 | Hydrolase | C00001180 | 0 / 0 |
| Q03181 | Peroxisome proliferator-activated receptor delta | NR1C2 | C00002903 | 0 / 0 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | C00002903 | 0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | C00002903 | 1 / 4 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | Enzyme | C00002647 | 0 / 0 |
| P27361 | Mitogen-activated protein kinase 3 | Erk | C00002903 | 0 / 0 |
| P29466 | Caspase-1 | C14 | C00002903 | 0 / 0 |
| Q16773 | Kynurenine--oxoglutarate transaminase 1 | Enzyme | C00000100 | 0 / 0 |
| P22894 | Neutrophil collagenase | M10A | C00002648 | 0 / 0 |
| P49146 | Neuropeptide Y receptor type 2 | Neuropeptide Y receptor | C00002903 | 0 / 0 |
| P33765 | Adenosine receptor A3 | Adenosine receptor | C00002903 | 0 / 0 |
| P27338 | Amine oxidase [flavin-containing] B | Oxidoreductase | C00002903 | 0 / 0 |
| P21728 | D(1A) dopamine receptor | Dopamine receptor | C00002903 | 0 / 0 |
| P09917 | Arachidonate 5-lipoxygenase | Oxidoreductase | C00002648 | 0 / 0 |
| P49798 | Regulator of G-protein signaling 4 | Unclassified protein | C00002903 | 2 / 0 |
| Q99700 | Ataxin-2 | Unclassified protein | C00002903 | 1 / 1 |
| Q00653 | Nuclear factor NF-kappa-B p100 subunit | Transcription Factor | C00002903 | 0 / 0 |
| Q04206 | Transcription factor p65 | Transcription Factor | C00002903 | 0 / 0 |
| P68400 | Casein kinase II subunit alpha | Ck2 | C00008703 | 0 / 0 |
| P19784 | Casein kinase II subunit alpha' | Ck2 | C00008703 | 0 / 0 |
| P67870 | Casein kinase II subunit beta | REG serine/threonine protein kinase family | C00008703 | 0 / 0 |
| P33527 | Multidrug resistance-associated protein 1 | drugs | C00008866 | 0 / 0 |
| P42345 | Serine/threonine-protein kinase mTOR | Enzyme | C00002903 | 0 / 0 |
| Q13148 | TAR DNA-binding protein 43 | Unclassified protein | C00002902 | 1 / 1 |
3345 Gene in CTD interactions
| gene | gene name | gene description | KNApSAcK metabolite in interactions |
|---|---|---|---|
| 581 | BAX, BCL2L4 | BCL2-associated X protein |
C00000947
C00000958
C00001180
C00002647
C00002902 C00002903 C00009077 |
| 7124 | TNF, DIF, TNF-alpha, TNFA, TNFSF2 | tumor necrosis factor |
C00000947
C00000958
C00001180
C00002647
C00002902 C00002903 |
| 5595 | MAPK3, ERK-1, ERK1, ERT2, HS44KDAP, HUMKER1A, P44ERK1, P44MAPK, PRKM3, p44-ERK1, p44-MAPK | mitogen-activated protein kinase 3 (EC:2.7.11.24) |
C00000947
C00000958
C00002647
C00002902
C00002903 C00009077 |
| 5594 | MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2, p38, p40, p41, p41mapk | mitogen-activated protein kinase 1 (EC:2.7.11.24) |
C00000947
C00000958
C00002647
C00002902
C00002903 C00009077 |
| 5743 | PTGS2, COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00000947
C00000958
C00001180
C00002647
C00002902 C00002903 |
| 207 | AKT1, AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 (EC:2.7.11.1) |
C00000947
C00000958
C00002647
C00002902
C00002903 C00009077 |
| 54205 | CYCS, CYC, HCS, THC4 | cytochrome c, somatic |
C00000947
C00000958
C00002647
C00002902
C00002903 C00009077 |
| 836 | CASP3, CPP32, CPP32B, SCA-1 | caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) |
C00000947
C00000958
C00002647
C00002902
C00002903 C00009077 |
| 5970 | RELA, NFKB3, p65 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
C00000947
C00000958
C00002647
C00002902
C00002903 |
| 3949 | LDLR, FH, FHC, LDLCQ2 | low density lipoprotein receptor |
C00000958
C00002647
C00002902
C00002903
C00002905 |
| 596 | BCL2, Bcl-2, PPP1R50 | B-cell CLL/lymphoma 2 |
C00000947
C00000958
C00002647
C00002902
C00002903 |
| 842 | CASP9, APAF-3, APAF3, ICE-LAP6, MCH6, PPP1R56 | caspase 9, apoptosis-related cysteine peptidase (EC:3.4.22.62) |
C00000947
C00000958
C00002902
C00002903
C00009077 |
| 1543 | CYP1A1, AHH, AHRR, CP11, CYP1, P1-450, P450-C, P450DX | cytochrome P450, family 1, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00000947
C00000958
C00002902
C00002903
C00002905 |
| 1026 | CDKN1A, CAP20, CDKN1, CIP1, MDA-6, P21, SDI1, WAF1, p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
C00000958
C00001180
C00002647
C00002902
C00002903 |
| 6817 | SULT1A1, HAST1/HAST2, P-PST, PST, ST1A1, ST1A3, STP, STP1, TSPST1 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (EC:2.8.2.1) |
C00000856
C00000947
C00002647
C00002648
C00002903 |
| 4609 | MYC, MRTL, MYCC, bHLHe39, c-Myc | v-myc avian myelocytomatosis viral oncogene homolog |
C00000958
C00001180
C00002902
C00002903
|
| 7153 | TOP2A, TOP2, TP2A | topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) |
C00000947
C00000958
C00002903
C00009077
|
| 841 | CASP8, ALPS2B, CAP4, Casp-8, FLICE, MACH, MCH5 | caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) |
C00000947
C00000958
C00002647
C00002903
|
| 3162 | HMOX1, HMOX1D, HO-1, HSP32, bK286B10 | heme oxygenase (decycling) 1 (EC:1.14.99.3) |
C00000947
C00000958
C00002902
C00002903
|
| 3553 | IL1B, IL-1, IL1-BETA, IL1F2 | interleukin 1, beta |
C00000947
C00000958
C00002902
C00002903
|
| 4792 | NFKBIA, IKBA, MAD-3, NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
C00000947
C00000958
C00002902
C00002903
|
| 3576 | IL8, CXCL8, GCP-1, GCP1, LECT, LUCT, LYNAP, MDNCF, MONAP, NAF, NAP-1, NAP1 | interleukin 8 |
C00000958
C00001180
C00002647
C00002903
|
| 1027 | CDKN1B, CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
C00000958
C00002647
C00002902
C00002903
|
| 6648 | SOD2, IPOB, MNSOD, MVCD6 | superoxide dismutase 2, mitochondrial (EC:1.15.1.1) |
C00000958
C00001180
C00002902
C00002903
|
| 5879 | RAC1, Rac-1, TC-25, p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
C00000947
C00000958
C00002647
C00002903
|
| 5243 | ABCB1, ABC20, CD243, CLCS, GP170, MDR1, P-GP, PGY1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 (EC:3.6.3.44) |
C00000958
C00001180
C00002647
C00002903
|
| 595 | CCND1, BCL1, D11S287E, PRAD1, U21B31 | cyclin D1 |
C00000947
C00000958
C00002902
C00002903
|
| 6347 | CCL2, GDCF-2, HC11, HSMCR30, MCAF, MCP-1, MCP1, SCYA2, SMC-CF | chemokine (C-C motif) ligand 2 |
C00000947
C00000958
C00001180
C00002903
|
| 7157 | TP53, BCC7, LFS1, P53, TRP53 | tumor protein p53 |
C00000958
C00002647
C00002902
C00002903
|
| 5601 | MAPK9, JNK-55, JNK2, JNK2A, JNK2ALPHA, JNK2B, JNK2BETA, PRKM9, SAPK, SAPK1a, p54a, p54aSAPK | mitogen-activated protein kinase 9 (EC:2.7.11.24) |
C00000958
C00002647
C00002902
C00002903
|
| 5599 | MAPK8, JNK, JNK-46, JNK1, JNK1A2, JNK21B1/2, PRKM8, SAPK1, SAPK1c | mitogen-activated protein kinase 8 (EC:2.7.11.24) |
C00000958
C00002647
C00002902
C00002903
|
| 3569 | IL6, BSF2, HGF, HSF, IFNB2, IL-6 | interleukin 6 (interferon, beta 2) |
C00000958
C00001180
C00002647
C00002903
|
| 4170 | MCL1, BCL2L3, EAT, MCL1-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT | myeloid cell leukemia sequence 1 (BCL2-related) |
C00000958
C00002902
C00002903
|
| 4318 | MMP9, CLG4B, GELB, MANDP2, MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) |
C00002647
C00002902
C00002903
|
| 1432 | MAPK14, CSBP, CSBP1, CSBP2, CSPB1, EXIP, Mxi2, PRKM14, PRKM15, RK, SAPK2A, p38, p38ALPHA | mitogen-activated protein kinase 14 (EC:2.7.11.24) |
C00000958
C00002902
C00002903
|
| 5604 | MAP2K1, CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | mitogen-activated protein kinase kinase 1 (EC:2.7.12.2) |
C00000947
C00002647
C00002903
|
| 331 | XIAP, API3, BIRC4, IAP-3, ILP1, MIHA, XLP2, hIAP-3, hIAP3 | X-linked inhibitor of apoptosis |
C00000958
C00002647
C00002903
|
| 4255 | MGMT | O-6-methylguanine-DNA methyltransferase (EC:2.1.1.63) |
C00000958
C00002647
C00002903
|
| 3725 | JUN, AP-1, AP1, c-Jun | jun proto-oncogene |
C00000958
C00001180
C00002903
|
| 9429 | ABCG2, ABC15, ABCP, BCRP, BCRP1, BMDP, CD338, CDw338, EST157481, GOUT1, MRX, MXR, MXR1, UAQTL1 | ATP-binding cassette, sub-family G (WHITE), member 2 |
C00000947
C00000958
C00002903
|
| 2932 | GSK3B | glycogen synthase kinase 3 beta (EC:2.7.11.1 2.7.11.26) |
C00000958
C00002903
C00009077
|
| 2896 | GRN, CLN11, GEP, GP88, PCDGF, PEPI, PGRN | granulin |
C00000947
C00000958
C00002903
|
| 4780 | NFE2L2, NRF2 | nuclear factor, erythroid 2-like 2 |
C00000947
C00000958
C00002903
|
| 4790 | NFKB1, EBP-1, KBF1, NF-kB1, NF-kappa-B, NF-kappaB, NFKB-p105, NFKB-p50, NFkappaB, p105, p50 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
C00000958
C00002902
C00002903
|
| 2100 | ESR2, ER-BETA, ESR-BETA, ESRB, ESTRB, Erb, NR3A2 | estrogen receptor 2 (ER beta) |
C00000958
C00001180
C00002903
|
| 4793 | NFKBIB, IKBB, TRIP9 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
C00000958
C00002647
C00002903
|
| 142 | PARP1, ADPRT, ADPRT_1, ADPRT1, ARTD1, PARP, PARP-1, PPOL, pADPRT-1 | poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) |
C00000958
C00002902
C00002903
|
| 5327 | PLAT, T-PA, TPA | plasminogen activator, tissue (EC:3.4.21.68) |
C00000947
C00000958
C00002903
|
| 5328 | PLAU, ATF, BDPLT5, QPD, UPA, URK, u-PA | plasminogen activator, urokinase (EC:3.4.21.73) |
C00000947
C00000958
C00002903
|
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative |
C00000947
C00000958
C00002903
|
| 5468 | PPARG, CIMT1, GLM1, NR1C3, PPARG1, PPARG2, PPARgamma | peroxisome proliferator-activated receptor gamma |
C00000958
C00001180
C00002903
|
| 5742 | PTGS1, COX1, COX3, PCOX1, PES-1, PGG/HS, PGHS-1, PGHS1, PHS1, PTGHS | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (EC:1.14.99.1) |
C00000958
C00001180
C00002903
|
| 2002 | ELK1 | ELK1, member of ETS oncogene family |
C00000958
C00001180
C00002902
|
| 1956 | EGFR, ERBB, ERBB1, HER1, PIG61, mENA | epidermal growth factor receptor (EC:2.7.10.1) |
C00000958
C00002902
C00002903
|
| 5747 | PTK2, FADK, FAK, FAK1, FRNK, PPP1R71, p125FAK, pp125FAK | protein tyrosine kinase 2 (EC:2.7.10.2) |
C00000947
C00000958
C00002903
|
| 1950 | EGF, HOMG4, URG | epidermal growth factor |
C00000958
C00002902
C00002903
|
| 598 | BCL2L1, BCL-XL/S, BCL2L, BCLX, BCLXL, BCLXS, Bcl-X, PPP1R52, bcl-xL, bcl-xS | BCL2-like 1 |
C00000958
C00002902
C00002903
|
| 1786 | DNMT1, ADCADN, AIM, CXXC9, DNMT, HSN1E, MCMT | DNA (cytosine-5-)-methyltransferase 1 (EC:2.1.1.37) |
C00000947
C00000958
C00002903
|
| 3627 | CXCL10, C7, IFI10, INP10, IP-10, SCYB10, crg-2, gIP-10, mob-1 | chemokine (C-X-C motif) ligand 10 |
C00000947
C00000958
C00002903
|
| 578 | BAK1, BAK, BAK-LIKE, BCL2L7, CDN1 | BCL2-antagonist/killer 1 |
C00000958
C00002902
C00002903
|
| 56616 | DIABLO, DFNA64, SMAC | diablo, IAP-binding mitochondrial protein |
C00000958
C00002902
C00002903
|
| 472 | ATM, AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | ataxia telangiectasia mutated (EC:2.7.11.1) |
C00000947
C00002647
C00002903
|
| 545 | ATR, FCTCS, FRP1, MEC1, SCKL, SCKL1 | ataxia telangiectasia and Rad3 related (EC:2.7.11.1) |
C00000958
C00002647
C00002903
|
| 1545 | CYP1B1, CP1B, CYPIB1, GLC3A, P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 (EC:1.14.14.1) |
C00000958
C00002903
C00002905
|
| 6772 | STAT1, CANDF7, ISGF-3, STAT91 | signal transducer and activator of transcription 1, 91kDa |
C00000947
C00000958
C00002903
|
| 6774 | STAT3, APRF, HIES | signal transducer and activator of transcription 3 (acute-phase response factor) |
C00000958
C00002902
C00002903
|
| 405 | ARNT, HIF-1-beta, HIF-1beta, HIF1-beta, HIF1B, HIF1BETA, TANGO, bHLHe2 | aryl hydrocarbon receptor nuclear translocator |
C00000100
C00000958
C00002903
|
| 4363 | ABCC1, ABC29, ABCC, GS-X, MRP, MRP1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
C00002903
C00002905
C00015693
|
| 196 | AHR, bHLHe76 | aryl hydrocarbon receptor |
C00000100
C00000958
C00002903
|
| 1649 | DDIT3, CEBPZ, CHOP, CHOP-10, CHOP10, GADD153 | DNA-damage-inducible transcript 3 |
C00000958
C00002902
C00002903
|
| 7422 | VEGFA, MVCD1, VEGF, VPF | vascular endothelial growth factor A |
C00000958
C00002647
C00002903
|
| 4240 | MFGE8, BA46, EDIL1, HMFG, HsT19888, MFG-E8, MFGM, OAcGD3S, SED1, SPAG10, hP47 | milk fat globule-EGF factor 8 protein |
C00000958
C00002903
C00009077
|
| 51101 | ZC2HC1A, C8orf70, FAM164A | zinc finger, C2HC-type containing 1A |
C00000958
C00002903
|
| 11169 | WDHD1, AND-1, CHTF4, CTF4 | WD repeat and HMG-box DNA binding protein 1 |
C00000958
C00002903
|
| 7453 | WARS, GAMMA-2, IFI53, IFP53 | tryptophanyl-tRNA synthetase (EC:6.1.1.2) |
C00000958
C00002903
|
| 7443 | VRK1, PCH1, PCH1A | vaccinia related kinase 1 (EC:2.7.11.1) |
C00000958
C00002903
|
| 7586 | ZKSCAN1, 9130423L19Rik, KOX18, PHZ-37, ZNF139, ZNF36, ZSCAN33 | zinc finger with KRAB and SCAN domains 1 |
C00000958
C00002903
|
| 317 | APAF1, APAF-1, CED4 | apoptotic peptidase activating factor 1 |
C00000958
C00002903
|
| 79609 | VCPKMT, C14orf138, METTL21D, VCP-KMT | valosin containing protein lysine (K) methyltransferase |
C00000958
C00002903
|
| 7402 | UTRN, DMDL, DRP, DRP1 | utrophin |
C00000958
C00002903
|
| 7357 | UGCG, GCS, GLCT1 | UDP-glucose ceramide glucosyltransferase (EC:2.4.1.80) |
C00000958
C00002903
|
| 55075 | UACA, NUCLING | uveal autoantigen with coiled-coil domains and ankyrin repeats |
C00000958
C00002903
|
| 358 | AQP1, AQP-CHIP, CHIP28, CO | aquaporin 1 |
C00000958
C00002903
|
| 360 | AQP3, AQP-3, GIL | aquaporin 3 (Gill blood group) |
C00000958
C00002903
|
| 7306 | TYRP1, CAS2, CATB, GP75, OCA3, TRP, TRP1, TYRP, b-PROTEIN | tyrosinase-related protein 1 (EC:1.14.18.-) |
C00000958
C00002903
|
| 367 | AR, AIS, DHTR, HUMARA, HYSP1, KD, NR3C4, SBMA, SMAX1, TFM | androgen receptor |
C00000958
C00002903
|
| 7295 | TXN, TRDX, TRX, TRX1 | thioredoxin |
C00000958
C00002903
|
| 7272 | TTK, CT96, ESK, MPH1, MPS1, MPS1L1, PYT | TTK protein kinase (EC:2.7.12.1) |
C00000958
C00002903
|
| 9319 | TRIP13, 16E1BP | thyroid hormone receptor interactor 13 |
C00000958
C00002903
|
| 286827 | TRIM59, MRF1, RNF104, TRIM57, TSBF1 | tripartite motif containing 59 |
C00000958
C00002903
|
| 55809 | TRERF1, BCAR2, HSA277276, RAPA, RP1-139D8.5, TREP132, TReP-132, dJ139D8.5 | transcriptional regulating factor 1 |
C00000958
C00002903
|
| 7164 | TPD52L1, D53, hD53 | tumor protein D52-like 1 |
C00000958
C00002903
|
| 7750 | ZMYM2, FIM, MYM, RAMP, SCLL, ZNF198 | zinc finger, MYM-type 2 |
C00000958
C00002903
|
| 195828 | ZNF367, AFF29, CDC14B, ZFF29 | zinc finger protein 367 |
C00000958
C00002903
|
| 8743 | TNFSF10, APO2L, Apo-2L, CD253, TL2, TRAIL | tumor necrosis factor (ligand) superfamily, member 10 |
C00000958
C00002903
|
| 25 | ABL1, ABL, JTK7, bcr/abl, c-ABL, p150, v-abl | c-abl oncogene 1, non-receptor tyrosine kinase (EC:2.7.10.2) |
C00002647
C00002903
|
| 56255 | TMX4, DJ971N18.2, PDIA14, TXNDC13 | thioredoxin-related transmembrane protein 4 |
C00000958
C00002903
|
| 84187 | TMEM164, bB360B22.3 | transmembrane protein 164 |
C00000958
C00002903
|
| 26277 | TINF2, DKCA3, TIN2 | TERF1 (TRF1)-interacting nuclear factor 2 |
C00000958
C00002903
|
| 7078 | TIMP3, HSMRK222, K222, K222TA2, SFD | TIMP metallopeptidase inhibitor 3 |
C00000958
C00002903
|
| 7050 | TGIF1, HPE4, TGIF | TGFB-induced factor homeobox 1 |
C00000958
C00002903
|
| 7046 | TGFBR1, AAT5, ACVRLK4, ALK-5, ALK5, LDS1A, LDS2A, MSSE, SKR4, TGFR-1 | transforming growth factor, beta receptor 1 (EC:2.7.11.30) |
C00000958
C00002903
|
| 7019 | TFAM, MTTF1, MTTFA, TCF6, TCF6L1, TCF6L2, TCF6L3 | transcription factor A, mitochondrial |
C00000958
C00002903
|
| 93081 | TEX30, C13orf27 | testis expressed 30 |
C00000958
C00002903
|
| 7015 | TERT, CMM9, DKCA2, DKCB4, EST2, PFBMFT1, TCS1, TP2, TRT, hEST2, hTRT | telomerase reverse transcriptase (EC:2.7.7.49) |
C00000958
C00002903
|
| 55775 | TDP1 | tyrosyl-DNA phosphodiesterase 1 (EC:3.1.4.-) |
C00000958
C00002903
|
| 6925 | TCF4, E2-2, ITF-2, ITF2, PTHS, SEF-2, SEF2, SEF2-1, SEF2-1A, SEF2-1B, TCF-4, bHLHb19 | transcription factor 4 |
C00000958
C00002903
|
| 94121 | SYTL4, SLP4 | synaptotagmin-like 4 |
C00000958
C00002903
|
| 23012 | STK38L, NDR2 | serine/threonine kinase 38 like (EC:2.7.11.1) |
C00000958
C00002903
|
| 9263 | STK17A, DRAK1 | serine/threonine kinase 17a (EC:2.7.11.1) |
C00000958
C00002903
|
| 302 | ANXA2, ANX2, ANX2L4, CAL1H, LIP2, LPC2, LPC2D, P36, PAP-IV | annexin A2 |
C00000947
C00002903
|
| 338 | APOB, FLDB, LDLCQ4 | apolipoprotein B |
C00002647
C00002903
|
| 6749 | SSRP1, FACT, FACT80, T160 | structure specific recognition protein 1 |
C00000958
C00002903
|
| 6733 | SRPK2, SFRSK2 | SRSF protein kinase 2 (EC:2.7.11.1) |
C00000958
C00001180
|
| 6720 | SREBF1, SREBP-1c, SREBP1, bHLHd1 | sterol regulatory element binding transcription factor 1 |
C00000958
C00002903
|
| 6716 | SRD5A2 | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) (EC:1.3.1.22) |
C00000958
C00002903
|
| 58472 | SQRDL, PRO1975 | sulfide quinone reductase-like (yeast) |
C00000958
C00002903
|
| 259266 | ASPM, ASP, Calmbp1, MCPH5 | asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
C00000958
C00002903
|
| 6713 | SQLE | squalene epoxidase (EC:1.14.13.132) |
C00000958
C00002903
|
| 147841 | SPC24, SPBC24 | SPC24, NDC80 kinetochore complex component |
C00000958
C00002903
|
| 467 | ATF3 | activating transcription factor 3 |
C00000958
C00002903
|
| 468 | ATF4, CREB-2, CREB2, TAXREB67, TXREB | activating transcription factor 4 |
C00000947
C00000958
|
| 6672 | SP100, lysp100b | SP100 nuclear antigen |
C00000958
C00002903
|
| 6649 | SOD3, EC-SOD | superoxide dismutase 3, extracellular (EC:1.15.1.1) |
C00000958
C00002903
|
| 1386 | ATF2, CRE-BP1, CREB2, HB16, TREB7 | activating transcription factor 2 (EC:2.3.1.48) |
C00002902
C00002903
|
| 6615 | SNAI1, SLUGH2, SNA, SNAH, SNAIL, SNAIL1, dJ710H13.1 | snail family zinc finger 1 |
C00000958
C00002903
|
| 9474 | ATG5, APG5, APG5-LIKE, APG5L, ASP, hAPG5 | autophagy related 5 |
C00000958
C00002903
|
| 6607 | SMN2, BCD541, C-BCD541, GEMIN1, SMNC, TDRD16B | survival of motor neuron 2, centromeric |
C00000958
C00002903
|
| 10592 | SMC2, CAP-E, CAPE, SMC-2, SMC2L1 | structural maintenance of chromosomes 2 |
C00000958
C00002903
|
| 4088 | SMAD3, HSPC193, HsT17436, JV15-2, LDS1C, LDS3, MADH3 | SMAD family member 3 |
C00000958
C00002903
|
| 4086 | SMAD1, BSP-1, BSP1, JV4-1, JV41, MADH1, MADR1 | SMAD family member 1 |
C00000958
C00002903
|
| 11001 | SLC27A2, ACSVL1, FACVL1, FATP2, HsT17226, VLACS, VLCS, hFACVL1 | solute carrier family 27 (fatty acid transporter), member 2 (EC:6.2.1.3) |
C00000958
C00002903
|
| 1836 | SLC26A2, D5S1708, DTD, DTDST, EDM4, MST153, MSTP157 | solute carrier family 26 (anion exchanger), member 2 |
C00000958
C00002903
|
| 26503 | SLC17A5, AST, ISSD, NSD, SD, SIALIN, SIASD, SLD | solute carrier family 17 (acidic sugar transporter), member 5 |
C00000958
C00002903
|
| 6502 | SKP2, FBL1, FBXL1, FLB1, p45 | S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
C00000958
C00002903
|
| 6470 | SHMT1, CSHMT, SHMT | serine hydroxymethyltransferase 1 (soluble) (EC:2.1.2.1) |
C00000958
C00002903
|
| 151246 | SGOL2, SGO2, TRIPIN | shugoshin-like 2 (S. pombe) |
C00000958
C00002903
|
| 7536 | SF1, BBP, D11S636, MBBP, ZCCHC25, ZFM1, ZNF162 | splicing factor 1 |
C00000947
C00000958
|
| 9949 | AMMECR1, AMMERC1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
C00000958
C00002903
|
| 6401 | SELE, CD62E, ELAM, ELAM1, ESEL, LECAM2 | selectin E |
C00000958
C00002903
|
| 6319 | SCD, FADS5, MSTP008, SCD1, SCDOS | stearoyl-CoA desaturase (delta-9-desaturase) (EC:1.14.19.1) |
C00000958
C00002903
|
| 4287 | ATXN3, AT3, ATX3, JOS, MJD, MJD1, SCA3 | ataxin 3 (EC:3.4.19.12) |
C00000958
C00002903
|
| 6256 | RXRA, NR2B1 | retinoid X receptor, alpha |
C00000958
C00002903
|
| 6790 | AURKA, AIK, ARK1, AURA, AURORA2, BTAK, PPP1R47, STK15, STK6, STK7 | aurora kinase A (EC:2.7.11.1) |
C00000958
C00002903
|
| 860 | RUNX2, AML3, CBF-alpha-1, CBFA1, CCD, CCD1, CLCD, OSF-2, OSF2, PEA2aA, PEBP2aA | runt-related transcription factor 2 |
C00000958
C00002903
|
| 9252 | RPS6KA5, MSK1, MSPK1, RLPK | ribosomal protein S6 kinase, 90kDa, polypeptide 5 (EC:2.7.11.1) |
C00000958
C00002647
|
| 6189 | RPS3A, FTE1, MFTL, S3A | ribosomal protein S3A |
C00000958
C00002903
|
| 79102 | RNF26 | ring finger protein 26 |
C00000958
C00002903
|
| 10535 | RNASEH2A, AGS4, JUNB, RNASEHI, RNHIA, RNHL | ribonuclease H2, subunit A (EC:3.1.26.4) |
C00000958
C00002903
|
| 8780 | RIOK3, SUDD | RIO kinase 3 (EC:2.7.11.1) |
C00000958
C00002903
|
| 9886 | RHOBTB1 | Rho-related BTB domain containing 1 |
C00000958
C00002903
|
| 9532 | BAG2, BAG-2, dJ417I1.2 | BCL2-associated athanogene 2 |
C00000958
C00002903
|
| 567 | B2M | beta-2-microglobulin |
C00001180
C00002903
|
| 8434 | RECK, ST15 | reversion-inducing-cysteine-rich protein with kazal motifs |
C00000958
C00002903
|
| 5937 | RBMS1, C2orf12, HCC-4, MSSP, MSSP-1, MSSP-2, MSSP-3, SCR2, YC1 | RNA binding motif, single stranded interacting protein 1 |
C00000958
C00002903
|
| 613 | BCR, ALL, BCR1, CML, D22S11, D22S662, PHL | breakpoint cluster region (EC:2.7.11.1) |
C00002647
C00002903
|
| 637 | BID, FP497 | BH3 interacting domain death agonist |
C00000947
C00002903
|
| 5925 | RB1, OSRC, RB, p105-Rb, pRb, pp110 | retinoblastoma 1 |
C00000958
C00002903
|
| 5915 | RARB, HAP, NR1B2, RRB2 | retinoic acid receptor, beta |
C00000947
C00000958
|
| 5914 | RARA, NR1B1, RAR | retinoic acid receptor, alpha |
C00000958
C00002903
|
| 5889 | RAD51C, BROVCA3, FANCO, R51H3, RAD51L2 | RAD51 paralog C |
C00000958
C00002903
|
| 5888 | RAD51, BRCC5, HRAD51, HsRad51, HsT16930, MRMV2, RAD51A, RECA | RAD51 recombinase |
C00000958
C00002903
|
| 10111 | RAD50, NBSLD, RAD502, hRad50 | RAD50 homolog (S. cerevisiae) |
C00000958
C00002903
|
| 840 | CASP7, CASP-7, CMH-1, ICE-LAP3, LICE2, MCH3 | caspase 7, apoptosis-related cysteine peptidase (EC:3.4.22.60) |
C00000947
C00002903
|
| 898 | CCNE1, CCNE | cyclin E1 |
C00002647
C00002903
|
| 602 | BCL3, BCL4, D19S37 | B-cell CLL/lymphoma 3 |
C00000958
C00002903
|
| 604 | BCL6, BCL5, BCL6A, LAZ3, ZBTB27, ZNF51 | B-cell CLL/lymphoma 6 |
C00000958
C00002903
|
| 998 | CDC42, CDC42Hs, G25K | cell division cycle 42 |
C00002647
C00002903
|
| 11031 | RAB31, Rab22B | RAB31, member RAS oncogene family |
C00000958
C00002903
|
| 5782 | PTPN12, PTP-PEST, PTPG1 | protein tyrosine phosphatase, non-receptor type 12 (EC:3.1.3.48) |
C00000958
C00002903
|
| 1147 | CHUK, IKBKA, IKK-alpha, IKK1, IKKA, NFKBIKA, TCF16 | conserved helix-loop-helix ubiquitous kinase (EC:2.7.11.10) |
C00002647
C00002903
|
| 330 | BIRC3, AIP1, API2, CIAP2, HAIP1, HIAP1, MALT2, MIHC, RNF49, c-IAP2 | baculoviral IAP repeat containing 3 |
C00000958
C00002903
|
| 64581 | CLEC7A, BGR, CANDF4, CLECSF12, DECTIN1 | C-type lectin domain family 7, member A |
C00001180
C00002903
|
| 7852 | CXCR4, CD184, D2S201E, FB22, HM89, HSY3RR, LAP3, LCR1, LESTR, NPY3R, NPYR, NPYRL, NPYY3R, WHIM | chemokine (C-X-C motif) receptor 4 |
C00002647
C00002903
|
| 5731 | PTGER1, EP1 | prostaglandin E receptor 1 (subtype EP1), 42kDa |
C00000958
C00002903
|
| 84722 | PSRC1, DDA3, FP3214 | proline/serine-rich coiled-coil 1 |
C00000958
C00002903
|
| 11168 | PSIP1, DFS70, LEDGF, PAIP, PSIP2, p52, p75 | PC4 and SFRS1 interacting protein 1 |
C00000958
C00002903
|
| 11098 | PRSS23, SIG13, SPUVE, ZSIG13 | protease, serine, 23 |
C00000958
C00002903
|
| 5587 | PRKD1, PKC-MU, PKCM, PKD, PRKCM | protein kinase D1 (EC:2.7.11.13) |
C00000958
C00002903
|
| 79866 | BORA, C13orf34 | bora, aurora kinase A activator |
C00000958
C00002903
|
| 5558 | PRIM2, PRIM2A, p58 | primase, DNA, polypeptide 2 (58kDa) (EC:2.7.7.-) |
C00000958
C00002903
|
| 672 | BRCA1, BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | breast cancer 1, early onset |
C00000958
C00002903
|
| 675 | BRCA2, BRCC2, BROVCA2, FACD, FAD, FAD1, FANCB, FANCD, FANCD1, GLM3, PNCA2 | breast cancer 2, early onset |
C00000958
C00002903
|
| 5557 | PRIM1, p49 | primase, DNA, polypeptide 1 (49kDa) (EC:2.7.7.7) |
C00000958
C00002903
|
| 1544 | CYP1A2, CP12, P3-450, P450(PA) | cytochrome P450, family 1, subfamily A, polypeptide 2 (EC:1.14.14.1) |
C00002903
C00002905
|
| 11201 | POLI, RAD30B, RAD3OB | polymerase (DNA directed) iota (EC:2.7.7.7) |
C00000958
C00002903
|
| 5427 | POLE2, DPE2 | polymerase (DNA directed), epsilon 2, accessory subunit (EC:2.7.7.7) |
C00000958
C00002903
|
| 23649 | POLA2 | polymerase (DNA directed), alpha 2, accessory subunit |
C00000958
C00002903
|
| 4860 | PNP, NP, PRO1837, PUNP | purine nucleoside phosphorylase (EC:2.4.2.1) |
C00000958
C00002903
|
| 1576 | CYP3A4, CP33, CP34, CYP3A, CYP3A3, CYPIIIA3, CYPIIIA4, HLP, NF-25, P450C3, P450PCN1 | cytochrome P450, family 3, subfamily A, polypeptide 4 (EC:1.14.13.67 1.14.13.97 1.14.13.32 1.14.13.157) |
C00002903
C00002905
|
| 5378 | PMS1, HNPCC3, PMSL1, hPMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
C00000958
C00002903
|
| 5376 | PMP22, CMT1A, CMT1E, DSS, GAS-3, HMSNIA, HNPP, Sp110 | peripheral myelin protein 22 |
C00000958
C00002903
|
| 5366 | PMAIP1, APR, NOXA | phorbol-12-myristate-13-acetate-induced protein 1 |
C00000958
C00002903
|
| 8985 | PLOD3, LH3 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 (EC:1.14.11.4) |
C00000958
C00002903
|
| 699 | BUB1, BUB1A, BUB1L, hBUB1 | BUB1 mitotic checkpoint serine/threonine kinase (EC:2.7.11.1) |
C00000958
C00002903
|
| 54477 | PLEKHA5, PEPP-2, PEPP2 | pleckstrin homology domain containing, family A member 5 |
C00000958
C00002903
|
| 5337 | PLD1 | phospholipase D1, phosphatidylcholine-specific (EC:3.1.4.4) |
C00000958
C00002903
|
| 7913 | DEK, D6S231E | DEK oncogene |
C00000947
C00002903
|
| 355 | FAS, ALPS1A, APO-1, APT1, CD95, FAS1, FASTM, TNFRSF6 | Fas cell surface death receptor |
C00000947
C00002903
|
| 5586 | PKN2, PAK2, PRK2, PRKCL2, PRO2042, Pak-2 | protein kinase N2 (EC:2.7.11.13) |
C00000958
C00002903
|
| 5291 | PIK3CB, P110BETA, PI3K, PI3KBETA, PIK3C1 | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta (EC:2.7.1.153) |
C00000958
C00002903
|
| 84419 | C15orf48, FOAP-11, NMES1 | chromosome 15 open reading frame 48 |
C00000958
C00002903
|
| 26147 | PHF19, MTF2L1, PCL3, TDRD19B | PHD finger protein 19 |
C00000958
C00002903
|
| 5241 | PGR, NR3C3, PR | progesterone receptor |
C00000958
C00002903
|
| 5159 | PDGFRB, CD140B, IBGC4, IMF1, JTK12, PDGFR, PDGFR-1, PDGFR1 | platelet-derived growth factor receptor, beta polypeptide (EC:2.7.10.1) |
C00000958
C00002903
|
| 55732 | C1orf112, RP1-97P20.1 | chromosome 1 open reading frame 112 |
C00000958
C00002903
|
| 301164 |
C00000958
C00002903
|
||
| 9131 | AIFM1, AIF, CMTX4, COWCK, COXPD6, PDCD8 | apoptosis-inducing factor, mitochondrion-associated, 1 |
C00000958
C00002903
|
| 4973 | OLR1, CLEC8A, LOX1, LOXIN, SCARE1, SLOX1 | oxidized low density lipoprotein (lectin-like) receptor 1 |
C00000958
C00002903
|
| 11339 | OIP5, 5730547N13Rik, CT86, LINT-25, MIS18B, MIS18beta, hMIS18beta | Opa interacting protein 5 |
C00000958
C00002903
|
| 4953 | ODC1, ODC | ornithine decarboxylase 1 (EC:4.1.1.17) |
C00000958
C00002903
|
| 57122 | NUP107, NUP84 | nucleoporin 107kDa |
C00000958
C00002903
|
| 3084 | NRG1, ARIA, GGF, GGF2, HGL, HRG, HRG1, HRGA, MST131, NDF, SMDF | neuregulin 1 |
C00000958
C00002903
|
| 2908 | NR3C1, GCCR, GCR, GR, GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
C00000958
C00002903
|
| 4846 | NOS3, ECNOS, eNOS | nitric oxide synthase 3 (endothelial cell) (EC:1.14.13.39) |
C00000958
C00002903
|
| 356 | FASLG, ALPS1B, APT1LG1, APTL, CD178, CD95-L, CD95L, FASL, TNFSF6 | Fas ligand (TNF superfamily, member 6) |
C00002647
C00002903
|
| 191 | AHCY, SAHH, adoHcyase | adenosylhomocysteinase (EC:3.3.1.1) |
C00000958
C00002903
|
| 9518 | GDF15, GDF-15, MIC-1, MIC1, NAG-1, PDF, PLAB, PTGFB | growth differentiation factor 15 |
C00002902
C00002903
|
| 2885 | GRB2, ASH, EGFRBP-GRB2, Grb3-3, MST084, MSTP084, NCKAP2 | growth factor receptor-bound protein 2 |
C00002647
C00002903
|
| 55247 | NEIL3, FGP2, FPG2, NEI3, hFPG2, hNEI3 | nei endonuclease VIII-like 3 (E. coli) (EC:4.2.99.18) |
C00000958
C00002903
|
| 4739 | NEDD9, CAS-L, CAS2, CASL, CASS2, HEF1 | neural precursor cell expressed, developmentally down-regulated 9 |
C00000958
C00002903
|
| 10397 | NDRG1, CAP43, CMT4D, DRG-1, DRG1, GC4, HMSNL, NDR1, NMSL, PROXY1, RIT42, RTP, TARG1, TDD5 | N-myc downstream regulated 1 |
C00000958
C00002903
|
| 50854 | C6orf48, D6S57, G8 | chromosome 6 open reading frame 48 |
C00000958
C00002903
|
| 10403 | NDC80, HEC, HEC1, HsHec1, KNTC2, TID3, hsNDC80 | NDC80 kinetochore complex component |
C00000958
C00002903
|
| 135112 | NCOA7, ERAP140, ESNA1, Nbla00052, Nbla10993, TLDC4, dJ187J11.3 | nuclear receptor coactivator 7 |
C00000958
C00002903
|
| 10499 | NCOA2, GRIP1, KAT13C, NCoA-2, SRC2, TIF2, bHLHe75 | nuclear receptor coactivator 2 |
C00000958
C00002903
|
| 8648 | NCOA1, F-SRC-1, KAT13A, RIP160, SRC1, bHLHe42, bHLHe74 | nuclear receptor coactivator 1 (EC:2.3.1.48) |
C00000958
C00002903
|
| 23397 | NCAPH, BRRN1, CAP-H | non-SMC condensin I complex, subunit H |
C00000958
C00002903
|
| 4683 | NBN, AT-V1, AT-V2, ATV, NBS, NBS1, P95 | nibrin |
C00000958
C00002903
|
| 133686 | NADK2, C5orf33, MNADK, NADKD1 | NAD kinase 2, mitochondrial (EC:2.7.1.23) |
C00000958
C00002903
|
| 4430 | MYO1B, myr1 | myosin IB |
C00000958
C00002903
|
| 3014 | H2AFX, H2A.X, H2A/X, H2AX | H2A histone family, member X |
C00000947
C00002903
|
| 9961 | MVP, LRP, VAULT1 | major vault protein |
C00000958
C00002903
|
| 51537 | MTFP1, MTP18 | mitochondrial fission process 1 |
C00000958
C00002903
|
| 4361 | MRE11A, ATLD, HNGS1, MRE11, MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
C00000958
C00002903
|
| 834 | CASP1, ICE, IL1BC, P45 | caspase 1, apoptosis-related cysteine peptidase (EC:3.4.22.36) |
C00000958
C00002903
|
| 3329 | HSPD1, CPN60, GROEL, HLD4, HSP-60, HSP60, HSP65, HuCHA60, SPG13 | heat shock 60kDa protein 1 (chaperonin) |
C00000947
C00002903
|
| 839 | CASP6, MCH2 | caspase 6, apoptosis-related cysteine peptidase (EC:3.4.22.59) |
C00000958
C00002903
|
| 3383 | ICAM1, BB2, CD54, P3.58 | intercellular adhesion molecule 1 |
C00000947
C00002903
|
| 3458 | IFNG, IFG, IFI | interferon, gamma |
C00002902
C00002903
|
| 847 | CAT | catalase (EC:1.11.1.6) |
C00000958
C00002903
|
| 10198 | MPHOSPH9, MPP-9, MPP9 | M-phase phosphoprotein 9 |
C00000958
C00002903
|
| 4316 | MMP7, MMP-7, MPSL1, PUMP-1 | matrix metallopeptidase 7 (matrilysin, uterine) (EC:3.4.24.23) |
C00000958
C00002647
|
| 4312 | MMP1, CLG, CLGN | matrix metallopeptidase 1 (interstitial collagenase) (EC:3.4.24.7) |
C00000958
C00002903
|
| 3586 | IL10, CSIF, GVHDS, IL-10, IL10A, TGIF | interleukin 10 |
C00000947
C00002903
|
| 55669 | MFN1, hfzo1, hfzo2 | mitofusin 1 |
C00000958
C00002903
|
| 3606 | IL18, IGIF, IL-18, IL-1g, IL1F4 | interleukin 18 (interferon-gamma-inducing factor) |
C00001180
C00002903
|
| 92342 | METTL18, AsTP2, C1orf156, HPM1 | methyltransferase like 18 |
C00000958
C00002903
|
| 4233 | MET, AUTS9, HGFR, RCCP2, c-Met | met proto-oncogene (EC:2.7.10.1) |
C00000958
C00002903
|
| 9833 | MELK, HPK38 | maternal embryonic leucine zipper kinase (EC:2.7.11.1 2.7.10.2) |
C00000958
C00002903
|
| 3683 | ITGAL, CD11A, LFA-1, LFA1A | integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
C00000947
C00002903
|
| 4157 | MC1R, CMM5, MSH-R, SHEP2 | melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
C00000958
C00002903
|
| 4144 | MAT2A, MATA2, MATII, SAMS2 | methionine adenosyltransferase II, alpha (EC:2.5.1.6) |
C00000958
C00002903
|
| 133 | ADM, AM | adrenomedullin |
C00000958
C00002903
|
| 3689 | ITGB2, CD18, LAD, LCAMB, LFA-1, MAC-1, MF17, MFI7 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
C00000947
C00002903
|
| 4313 | MMP2, CLG4, CLG4A, MMP-II, MONA, TBE-1 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) |
C00002647
C00002903
|
| 653361 | NCF1, NCF1A, NOXO2, SH3PXD1A, p47phox | neutrophil cytosolic factor 1 |
C00000947
C00002903
|
| 100286684 |
C00000958
C00002903
|
||
| 4843 | NOS2, HEP-NOS, INOS, NOS, NOS2A | nitric oxide synthase 2, inducible (EC:1.14.13.39) |
C00002902
C00002903
|
| 79694 | MANEA, ENDO, hEndo | mannosidase, endo-alpha (EC:3.2.1.130) |
C00000958
C00002903
|
| 130574 | LYPD6 | LY6/PLAUR domain containing 6 |
C00000958
C00002903
|
| 9208 | LRRFIP1, FLAP-1, FLAP1, FLIIAP1, GCF-2, GCF2, HUFI-1, TRIP | leucine rich repeat (in FLII) interacting protein 1 |
C00000958
C00002903
|
| 54839 | LRRC49, PGs4 | leucine rich repeat containing 49 |
C00000958
C00002903
|
| 122769 | LRR1, 4-1BBLRR, LRR-1, PPIL5 | leucine rich repeat protein 1 |
C00000958
C00002903
|
| 4851 | NOTCH1, TAN1, hN1 | notch 1 |
C00000947
C00002903
|
| 891 | CCNB1, CCNB | cyclin B1 |
C00000958
C00002903
|
| 4001 | LMNB1, ADLD, LMN, LMN2, LMNB | lamin B1 |
C00000958
C00002903
|
| 8856 | NR1I2, BXR, ONR1, PAR, PAR1, PAR2, PARq, PRR, PXR, SAR, SXR | nuclear receptor subfamily 1, group I, member 2 |
C00002902
C00002903
|
| 9970 | NR1I3, CAR, CAR1, MB67 | nuclear receptor subfamily 1, group I, member 3 |
C00002902
C00002903
|
| 9735 | KNTC1, ROD | kinetochore associated 1 |
C00000958
C00002903
|
| 90417 | KNSTRN, C15orf23, SKAP, TRAF4AF1 | kinetochore-localized astrin/SPAG5 binding protein |
C00000958
C00002903
|
| 9314 | KLF4, EZF, GKLF | Kruppel-like factor 4 (gut) |
C00000958
C00002903
|
| 9365 | KL | klotho (EC:3.2.1.31) |
C00000958
C00002903
|
| 81930 | KIF18A, MS-KIF18A | kinesin family member 18A |
C00000958
C00002903
|
| 9928 | KIF14 | kinesin family member 14 |
C00000958
C00002903
|
| 3791 | KDR, CD309, FLK1, VEGFR, VEGFR2 | kinase insert domain receptor (a type III receptor tyrosine kinase) (EC:2.7.10.1) |
C00000958
C00002903
|
| 929 | CD14 | CD14 molecule |
C00000958
C00002903
|
| 84056 | KATNAL1 | katanin p60 subunit A-like 1 (EC:3.6.4.3) |
C00000958
C00002903
|
| 8654 | PDE5A, CGB-PDE, CN5A, PDE5 | phosphodiesterase 5A, cGMP-specific (EC:3.1.4.35) |
C00002903
C00006735
|
| 993 | CDC25A, CDC25A2 | cell division cycle 25A (EC:3.1.3.48) |
C00000958
C00002903
|
| 995 | CDC25C, CDC25, PPP1R60 | cell division cycle 25C (EC:3.1.3.48) |
C00000958
C00002903
|
| 996 | CDC27, ANAPC3, APC3, CDC27Hs, D0S1430E, D17S978E, HNUC, NUC2 | cell division cycle 27 |
C00000958
C00002903
|
| 3717 | JAK2, JTK10, THCYT3 | Janus kinase 2 (EC:2.7.10.2) |
C00000958
C00002902
|
| 23421 | ITGB3BP, CENP-R, CENPR, HSU37139, NRIF3, TAP20 | integrin beta 3 binding protein (beta3-endonexin) |
C00000958
C00002903
|
| 3685 | ITGAV, CD51, MSK8, VNRA, VTNR | integrin, alpha V |
C00000958
C00002903
|
| 83461 | CDCA3, GRCC8, TOME-1 | cell division cycle associated 3 |
C00000958
C00002903
|
| 3638 | INSIG1, CL-6, CL6 | insulin induced gene 1 |
C00000958
C00002903
|
| 1019 | CDK4, CMM3, PSK-J3 | cyclin-dependent kinase 4 (EC:2.7.11.22) |
C00000958
C00002903
|
| 196294 | IMMP1L, IMP1, IMP1-LIKE | IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
C00000958
C00002903
|
| 55755 | CDK5RAP2, C48, Cep215, MCPH3 | CDK5 regulatory subunit associated protein 2 |
C00000958
C00002903
|
| 1021 | CDK6, PLSTIRE | cyclin-dependent kinase 6 (EC:2.7.11.22) |
C00000958
C00002903
|
| 6792 | CDKL5, EIEE2, ISSX, STK9 | cyclin-dependent kinase-like 5 (EC:2.7.11.22) |
C00000958
C00002903
|
| 47 | ACLY, ACL, ATPCL, CLATP | ATP citrate lyase (EC:2.3.3.8) |
C00000958
C00002903
|
| 5175 | PECAM1, CD31, CD31/EndoCAM, GPIIA', PECA1, PECAM-1, endoCAM | platelet/endothelial cell adhesion molecule 1 |
C00000947
C00002903
|
| 1029 | CDKN2A, ARF, CDK4I, CDKN2, CMM2, INK4, INK4A, MLM, MTS-1, MTS1, P14, P14ARF, P16, P16-INK4A, P16INK4, P16INK4A, P19, P19ARF, TP16 | cyclin-dependent kinase inhibitor 2A |
C00000958
C00002903
|
| 5578 | PRKCA, AAG6, PKC-alpha, PKCA, PRKACA | protein kinase C, alpha (EC:2.7.11.13) |
C00002647
C00002903
|
| 3575 | IL7R, CD127, CDW127, IL-7R-alpha, IL7RA, ILRA | interleukin 7 receptor |
C00000958
C00002903
|
| 1052 | CEBPD, C/EBP-delta, CELF, CRP3, NF-IL6-beta | CCAAT/enhancer binding protein (C/EBP), delta |
C00000958
C00002903
|
| 7916 | PRRC2A, BAT2, D6S51, D6S51E, G2 | proline-rich coiled-coil 2A |
C00000947
C00002903
|
| 1058 | CENPA, CENP-A, CenH3 | centromere protein A |
C00000958
C00002903
|
| 1062 | CENPE, CENP-E, KIF10, PPP1R61 | centromere protein E, 312kDa |
C00000958
C00002903
|
| 3558 | IL2, IL-2, TCGF, lymphokine | interleukin 2 |
C00000958
C00002903
|
| 6195 | RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1) |
C00002647
C00002903
|
| 3552 | IL1A, IL-1A, IL1, IL1-ALPHA, IL1F1 | interleukin 1, alpha |
C00000958
C00002903
|
| 145508 | CEP128, C14orf145, C14orf61, LEDP/132 | centrosomal protein 128kDa |
C00000958
C00002903
|
| 8517 | IKBKG, AMCBX1, FIP-3, FIP3, Fip3p, IKK-gamma, IP, IP1, IP2, IPD2, NEMO, ZC2HC9 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
C00000958
C00002903
|
| 3551 | IKBKB, IKK-beta, IKK2, IKKB, NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (EC:2.7.11.10) |
C00000958
C00002647
|
| 3481 | IGF2, C11orf43, IGF-II, PP9974 | insulin-like growth factor 2 (somatomedin A) |
C00000958
C00002903
|
| 3480 | IGF1R, CD221, IGFIR, IGFR, JTK13 | insulin-like growth factor 1 receptor (EC:2.7.10.1) |
C00000958
C00002903
|
| 3479 | IGF1, IGF-I, IGF1A, IGFI | insulin-like growth factor 1 (somatomedin C) |
C00000958
C00002903
|
| 9592 | IER2, ETR101 | immediate early response 2 |
C00000958
C00002903
|
| 219844 | HYLS1, HLS | hydrolethalus syndrome 1 |
C00000958
C00002903
|
| 8837 | CFLAR, CASH, CASP8AP1, CLARP, Casper, FLAME, FLAME-1, FLAME1, FLIP, I-FLICE, MRIT, c-FLIP, c-FLIPL, c-FLIPR, c-FLIPS | CASP8 and FADD-like apoptosis regulator |
C00000958
C00002903
|
| 3304 | HSPA1B, HSP70-1B, HSP70-2 | heat shock 70kDa protein 1B |
C00000958
C00002903
|
| 494143 | CHAC2 | ChaC, cation transport regulator homolog 2 (E. coli) |
C00000958
C00002903
|
| 6301 | SARS, SERRS, SERS | seryl-tRNA synthetase (EC:6.1.1.11) |
C00002902
C00002903
|
| 3161 | HMMR, CD168, IHABP, RHAMM | hyaluronan-mediated motility receptor (RHAMM) |
C00000958
C00002903
|
| 3157 | HMGCS1, HMGCS | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) (EC:2.3.3.10) |
C00000958
C00002903
|
| 3146 | HMGB1, HMG1, HMG3, SBP-1 | high mobility group box 1 |
C00000958
C00002903
|
| 3099 | HK2, HKII, HXK2 | hexokinase 2 (EC:2.7.1.1) |
C00000958
C00002903
|
| 3096 | HIVEP1, CIRIP, CRYBP1, MBP-1, PRDII-BF1, Schnurri-1, ZAS1, ZNF40, ZNF40A | human immunodeficiency virus type I enhancer binding protein 1 |
C00000958
C00002903
|
| 8334 | HIST1H2AC, H2A/l, H2AFL, dJ221C16.4 | histone cluster 1, H2ac |
C00000958
C00002903
|
| 3091 | HIF1A, HIF-1A, HIF-1alpha, HIF1, HIF1-ALPHA, MOP1, PASD8, bHLHe78 | hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
C00000958
C00002903
|
| 3082 | HGF, DFNB39, F-TCF, HGFB, HPTA, SF | hepatocyte growth factor (hepapoietin A; scatter factor) |
C00000958
C00002903
|
| 3280 | HES1, HES-1, HHL, HRY, bHLHb39 | hes family bHLH transcription factor 1 |
C00000958
C00002903
|
| 1164 | CKS2, CKSHS2 | CDC28 protein kinase regulatory subunit 2 |
C00000958
C00002903
|
| 3036 | HAS1, HAS | hyaluronan synthase 1 (EC:2.4.1.212) |
C00000958
C00002903
|
| 2950 | GSTP1, DFN7, FAEES3, GST3, GSTP, PI | glutathione S-transferase pi 1 (EC:2.5.1.18) |
C00000958
C00002903
|
| 2941 | GSTA4, GSTA4-4, GTA4 | glutathione S-transferase alpha 4 (EC:2.5.1.18) |
C00000958
C00002903
|
| 9619 | ABCG1, ABC8, WHITE1 | ATP-binding cassette, sub-family G (WHITE), member 1 |
C00000958
C00002903
|
| 2931 | GSK3A | glycogen synthase kinase 3 alpha (EC:2.7.11.1 2.7.11.26) |
C00000958
C00002903
|
| 5054 | SERPINE1, PAI, PAI-1, PAI1, PLANH1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
C00000947
C00002903
|
| 2876 | GPX1, GPXD, GSHPX1 | glutathione peroxidase 1 (EC:1.11.1.9) |
C00000958
C00002903
|
| 10978 | CLP1, HEAB, hClp1 | cleavage and polyadenylation factor I subunit 1 (EC:2.7.1.78) |
C00000947
C00000958
|
| 29899 | GPSM2, CMCS, DFNB82, LGN, PINS | G-protein signaling modulator 2 |
C00000958
C00002903
|
| 51022 | GLRX2, CGI-133, GRX2 | glutaredoxin 2 |
C00000958
C00002903
|
| 9815 | GIT2, CAT-2, CAT2 | G protein-coupled receptor kinase interacting ArfGAP 2 |
C00000958
C00002903
|
| 9837 | GINS1, PSF1 | GINS complex subunit 1 (Psf1 homolog) |
C00000958
C00002903
|
| 1647 | GADD45A, DDIT1, GADD45 | growth arrest and DNA-damage-inducible, alpha |
C00000958
C00002903
|
| 55632 | G2E3, KIAA1333, PHF7B | G2/M-phase specific E3 ubiquitin protein ligase |
C00000958
C00002903
|
| 2535 | FZD2, Fz2, fz-2, fzE2, hFz2 | frizzled family receptor 2 |
C00000958
C00002903
|
| 2534 | FYN, SLK, SYN, p59-FYN | FYN oncogene related to SRC, FGR, YES (EC:2.7.10.2) |
C00000958
C00002903
|
| 2395 | FXN, CyaY, FA, FARR, FRDA, X25 | frataxin (EC:1.16.3.1) |
C00000958
C00002903
|
| 122786 | FRMD6, C14orf31, EX1, Willin, c14_5320 | FERM domain containing 6 |
C00000958
C00002903
|
| 2353 | FOS, AP-1, C-FOS, p55 | FBJ murine osteosarcoma viral oncogene homolog |
C00000958
C00002903
|
| 114793 | FMNL2, FHOD2 | formin-like 2 |
C00000958
C00002903
|
| 2247 | FGF2, BFGF, FGF-2, FGFB, HBGF-2 | fibroblast growth factor 2 (basic) |
C00000958
C00002903
|
| 2246 | FGF1, AFGF, ECGF, ECGF-beta, ECGFA, ECGFB, FGF-1, FGF-alpha, FGFA, GLIO703, HBGF-1, HBGF1 | fibroblast growth factor 1 (acidic) |
C00000958
C00002903
|
| 2194 | FASN, FAS, OA-519, SDR27X1 | fatty acid synthase (EC:2.3.1.85 2.3.1.38 2.3.1.39 2.3.1.41 3.1.2.14 4.2.1.59 1.1.1.100 1.3.1.39) |
C00000958
C00002903
|
| 90362 | FAM110B, C8orf72 | family with sequence similarity 110, member B |
C00000958
C00002903
|
| 1277 | COL1A1, OI4 | collagen, type I, alpha 1 |
C00000958
C00002903
|
| 8772 | FADD, MORT1 | Fas (TNFRSF6)-associated via death domain |
C00000958
C00002903
|
| 2152 | F3, CD142, TF, TFA | coagulation factor III (thromboplastin, tissue factor) |
C00000958
C00002903
|
| 27235 | COQ2, CL640, COQ10D1, MSA1 | coenzyme Q2 4-hydroxybenzoate polyprenyltransferase (EC:2.5.1.39) |
C00000958
C00002903
|
| 2147 | F2, PT, RPRGL2, THPH1 | coagulation factor II (thrombin) (EC:3.4.21.5) |
C00000958
C00002903
|
| 6622 | SNCA, NACP, PARK1, PARK4, PD1 | synuclein, alpha (non A4 component of amyloid precursor) |
C00002647
C00002903
|
| 54206 | ERRFI1, GENE-33, MIG-6, MIG6, RALT | ERBB receptor feedback inhibitor 1 |
C00000958
C00002903
|
| 4512 | COX1, COI, MTCO1, MT-CO1 | cytochrome c oxidase subunit I |
C00000958
C00002903
|
| 10063 | COX17 | COX17 cytochrome c oxidase copper chaperone |
C00000947
C00000958
|
| 2043 | EPHA4, HEK8, SEK, TYRO1 | EPH receptor A4 (EC:2.7.10.1) |
C00000958
C00002903
|
| 54898 | ELOVL2, SSC2 | ELOVL fatty acid elongase 2 (EC:2.3.1.199) |
C00000958
C00002903
|
| 132864 | CPEB2, CPE-BP2, CPEB-2, hCPEB-2 | cytoplasmic polyadenylation element binding protein 2 |
C00000958
C00002903
|
| 10057 | ABCC5, ABC33, EST277145, MOAT-C, MOATC, MRP5, SMRP, pABC11 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
C00000958
C00002903
|
| 1974 | EIF4A2, BM-010, DDX2B, EIF4A, EIF4F, eIF-4A-II, eIF4A-II | eukaryotic translation initiation factor 4A2 (EC:3.6.4.13) |
C00000958
C00002903
|
| 1958 | EGR1, AT225, G0S30, KROX-24, NGFI-A, TIS8, ZIF-268, ZNF225 | early growth response 1 |
C00000958
C00002903
|
| 1385 | CREB1, CREB | cAMP responsive element binding protein 1 |
C00000958
C00002903
|
| 6647 | SOD1, ALS, ALS1, IPOA, SOD, hSod1, homodimer | superoxide dismutase 1, soluble (EC:1.15.1.1) |
C00001180
C00002903
|
| 6667 | SP1 | Sp1 transcription factor |
C00002902
C00002903
|
| 1948 | EFNB2, EPLG5, HTKL, Htk-L, LERK5 | ephrin-B2 |
C00000958
C00002903
|
| 1942 | EFNA1, B61, ECKLG, EFL1, EPLG1, LERK-1, LERK1, TNFAIP4 | ephrin-A1 |
C00000958
C00002903
|
| 1906 | EDN1, ET1, HDLCQ7, PPET1 | endothelin 1 |
C00000958
C00002903
|
| 1894 | ECT2, ARHGEF31 | epithelial cell transforming sequence 2 oncogene |
C00000958
C00002903
|
| 1889 | ECE1, ECE | endothelin converting enzyme 1 (EC:3.4.24.71) |
C00000958
C00002903
|
| 1859 | DYRK1A, DYRK, DYRK1, HP86, MNB, MNBH, MRD7 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (EC:2.7.12.1) |
C00000958
C00002903
|
| 1459 | CSNK2A2, CK2A2, CSNK2A1 | casein kinase 2, alpha prime polypeptide (EC:2.7.11.1) |
C00000958
C00002903
|
| 1850 | DUSP8, C11orf81, HB5, HVH-5, HVH8 | dual specificity phosphatase 8 (EC:3.1.3.16 3.1.3.48) |
C00000958
C00002903
|
| 1789 | DNMT3B, ICF, ICF1, M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta (EC:2.1.1.37) |
C00000958
C00002903
|
| 6732 | SRPK1, SFRSK1 | SRSF protein kinase 1 (EC:2.7.11.1) |
C00001180
C00002903
|
| 1499 | CTNNB1, CTNNB, MRD19, armadillo | catenin (cadherin-associated protein), beta 1, 88kDa |
C00000958
C00002903
|
| 1763 | DNA2, DNA2L, PEOA6, hDNA2 | DNA replication helicase/nuclease 2 (EC:3.6.4.12) |
C00000958
C00002903
|
| 51797 |
C00000958
C00002903
|
||
| 9787 | DLGAP5, DLG7, HURP | discs, large (Drosophila) homolog-associated protein 5 |
C00000958
C00002903
|
| 1738 | DLD, DLDD, DLDH, E3, GCSL, LAD, PHE3 | dihydrolipoamide dehydrogenase (EC:1.8.1.4) |
C00000958
C00002903
|
| 6778 | STAT6, D12S1644, IL-4-STAT, STAT6B, STAT6C | signal transducer and activator of transcription 6, interleukin-4 induced |
C00000947
C00002903
|
| 1734 | DIO2, 5DII, D2, DIOII, SelY, TXDI2 | deiodinase, iodothyronine, type II (EC:1.97.1.10) |
C00000958
C00002903
|
| 81624 | DIAPH3, AN, AUNA1, DIA2, DRF3, NSDAN, diap3, mDia2 | diaphanous-related formin 3 |
C00000958
C00002903
|
| 6932 | TCF7, TCF-1 | transcription factor 7 (T-cell specific, HMG-box) |
C00000947
C00002903
|
| 7040 | TGFB1, CED, DPD1, LAP, TGFB, TGFbeta | transforming growth factor, beta 1 |
C00001180
C00002903
|
| 114757 | CYGB, HGB, STAP | cytoglobin |
C00000958
C00002903
|
| 7083 | TK1, TK2 | thymidine kinase 1, soluble (EC:2.7.1.21) |
C00000947
C00002903
|
| 3952 | LEP, LEPD, OB, OBS | leptin |
C00000856
C00002903
|
| 1719 | DHFR, DHFRP1, DYR | dihydrofolate reductase (EC:1.5.1.3) |
C00000958
C00002903
|
| 7224 | TRPC5, TRP5 | transient receptor potential cation channel, subfamily C, member 5 |
C00002647
C00002903
|
| 9937 | DCLRE1A, PSO2, SNM1, SNM1A | DNA cross-link repair 1A |
C00000958
C00002903
|
| 55170 | PRMT6, HRMT1L6 | protein arginine methyltransferase 6 (EC:2.1.1.125) |
C00002903
|
| 1612 | DAPK1, DAPK | death-associated protein kinase 1 (EC:2.7.11.1) |
C00000958
|
| 1613 | DAPK3, ZIP, ZIPK | death-associated protein kinase 3 (EC:2.7.11.1) |
C00000958
|
| 55157 | DARS2, ASPRS, LBSL, MT-ASPRS, RP3-383J4.2 | aspartyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.12) |
C00000958
|
| 285761 | DCBLD1, dJ94G16.1 | discoidin, CUB and LCCL domain containing 1 |
C00000958
|
| 166614 | DCLK2, CL2, CLICK-II, CLICK2, CLIK2, DCAMKL2, DCDC3, DCDC3B, DCK2 | doublecortin-like kinase 2 (EC:2.7.11.1) |
C00000958
|
| 23002 | DAAM1 | dishevelled associated activator of morphogenesis 1 |
C00000958
|
| 23142 | DCUN1D4 | DCN1, defective in cullin neddylation 1, domain containing 4 |
C00000958
|
| 100286728 |
C00000958
|
||
| 80821 | DDHD1, PA-PLA1, PAPLA1, SPG28 | DDHD domain containing 1 |
C00000958
|
| 9265 | CYTH3, ARNO3, GRP1, PSCD3 | cytohesin 3 |
C00000958
|
| 1662 | DDX10, HRH-J8 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 (EC:3.6.4.13) |
C00000958
|
| 23586 | DDX58, RIG-I, RIGI, RLR-1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (EC:3.6.4.13) |
C00000958
|
| 55601 | DDX60 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (EC:3.6.4.13) |
C00000958
|
| 163259 | DENND2C, dJ1156J9.1 | DENN/MADD domain containing 2C |
C00000958
|
| 79961 | DENND2D | DENN/MADD domain containing 2D |
C00000958
|
| 22898 | DENND3 | DENN/MADD domain containing 3 |
C00000958
|
| 10260 | DENND4A, IRLB, MYCPBP | DENN/MADD domain containing 4A |
C00000958
|
| 55667 | DENND4C, C9orf55, C9orf55B, bA513M16.3 | DENN/MADD domain containing 4C |
C00000958
|
| 201627 | DENND6A, AFI1A, FAM116A | DENN/MADD domain containing 6A |
C00000958
|
| 8562 | DENR, DRP, DRP1, SMAP-3 | density-regulated protein |
C00000958
|
| 120863 | DEPDC4, DEP.4 | DEP domain containing 4 |
C00000958
|
| 91614 | DEPDC7, TR2, dJ85M6.4 | DEP domain containing 7 |
C00000958
|
| 55070 | DET1 | de-etiolated homolog 1 (Arabidopsis) |
C00000958
|
| 1687 | DFNA5, ICERE-1 | deafness, autosomal dominant 5 |
C00000958
|
| 8527 | DGKD, DGKdelta, dgkd-2 | diacylglycerol kinase, delta 130kDa (EC:2.7.1.107) |
C00000958
|
| 160851 | DGKH, DGKeta | diacylglycerol kinase, eta (EC:2.7.1.107) |
C00000958
|
| 1594 | CYP27B1, CP2B, CYP1, CYP1alpha, CYP27B, P450c1, PDDR, VDD1, VDDR, VDDRI, VDR | cytochrome P450, family 27, subfamily B, polypeptide 1 (EC:1.14.13.13) |
C00000958
|
| 55760 | DHX32, DDX32, DHLP1 | DEAH (Asp-Glu-Ala-His) box polypeptide 32 (EC:3.6.4.13) |
C00000958
|
| 60625 | DHX35, C20orf15, DDX35, KAIA0875 | DEAH (Asp-Glu-Ala-His) box polypeptide 35 (EC:3.6.4.13) |
C00000958
|
| 90957 | DHX57, DDX57 | DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 (EC:3.6.4.13) |
C00000958
|
| 124637 | CYB5D1 | cytochrome b5 domain containing 1 |
C00000958
|
| 11068 | CYB561D2, 101F6, TSP10 | cytochrome b561 family, member D2 |
C00000958
|
| 445162 |
C00000958
|
||
| 51523 | CXXC5, CF5, RINF, WID | CXXC finger protein 5 |
C00000958
|
| 23181 | DIP2A, C21orf106, DIP2 | DIP2 disco-interacting protein 2 homolog A (Drosophila) |
C00000958
|
| 57609 | DIP2B | DIP2 disco-interacting protein 2 homolog B (Drosophila) |
C00000958
|
| 115752 | DIS3L, DIS3L1 | DIS3 mitotic control homolog (S. cerevisiae)-like (EC:3.1.13.-) |
C00000958
|
| 84976 | DISP1, DISPA | dispatched homolog 1 (Drosophila) |
C00000958
|
| 85458 | DIXDC1, CCD1 | DIX domain containing 1 |
C00000958
|
| 22943 | DKK1, DKK-1, SK | dickkopf WNT signaling pathway inhibitor 1 |
C00000958
|
| 2919 | CXCL1, FSP, GRO1, GROa, MGSA, MGSA-a, NAP-3, SCYB1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
C00000958
|
| 8847 | DLEU2, 1B4, BCMSUN, DLB2, LEU2, LINC00022, MIR15AHG, NCRNA00022, RFP2OS, TRIM13OS | deleted in lymphocytic leukemia 2 (non-protein coding) |
C00000958
|
| 1739 | DLG1, DLGH1, SAP-97, SAP97, dJ1061C18.1.1, hdlg | discs, large homolog 1 (Drosophila) |
C00000958
|
| 56474 | CTPS2 | CTP synthase 2 (EC:6.3.4.2) |
C00000958
|
| 65989 | DLK2, DLK-2, EGFL9 | delta-like 2 homolog (Drosophila) |
C00000958
|
| 54567 | DLL4, hdelta2 | delta-like 4 (Drosophila) |
C00000958
|
| 55929 | DMAP1, DNMAP1, DNMTAP1, EAF2, MEAF2, SWC4 | DNA methyltransferase 1 associated protein 1 |
C00000958
|
| 1657 | DMXL1 | Dmx-like 1 |
C00000958
|
| 23312 | DMXL2, RC3 | Dmx-like 2 |
C00000958
|
| 1497 | CTNS, CTNS-LSB, PQLC4 | cystinosin, lysosomal cystine transporter |
C00000958
|
| 55172 | DNAAF2, C14orf104, CILD10, KTU, PF13 | dynein, axonemal, assembly factor 2 |
C00000958
|
| 11080 | DNAJB4, DNAJW, DjB4, HLJ1 | DnaJ (Hsp40) homolog, subfamily B, member 4 |
C00000958
|
| 25822 | DNAJB5, Hsc40 | DnaJ (Hsp40) homolog, subfamily B, member 5 |
C00000958
|
| 4189 | DNAJB9, ERdj4, MDG-1, MDG1, MST049, MSTP049 | DnaJ (Hsp40) homolog, subfamily B, member 9 |
C00000958
|
| 23317 | DNAJC13, RME8 | DnaJ (Hsp40) homolog, subfamily C, member 13 |
C00000958
|
| 85406 | DNAJC14, DNAJ, DRIP78, HDJ3, LIP6 | DnaJ (Hsp40) homolog, subfamily C, member 14 |
C00000958
|
| 131118 | DNAJC19, PAM18, TIM14, TIMM14 | DnaJ (Hsp40) homolog, subfamily C, member 19 |
C00000958
|
| 54943 | DNAJC28, C21orf55, C21orf78 | DnaJ (Hsp40) homolog, subfamily C, member 28 |
C00000958
|
| 144132 | DNHD1, C11orf47, CCDC35, DHCD1, DNHD1L | dynein heavy chain domain 1 |
C00000958
|
| 26052 | DNM3, Dyna_III | dynamin 3 (EC:3.6.5.5) |
C00000958
|
| 9811 | CTIF, Gm672, KIAA0427 | CBP80/20-dependent translation initiation factor |
C00000958
|
| 57325 | CSRP2BP, ATAC2, CRP2BP, KAT14, PRO1194, dJ717M23.1 | CSRP2 binding protein |
C00000958
|
| 23549 | DNPEP, ASPEP, DAP | aspartyl aminopeptidase (EC:3.4.11.21) |
C00000958
|
| 116092 | DNTTIP1, C20orf167, Tdif1, dJ447F3.4 | deoxynucleotidyltransferase, terminal, interacting protein 1 |
C00000958
|
| 30836 | DNTTIP2, ERBP, FCF2, HSU15552, LPTS-RP2, TdIF2 | deoxynucleotidyltransferase, terminal, interacting protein 2 |
C00000958
|
| 55619 | DOCK10, DRIP2, Nbla10300, ZIZ3 | dedicator of cytokinesis 10 |
C00000958
|
| 139818 | DOCK11, ACG, ZIZ2, bB128O4.1 | dedicator of cytokinesis 11 |
C00000958
|
| 85440 | DOCK7, ZIR2 | dedicator of cytokinesis 7 |
C00000958
|
| 23348 | DOCK9, ZIZ1, ZIZIMIN1 | dedicator of cytokinesis 9 |
C00000958
|
| 1796 | DOK1, P62DOK | docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
C00000958
|
| 9980 | DOPEY2, 21orf5, C21orf5 | dopey family member 2 |
C00000958
|
| 89978 | DPH6, ATPBD4 | diphthamine biosynthesis 6 (EC:6.3.1.14) |
C00000958
|
| 1803 | DPP4, ADABP, ADCP2, CD26, DPPIV, TP103 | dipeptidyl-peptidase 4 (EC:3.4.14.5) |
C00000958
|
| 55332 | DRAM1, DRAM | DNA-damage regulated autophagy modulator 1 |
C00000958
|
| 1813 | DRD2, D2DR, D2R | dopamine receptor D2 |
C00000958
|
| 79075 | DSCC1, DCC1 | DNA replication and sister chromatid cohesion 1 |
C00000958
|
| 29940 | DSE, DSEP, DSEPI, SART-2, SART2 | dermatan sulfate epimerase (EC:5.1.3.19) |
C00000958
|
| 1830 | DSG3, CDHF6, PVA | desmoglein 3 |
C00000958
|
| 667 | DST, BP240, BPA, BPAG1, CATX-15, CATX15, D6S1101, DMH, DT, EBSB2, HSAN6, MACF2 | dystonin |
C00000958
|
| 112487 | DTD2, C14orf126 | D-tyrosyl-tRNA deacylase 2 (putative) |
C00000958
|
| 84062 | DTNBP1, BLOC1S8, DBND, HPS7, My031, SDY | dystrobrevin binding protein 1 |
C00000958
|
| 285605 | DTWD2 | DTW domain containing 2 |
C00000958
|
| 1841 | DTYMK, CDC8, PP3731, TMPK, TYMK | deoxythymidylate kinase (thymidylate kinase) (EC:2.7.4.9) |
C00000958
|
| 54920 | DUS2, DUS2L, SMM1, URLC8 | dihydrouridine synthase 2 |
C00000958
|
| 1846 | DUSP4, HVH2, MKP-2, MKP2, TYP | dual specificity phosphatase 4 (EC:3.1.3.16 3.1.3.48) |
C00000958
|
| 1847 | DUSP5, DUSP, HVH3 | dual specificity phosphatase 5 (EC:3.1.3.16 3.1.3.48) |
C00000958
|
| 1848 | DUSP6, HH19, MKP3, PYST1 | dual specificity phosphatase 6 (EC:3.1.3.16 3.1.3.48) |
C00000958
|
| 1849 | DUSP7, MKPX, PYST2 | dual specificity phosphatase 7 (EC:3.1.3.16 3.1.3.48) |
C00000958
|
| 1464 | CSPG4, HMW-MAA, MCSP, MCSPG, MEL-CSPG, MSK16, NG2 | chondroitin sulfate proteoglycan 4 |
C00000958
|
| 6993 | DYNLT1, CW-1, TCTEL1, tctex-1 | dynein, light chain, Tctex-type 1 |
C00000958
|
| 9946 | CRYZL1, 4P11, QOH-1 | crystallin, zeta (quinone reductase)-like 1 |
C00000958
|
| 9666 | DZIP3, PPP1R66, UURF2, hRUL138 | DAZ interacting zinc finger protein 3 |
C00000958
|
| 85403 | EAF1 | ELL associated factor 1 |
C00000958
|
| 9166 | EBAG9, EB9, PDAF | estrogen receptor binding site associated, antigen, 9 |
C00000958
|
| 1415 | CRYBB2, CCA2, CRYB2, CRYB2A, CTRCT3, D22S665 | crystallin, beta B2 |
C00000958
|
| 1893 | ECM1, URBWD | extracellular matrix protein 1 |
C00000958
|
| 1410 | CRYAB, CMD1II, CRYA2, CTPP2, CTRCT16, HSPB5, MFM2 | crystallin, alpha B |
C00000958
|
| 80267 | EDEM3, C1orf22 | ER degradation enhancer, mannosidase alpha-like 3 (EC:3.2.1.113) |
C00000958
|
| 54677 | CROT, COT | carnitine O-octanoyltransferase (EC:2.3.1.137) |
C00000958
|
| 1910 | EDNRB, ABCDS, ET-B, ET-BR, ETB, ETBR, ETRB, HSCR, HSCR2, WS4A | endothelin receptor type B |
C00000958
|
| 8411 | EEA1, MST105, MSTP105, ZFYVE2 | early endosome antigen 1 |
C00000958
|
| 1915 | EEF1A1, CCS-3, CCS3, EE1A1, EEF-1, EEF1A, EF-Tu, EF1A, GRAF-1EF, HNGC:16303, LENG7, PTI1, eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 |
C00000958
|
| 29904 | EEF2K, HSU93850, eEF-2K | eukaryotic elongation factor-2 kinase (EC:2.7.11.20) |
C00000958
|
| 114327 | EFHC1, dJ304B14.2 | EF-hand domain (C-terminal) containing 1 |
C00000958
|
| 147081 | CRHR1-IT1, C17orf69 | CRHR1 intronic transcript 1 (non-protein coding) |
C00000958
|
| 1945 | EFNA4, EFL4, EPLG4, LERK4 | ephrin-A4 |
C00000958
|
| 1946 | EFNA5, AF1, EFL5, EPLG7, GLC1M, LERK7, RAGS | ephrin-A5 |
C00000958
|
| 58487 | CREBZF, SMILE, ZF | CREB/ATF bZIP transcription factor |
C00000958
|
| 23167 | EFR3A | EFR3 homolog A (S. cerevisiae) |
C00000958
|
| 22979 | EFR3B, KIAA0953 | EFR3 homolog B (S. cerevisiae) |
C00000958
|
| 79631 | EFTUD1, FAM42A, HsT19294, RIA1 | elongation factor Tu GTP binding domain containing 1 |
C00000958
|
| 153222 | CREBRF, C5orf41, LRF | CREB3 regulatory factor |
C00000958
|
| 90993 | CREB3L1, OASIS | cAMP responsive element binding protein 3-like 1 |
C00000958
|
| 8738 | CRADD, MRT34, RAIDD | CASP2 and RIPK1 domain containing adaptor with death domain |
C00000958
|
| 23301 | EHBP1, HPC12, NACSIN | EH domain binding protein 1 |
C00000958
|
| 1962 | EHHADH, ECHD, L-PBE, LBFP, LBP, PBFE | enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (EC:1.1.1.35 5.3.3.8 4.2.1.17) |
C00000958
|
| 79813 | EHMT1, EUHMTASE1, Eu-HMTase1, FP13812, GLP, GLP1, KMT1D, bA188C12.1 | euchromatic histone-lysine N-methyltransferase 1 (EC:2.1.1.43) |
C00000958
|
| 440275 | EIF2AK4, GCN2 | eukaryotic translation initiation factor 2 alpha kinase 4 (EC:2.7.11.1) |
C00000958
|
| 1968 | EIF2S3, EIF2, EIF2G, EIF2gamma, eIF-2gA | eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
C00000958
|
| 51692 | CPSF3, CPSF-73, CPSF73 | cleavage and polyadenylation specific factor 3, 73kDa |
C00000958
|
| 1975 | EIF4B, EIF-4B, PRO1843 | eukaryotic translation initiation factor 4B |
C00000958
|
| 55520 | ELAC1, D29 | elaC ribonuclease Z 1 (EC:3.1.26.11) |
C00000958
|
| 1993 | ELAVL2, HEL-N1, HELN1, HUB | ELAV like neuron-specific RNA binding protein 2 |
C00000958
|
| 1997 | ELF1 | E74-like factor 1 (ets domain transcription factor) |
C00000958
|
| 144402 | CPNE8 | copine VIII |
C00000958
|
| 22936 | ELL2 | elongation factor, RNA polymerase II, 2 |
C00000958
|
| 84337 | ELOF1, ELF1 | elongation factor 1 homolog (S. cerevisiae) |
C00000958
|
| 64834 | ELOVL1, Ssc1 | ELOVL fatty acid elongase 1 (EC:2.3.1.199) |
C00000958
|
| 1363 | CPE | carboxypeptidase E (EC:3.4.17.10) |
C00000958
|
| 26610 | ELP4, C11orf19, PAX6NEB, PAXNEB, dJ68P15A.1 | elongator acetyltransferase complex subunit 4 |
C00000958
|
| 9694 | EMC2, KIAA0103, TTC35 | ER membrane protein complex subunit 2 |
C00000958
|
| 2010 | EMD, EDMD, LEMD5, STA | emerin |
C00000958
|
| 10436 | EMG1, C2F, Grcc2f, NEP1 | EMG1 N1-specific pseudouridine methyltransferase |
C00000958
|
| 27436 | EML4, C2orf2, ELP120, EMAP-4, EMAPL4, ROPP120 | echinoderm microtubule associated protein like 4 |
C00000958
|
| 10495 | ENOX2, APK1, COVA1, tNOX | ecto-NOX disulfide-thiol exchanger 2 |
C00000958
|
| 114915 | EPB41L4A-AS1, C5orf26, NCRNA00219, TIGA1 | EPB41L4A antisense RNA 1 |
C00000958
|
| 54566 | EPB41L4B, CG1, EHM2 | erythrocyte membrane protein band 4.1 like 4B |
C00000958
|
| 57724 | EPG5, HEEW1, KIAA1632, VICIS, hEPG5 | ectopic P-granules autophagy protein 5 homolog (C. elegans) |
C00000958
|
| 1359 | CPA3, MC-CPA | carboxypeptidase A3 (mast cell) (EC:3.4.17.1) |
C00000958
|
| 9852 | EPM2AIP1 | EPM2A (laforin) interacting protein 1 |
C00000958
|
| 22905 | EPN2, EHB21 | epsin 2 |
C00000958
|
| 2059 | EPS8 | epidermal growth factor receptor pathway substrate 8 |
C00000958
|
| 94240 | EPSTI1, BRESI1 | epithelial stromal interaction 1 (breast) |
C00000958
|
| 23085 | ERC1, Cast2, ELKS, RAB6IP2 | ELKS/RAB6-interacting/CAST family member 1 |
C00000958
|
| 2072 | ERCC4, ERCC11, FANCQ, RAD1, XPF | excision repair cross-complementing rodent repair deficiency, complementation group 4 |
C00000958
|
| 375748 | ERCC6L2, C9orf102, RAD26L, SR278 | excision repair cross-complementing rodent repair deficiency, complementation group 6-like 2 |
C00000958
|
| 51290 | ERGIC2, Erv41, PTX1, cd002 | ERGIC and golgi 2 |
C00000958
|
| 157697 | ERICH1 | glutamate-rich 1 |
C00000958
|
| 27248 | ERLEC1, C2orf30, CIM, CL24936, CL25084, XTP3-B, XTP3TPB | endoplasmic reticulum lectin 1 |
C00000958
|
| 55780 | ERMARD, C6orf70, dJ266L20.3 | ER membrane-associated RNA degradation |
C00000958
|
| 10961 | ERP29, C12orf8, ERp28, ERp31, PDI-DB, PDIA9 | endoplasmic reticulum protein 29 |
C00000958
|
| 23406 | COTL1, CLP | coactosin-like 1 (Dictyostelium) |
C00000958
|
| 2098 | ESD, FGH | esterase D (EC:3.1.2.12 3.1.1.56) |
C00000958
|
| 284729 | ESPNP, RP1-163M9.1, dJ1182A14.1 | espin pseudogene |
C00000958
|
| 57017 | COQ9, C16orf49, COQ10D5 | coenzyme Q9 |
C00000958
|
| 54465 | ETAA1, ETAA16 | Ewing tumor-associated antigen 1 |
C00000958
|
| 2117 | ETV3, METS, PE-1, PE1, bA110J1.4 | ets variant 3 |
C00000958
|
| 2119 | ETV5, ERM | ets variant 5 |
C00000958
|
| 2120 | ETV6, TEL, TEL/ABL | ets variant 6 |
C00000958
|
| 55763 | EXOC1, BM-102, SEC3, SEC3L1, SEC3P | exocyst complex component 1 |
C00000958
|
| 55770 | EXOC2, SEC5, SEC5L1, Sec5p | exocyst complex component 2 |
C00000958
|
| 60412 | EXOC4, SEC8, SEC8L1, Sec8p | exocyst complex component 4 |
C00000958
|
| 54536 | EXOC6, EXOC6A, SEC15, SEC15L, SEC15L1, SEC15L3, Sec15p | exocyst complex component 6 |
C00000958
|
| 23086 | EXPH5, SLAC2-B, SLAC2B | exophilin 5 |
C00000958
|
| 2139 | EYA2, EAB1 | eyes absent homolog 2 (Drosophila) (EC:3.1.3.48) |
C00000958
|
| 84274 | COQ5 | coenzyme Q5 homolog, methyltransferase (S. cerevisiae) (EC:2.1.1.201) |
C00000958
|
| 1291 | COL6A1, OPLL | collagen, type VI, alpha 1 |
C00000958
|
| 1282 | COL4A1, HANAC, ICH, POREN1, arresten | collagen, type IV, alpha 1 |
C00000958
|
| 3995 | FADS3, CYB5RP, LLCDL3 | fatty acid desaturase 3 (EC:1.14.19.-) |
C00000958
|
| 55179 | FAIM, FAIM1 | Fas apoptotic inhibitory molecule |
C00000958
|
| 144347 | FAM101A | family with sequence similarity 101, member A |
C00000958
|
| 284611 | FAM102B, SYM-3B | family with sequence similarity 102, member B |
C00000958
|
| 91949 | COG7, CDG2E | component of oligomeric golgi complex 7 |
C00000958
|
| 92689 | FAM114A1, Noxp20 | family with sequence similarity 114, member A1 |
C00000958
|
| 285966 | FAM115C, FAM139A | family with sequence similarity 115, member C |
C00000958
|
| 79607 | FAM118B | family with sequence similarity 118, member B |
C00000958
|
| 26355 | FAM162A, C3orf28, E2IG5, HGTD-P | family with sequence similarity 162, member A |
C00000958
|
| 400823 | FAM177B | family with sequence similarity 177, member B |
C00000958
|
| 51313 | FAM198B, AD036, C4orf18 | family with sequence similarity 198, member B |
C00000958
|
| 85395 | FAM207A, C21orf70 | family with sequence similarity 207, member A |
C00000958
|
| 23272 | FAM208A, C3orf63, RAP140, se89-1 | family with sequence similarity 208, member A |
C00000958
|
| 54537 | FAM35A, FAM35A1, bA163M19.1 | family with sequence similarity 35, member A |
C00000958
|
| 10447 | FAM3C, ILEI | family with sequence similarity 3, member C |
C00000958
|
| 51571 | FAM49B, L1 | family with sequence similarity 49, member B |
C00000958
|
| 26240 | FAM50B, D6S2654E, X5L | family with sequence similarity 50, member B |
C00000958
|
| 199870 | FAM76A | family with sequence similarity 76, member A |
C00000958
|
| 22909 | FAN1, KIAA1018, KMIN, MTMR15 | FANCD2/FANCI-associated nuclease 1 (EC:3.1.4.1) |
C00000958
|
| 2176 | FANCC, FA3, FAC, FACC | Fanconi anemia, complementation group C |
C00000958
|
| 57697 | FANCM, FAAP250, KIAA1596 | Fanconi anemia, complementation group M (EC:3.6.4.13) |
C00000958
|
| 55711 | FAR2, MLSTD1, SDR10E2 | fatty acyl CoA reductase 2 (EC:1.2.1.n2) |
C00000958
|
| 10667 | FARS2, COXPD14, FARS1, HSPC320, PheRS, dJ520B18.2 | phenylalanyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.20) |
C00000958
|
| 57511 | COG6, CDG2L, COD2, SHNS | component of oligomeric golgi complex 6 |
C00000958
|
| 79675 | FASTKD1 | FAST kinase domains 1 |
C00000958
|
| 22868 | FASTKD2, KIAA0971 | FAST kinase domains 2 |
C00000958
|
| 2195 | FAT1, CDHF7, CDHR8, FAT, ME5, hFat1 | FAT atypical cadherin 1 |
C00000958
|
| 79633 | FAT4, CDHF14, CDHR11, FAT-J, FATJ, NBLA00548 | FAT atypical cadherin 4 |
C00000958
|
| 2200 | FBN1, ACMICD, ECTOL1, FBN, GPHYSD2, MASS, MFS1, OCTD, SGS, SSKS, WMS, WMS2 | fibrillin 1 |
C00000958
|
| 2201 | FBN2, CCA, DA9 | fibrillin 2 |
C00000958
|
| 54850 | FBXL12, Fbl12 | F-box and leucine-rich repeat protein 12 |
C00000958
|
| 25827 | FBXL2, FBL2, FBL3 | F-box and leucine-rich repeat protein 2 |
C00000958
|
| 26235 | FBXL4, FBL4, FBL5, MTDPS13 | F-box and leucine-rich repeat protein 4 |
C00000958
|
| 26234 | FBXL5, FBL4, FBL5, FLR1 | F-box and leucine-rich repeat protein 5 |
C00000958
|
| 23194 | FBXL7, FBL6, FBL7 | F-box and leucine-rich repeat protein 7 |
C00000958
|
| 84893 | FBXO18, FBH1, Fbx18 | F-box protein, helicase, 18 (EC:3.6.4.12) |
C00000958
|
| 554251 | FBXO48 | F-box protein 48 |
C00000958
|
| 115548 | FCHO2 | FCH domain only 2 |
C00000958
|
| 9873 | FCHSD2, NWK, SH3MD3 | FCH and double SH3 domains 2 |
C00000958
|
| 55527 | FEM1A, EPRAP | fem-1 homolog a (C. elegans) |
C00000958
|
| 56929 | FEM1C, EUROIMAGE686608, EUROIMAGE783647, FEM1A | fem-1 homolog c (C. elegans) |
C00000958
|
| 2241 | FER, FerT, PPP1R74, TYK3 | fer (fps/fes related) tyrosine kinase (EC:2.7.10.2) |
C00000958
|
| 10979 | FERMT2, KIND2, MIG2, PLEKHC1, UNC112, UNC112B, mig-2 | fermitin family member 2 |
C00000958
|
| 55785 | FGD6, ZFYVE24 | FYVE, RhoGEF and PH domain containing 6 |
C00000958
|
| 22796 | COG2, LDLC | component of oligomeric golgi complex 2 |
C00000958
|
| 2257 | FGF12, FGF12B, FHF1 | fibroblast growth factor 12 |
C00000958
|
| 8817 | FGF18, FGF-18, ZFGF5 | fibroblast growth factor 18 |
C00000958
|
| 388753 | COA6, C1orf31 | cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
C00000958
|
| 11259 | FILIP1L, DOC-1, DOC1, GIP130, GIP90 | filamin A interacting protein 1-like |
C00000958
|
| 60681 | FKBP10, FKBP65, OI11, OI6, PPIASE, hFKBP65 | FK506 binding protein 10, 65 kDa (EC:5.2.1.8) |
C00000958
|
| 11328 | FKBP9, FKBP60, FKBP63, PPIase | FK506 binding protein 9, 63 kDa (EC:5.2.1.8) |
C00000958
|
| 23768 | FLRT2 | fibronectin leucine rich transmembrane protein 2 |
C00000958
|
| 28958 | COA3, CCDC56, MITRAC12 | cytochrome c oxidase assembly factor 3 |
C00000958
|
| 54874 | FNBP1L, C1orf39, TOCA1 | formin binding protein 1-like |
C00000958
|
| 22862 | FNDC3A, FNDC3, HUGO, bA203I16.1, bA203I16.5 | fibronectin type III domain containing 3A |
C00000958
|
| 64778 | FNDC3B, FAD104, PRO4979, YVTM2421 | fibronectin type III domain containing 3B |
C00000958
|
| 163479 | FNDC7, RP11-293A10.2 | fibronectin type III domain containing 7 |
C00000958
|
| 54914 | FOCAD, KIAA1797 | focadhesin |
C00000958
|
| 54875 | CNTLN, C9orf101, C9orf39, RP11-340N12.1, bA340N12.1 | centlein, centrosomal protein |
C00000958
|
| 2354 | FOSB, AP-1, G0S3, GOS3, GOSB | FBJ murine osteosarcoma viral oncogene homolog B |
C00000958
|
| 27086 | FOXP1, 12CC4, QRF1, hFKH1B | forkhead box P1 |
C00000958
|
| 246175 | CNOT6L, CCR4b | CCR4-NOT transcription complex, subunit 6-like (EC:3.1.13.4) |
C00000958
|
| 10818 | FRS2, FRS2A, FRS2alpha, SNT, SNT-1, SNT1 | fibroblast growth factor receptor substrate 2 |
C00000958
|
| 10129 | FRY, 13CDNA73, 214K23.2, C13orf14, CG003, bA207N4.2, bA37E23.1 | furry homolog (Drosophila) |
C00000958
|
| 285527 | FRYL, KIAA0826 | FRY-like |
C00000958
|
| 2519 | FUCA2, dJ20N2.5 | fucosidase, alpha-L- 2, plasma (EC:3.2.1.51) |
C00000958
|
| 2530 | FUT8 | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) (EC:2.4.1.68) |
C00000958
|
| 4848 | CNOT2, CDC36, NOT2, NOT2H | CCR4-NOT transcription complex, subunit 2 |
C00000958
|
| 53827 | FXYD5, DYSAD, IWU1, KCT1, OIT2, PRO6241, RIC | FXYD domain containing ion transport regulator 5 |
C00000958
|
| 55571 | CNOT11, C2orf29 | CCR4-NOT transcription complex, subunit 11 |
C00000958
|
| 8321 | FZD1 | frizzled family receptor 1 |
C00000958
|
| 26507 | CNNM1, ACDP1, CLP-1 | cyclin M1 |
C00000958
|
| 154043 | CNKSR3, MAGI1 | CNKSR family member 3 |
C00000958
|
| 2548 | GAA, LYAG | glucosidase, alpha; acid (EC:3.2.1.20) |
C00000958
|
| 2549 | GAB1 | GRB2-associated binding protein 1 |
C00000958
|
| 9846 | GAB2 | GRB2-associated binding protein 2 |
C00000958
|
| 2564 | GABRE | gamma-aminobutyric acid (GABA) A receptor, epsilon |
C00000958
|
| 22866 | CNKSR2, CNK2, KSR2, MAGUIN | connector enhancer of kinase suppressor of Ras 2 |
C00000958
|
| 51083 | GAL, GAL-GMAP, GALN, GLNN, GMAP | galanin/GMAP prepropeptide |
C00000958
|
| 130589 | GALM, IBD1 | galactose mutarotase (aldose 1-epimerase) (EC:5.1.3.3) |
C00000958
|
| 2589 | GALNT1, GALNAC-T1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (EC:2.4.1.41) |
C00000958
|
| 2632 | GBE1, APBD, GBE, GSD4 | glucan (1,4-alpha-), branching enzyme 1 (EC:2.4.1.18) |
C00000958
|
| 2633 | GBP1 | guanylate binding protein 1, interferon-inducible |
C00000958
|
| 2634 | GBP2 | guanylate binding protein 2, interferon-inducible |
C00000958
|
| 2635 | GBP3 | guanylate binding protein 3 |
C00000958
|
| 115361 | GBP4, Mpa2 | guanylate binding protein 4 |
C00000958
|
| 25801 | GCA, GCL | grancalcin, EF-hand calcium binding protein |
C00000958
|
| 79571 | GCC1, GCC1P, GCC88 | GRIP and coiled-coil domain containing 1 |
C00000958
|
| 2650 | GCNT1, C2GNT, C2GNT-L, C2GNT1, G6NT, NACGT2, NAGCT2 | glucosaminyl (N-acetyl) transferase 1, core 2 (EC:2.4.1.102) |
C00000958
|
| 2651 | GCNT2, CCAT, CTRCT13, GCNT2C, GCNT5, IGNT, II, NACGT1, NAGCT1, ULG3, bA360O19.2, bA421M1.1 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) (EC:2.4.1.150) |
C00000958
|
| 2661 | GDF9 | growth differentiation factor 9 |
C00000958
|
| 284161 | GDPD1, GDE4 | glycerophosphodiester phosphodiesterase domain containing 1 |
C00000958
|
| 79833 | GEMIN6 | gem (nuclear organelle) associated protein 6 |
C00000958
|
| 9945 | GFPT2, GFAT2 | glutamine-fructose-6-phosphate transaminase 2 (EC:2.6.1.16) |
C00000958
|
| 26058 | GIGYF2, GYF2, PARK11, PERQ2, PERQ3, TNRC15 | GRB10 interacting GYF protein 2 |
C00000958
|
| 54826 | GIN1, GIN-1, TGIN1, ZH2C2 | gypsy retrotransposon integrase 1 |
C00000958
|
| 80790 | CMIP, TCMIP | c-Maf inducing protein |
C00000958
|
| 55907 | CMAS, CSS | cytidine monophosphate N-acetylneuraminic acid synthetase (EC:2.7.7.43) |
C00000958
|
| 2701 | GJA4, CX37 | gap junction protein, alpha 4, 37kDa |
C00000958
|
| 113263 | GLCCI1, FAM117C, GCTR, GIG18, TSSN1 | glucocorticoid induced transcript 1 |
C00000958
|
| 2736 | GLI2, HPE9, THP1, THP2 | GLI family zinc finger 2 |
C00000958
|
| 2737 | GLI3, ACLS, GCPS, GLI3-190, GLI3FL, PAP-A, PAPA, PAPA1, PAPB, PHS, PPDIV | GLI family zinc finger 3 |
C00000958
|
| 169792 | GLIS3, ZNF515 | GLIS family zinc finger 3 |
C00000958
|
| 11146 | GLMN, FAP, FAP48, FAP68, FKBPAP, GLML, GVM, VMGLOM | glomulin, FKBP associated protein |
C00000958
|
| 8218 | CLTCL1, CHC22, CLH22, CLTCL, CLTD | clathrin, heavy chain-like 1 |
C00000958
|
| 2744 | GLS, AAD20, GAC, GAM, GLS1, KGA | glutaminase (EC:3.5.1.2) |
C00000958
|
| 55830 | GLT8D1, MSTP139 | glycosyltransferase 8 domain containing 1 |
C00000958
|
| 80772 | GLTPD1, CPTP | glycolipid transfer protein domain containing 1 |
C00000958
|
| 29997 | GLTSCR2, PICT-1, PICT1 | glioma tumor suppressor candidate region gene 2 |
C00000958
|
| 2766 | GMPR, GMPR1 | guanosine monophosphate reductase (EC:1.7.1.7) |
C00000958
|
| 2770 | GNAI1, Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
C00000958
|
| 2776 | GNAQ, CMC1, G-ALPHA-q, GAQ, SWS | guanine nucleotide binding protein (G protein), q polypeptide |
C00000958
|
| 10020 | GNE, DMRV, GLCNE, IBM2, NM, Uae1 | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (EC:2.7.1.60 3.2.1.183) |
C00000958
|
| 26354 | GNL3, C77032, E2IG3, NNP47, NS | guanine nucleotide binding protein-like 3 (nucleolar) |
C00000958
|
| 79158 | GNPTAB, GNPTA, ICD | N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (EC:2.7.8.17) |
C00000958
|
| 2796 | GNRH1, GNRH, GRH, HH12, LHRH, LNRH | gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
C00000958
|
| 2800 | GOLGA1, golgin-97 | golgin A1 |
C00000958
|
| 27333 | GOLIM4, GIMPC, GOLPH4, GPP130, P138 | golgi integral membrane protein 4 |
C00000958
|
| 54865 | GPATCH4, GPATC4 | G patch domain containing 4 |
C00000958
|
| 10243 | GPHN, GEPH, GPH, GPHRYN, HKPX1, MOCODC | gephyrin (EC:2.10.1.1 2.7.7.75) |
C00000958
|
| 26996 | GPR160, GPCR1, GPCR150 | G protein-coupled receptor 160 |
C00000958
|
| 160897 | GPR180, ITR | G protein-coupled receptor 180 |
C00000958
|
| 2844 | GPR21 | G protein-coupled receptor 21 |
C00000958
|
| 2863 | GPR39 | G protein-coupled receptor 39 |
C00000958
|
| 53836 | GPR87, GPR95, KPG_002 | G protein-coupled receptor 87 |
C00000958
|
| 51704 | GPRC5B, RAIG-2, RAIG2 | G protein-coupled receptor, family C, group 5, member B |
C00000958
|
| 81570 | CLPB, HSP78, SKD3 | ClpB caseinolytic peptidase B homolog (E. coli) |
C00000958
|
| 9575 | CLOCK, KAT13D, bHLHe8 | clock circadian regulator (EC:2.3.1.48) |
C00000958
|
| 57476 | GRAMD1B | GRAM domain containing 1B |
C00000958
|
| 65983 | GRAMD3, NS3TP2 | GRAM domain containing 3 |
C00000958
|
| 80000 | GREB1L, C18orf6, KIAA1772 | growth regulation by estrogen in breast cancer-like |
C00000958
|
| 79827 | CLMP, ACAM, ASAM, CSBM, CSBS | CXADR-like membrane protein |
C00000958
|
| 54103 | GSAP, PION | gamma-secretase activating protein |
C00000958
|
| 79789 | CLMN | calmin (calponin-like, transmembrane) |
C00000958
|
| 1198 | CLK3, PHCLK3, PHCLK3/152 | CDC-like kinase 3 (EC:2.7.12.1) |
C00000958
|
| 2937 | GSS, GSHS | glutathione synthetase (EC:6.3.2.3) |
C00000958
|
| 1195 | CLK1, CLK, CLK/STY, STY | CDC-like kinase 1 (EC:2.7.12.1) |
C00000958
|
| 1366 | CLDN7, CEPTRL2, CLDN-7, CPETRL2, Hs.84359, claudin-1 | claudin 7 |
C00000958
|
| 404672 | GTF2H5, C6orf175, TFB5, TFIIH, TGF2H5, TTD, TTD-A, TTDA, bA120J8.2 | general transcription factor IIH, polypeptide 5 |
C00000958
|
| 9569 | GTF2IRD1, BEN, CREAM1, GTF3, MUSTRD1, RBAP2, WBS, WBSCR11, WBSCR12, hMusTRD1alpha1 | GTF2I repeat domain containing 1 |
C00000958
|
| 2976 | GTF3C2, TFIIIC-BETA, TFIIIC110 | general transcription factor IIIC, polypeptide 2, beta 110kDa |
C00000958
|
| 9329 | GTF3C4, KAT12, TFIII90, TFIIIC290, TFIIIC90, TFIIICDELTA | general transcription factor IIIC, polypeptide 4, 90kDa (EC:2.3.1.48) |
C00000958
|
| 54676 | GTPBP2 | GTP binding protein 2 |
C00000958
|
| 51454 | GULP1, CED-6, CED6, GULP | GULP, engulfment adaptor PTB domain containing 1 |
C00000958
|
| 2990 | GUSB, BG, MPS7 | glucuronidase, beta (EC:3.2.1.31) |
C00000958
|
| 3005 | H1F0, H10, H1FV | H1 histone family, member 0 |
C00000958
|
| 55506 | H2AFY2, macroH2A2 | H2A histone family, member Y2 |
C00000958
|
| 145864 | HAPLN3, EXLD1, HsT19883 | hyaluronan and proteoglycan link protein 3 |
C00000958
|
| 23332 | CLASP1, MAST1 | cytoplasmic linker associated protein 1 |
C00000958
|
| 1839 | HBEGF, DTR, DTS, DTSF, HEGFL | heparin-binding EGF-like growth factor |
C00000958
|
| 10014 | HDAC5, HD5, NY-CO-9 | histone deacetylase 5 (EC:3.5.1.98) |
C00000958
|
| 55869 | HDAC8, CDLS5, HD8, HDACL1, MRXS6, RPD3, WTS | histone deacetylase 8 (EC:3.5.1.98) |
C00000958
|
| 67365 |
C00000958
|
||
| 55027 | HEATR3, SYO1 | HEAT repeat containing 3 |
C00000958
|
| 25938 | HEATR5A, C14orf125 | HEAT repeat containing 5A |
C00000958
|
| 54497 | HEATR5B | HEAT repeat containing 5B |
C00000958
|
| 63897 | HEATR6, ABC1 | HEAT repeat containing 6 |
C00000958
|
| 51696 | HECA, HDC, HDCL, HHDC, dJ225E12.1 | headcase homolog (Drosophila) |
C00000958
|
| 57493 | HEG1, HEG, MST112, MSTP112 | heart development protein with EGF-like domains 1 |
C00000958
|
| 51409 | HEMK1, HEMK, MTQ1 | HemK methyltransferase family member 1 |
C00000958
|
| 8925 | HERC1, p532, p619 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
C00000958
|
| 26091 | HERC4 | HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
C00000958
|
| 55008 | HERC6 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
C00000958
|
| 9709 | HERPUD1, HERP, Mif1, SUP | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
C00000958
|
| 64224 | HERPUD2 | HERPUD family member 2 |
C00000958
|
| 9793 | CKAP5, CHTOG, MSPS, TOG, TOGp, ch-TOG | cytoskeleton associated protein 5 |
C00000958
|
| 10614 | HEXIM1, CLP1, EDG1, HIS1, MAQ1 | hexamethylene bis-acetamide inducible 1 |
C00000958
|
| 25792 | CIZ1, LSFR1, NP94, ZNF356 | CDKN1A interacting zinc finger protein 1 |
C00000958
|
| 138050 | HGSNAT, HGNAT, MPS3C, TMEM76 | heparan-alpha-glucosaminide N-acetyltransferase (EC:2.3.1.78) |
C00000958
|
| 3087 | HHEX, HEX, HMPH, HOX11L-PEN, PRH, PRHX | hematopoietically expressed homeobox |
C00000958
|
| 26275 | HIBCH, HIBYLCOAH | 3-hydroxyisobutyryl-CoA hydrolase (EC:3.1.2.4) |
C00000958
|
| 55847 | CISD1, C10orf70, ZCD1, mitoNEET | CDGSH iron sulfur domain 1 |
C00000958
|
| 55662 | HIF1AN, FIH1 | hypoxia inducible factor 1, alpha subunit inhibitor (EC:1.14.11.30 1.14.11.n4) |
C00000958
|
| 3092 | HIP1, HIP-I, ILWEQ | huntingtin interacting protein 1 |
C00000958
|
| 3024 | HIST1H1A, H1.1, H1A, H1F1, HIST1 | histone cluster 1, H1a |
C00000958
|
| 3009 | HIST1H1B, H1, H1.5, H1B, H1F5, H1s-3 | histone cluster 1, H1b |
C00000958
|
| 3006 | HIST1H1C, H1.2, H1C, H1F2, H1s-1 | histone cluster 1, H1c |
C00000958
|
| 3007 | HIST1H1D, H1.3, H1D, H1F3, H1s-2 | histone cluster 1, H1d |
C00000958
|
| 51363 | CHST15, BRAG, GALNAC4S-6ST, RP11-47G11.1 | carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 (EC:2.8.2.33) |
C00000958
|
| 8345 | HIST1H2BH, H2B/j, H2BFJ | histone cluster 1, H2bh |
C00000958
|
| 8342 | HIST1H2BM, H2B/e, H2BFE, dJ160A22.3 | histone cluster 1, H2bm |
C00000958
|
| 317772 | HIST2H2AB, H2AB | histone cluster 2, H2ab |
C00000958
|
| 50515 | CHST11, C4ST, C4ST-1, C4ST1, HSA269537 | carbohydrate (chondroitin 4) sulfotransferase 11 (EC:2.8.2.5) |
C00000958
|
| 3097 | HIVEP2, HIV-EP2, MBP-2, MIBP1, SHN2, ZAS2, ZNF40B | human immunodeficiency virus type I enhancer binding protein 2 |
C00000958
|
| 79643 | CHMP6, VPS20 | charged multivesicular body protein 6 |
C00000958
|
| 3141 | HLCS, HCS | holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) (EC:6.3.4.10 6.3.4.11 6.3.4.15 6.3.4.9) |
C00000958
|
| 6596 | HLTF, HIP116, HIP116A, HLTF1, RNF80, SMARCA3, SNF2L3, ZBU1 | helicase-like transcription factor |
C00000958
|
| 3159 | HMGA1, HMG-R, HMGA1A, HMGIY | high mobility group AT-hook 1 |
C00000958
|
| 80205 | CHD9, AD013, CReMM, KISH2, PRIC320 | chromodomain helicase DNA binding protein 9 (EC:3.6.4.12) |
C00000958
|
| 55636 | CHD7, HH5, IS3, KAL5 | chromodomain helicase DNA binding protein 7 (EC:3.6.4.12) |
C00000958
|
| 9324 | HMGN3, PNAS-25, TRIP7 | high mobility group nucleosomal binding domain 3 |
C00000958
|
| 84181 | CHD6, CHD-6, CHD5, RIGB | chromodomain helicase DNA binding protein 6 (EC:3.6.4.12) |
C00000958
|
| 118487 | CHCHD1, C10orf34, C2360, MRP-S37 | coiled-coil-helix-coiled-coil-helix domain containing 1 |
C00000958
|
| 3163 | HMOX2, HO-2 | heme oxygenase (decycling) 2 (EC:1.14.99.3) |
C00000958
|
| 340784 | HMX3, NKX-5.1, NKX5.1, Nkx5-1 | H6 family homeobox 3 |
C00000958
|
| 3187 | HNRNPH1, HNRPH, HNRPH1, hnRNPH | heterogeneous nuclear ribonucleoprotein H1 (H) |
C00000958
|
| 9456 | HOMER1, HOMER, HOMER1A, HOMER1B, HOMER1C, SYN47, Ves-1 | homer homolog 1 (Drosophila) |
C00000958
|
| 3219 | HOXB9, HOX-2.5, HOX2, HOX2E | homeobox B9 |
C00000958
|
| 84343 | HPS3, SUTAL | Hermansky-Pudlak syndrome 3 |
C00000958
|
| 11234 | HPS5, AIBP63 | Hermansky-Pudlak syndrome 5 |
C00000958
|
| 10247 | HRSP12, P14.5, PSP, UK114 | heat-responsive protein 12 |
C00000958
|
| 158160 | HSD17B7P2, HSD17B7, Hsd17b_2, bA291L22.1 | hydroxysteroid (17-beta) dehydrogenase 7 pseudogene 2 |
C00000958
|
| 84263 | HSDL2, C9orf99, SDR13C1 | hydroxysteroid dehydrogenase like 2 |
C00000958
|
| 79094 | CHAC1 | ChaC, cation transport regulator homolog 1 (E. coli) |
C00000958
|
| 3305 | HSPA1L, HSP70-1L, HSP70-HOM, HSP70T, hum70t | heat shock 70kDa protein 1-like |
C00000958
|
| 22824 | HSPA4L, APG-1, HSPH3, Osp94 | heat shock 70kDa protein 4-like |
C00000958
|
| 27129 | HSPB7, cvHSP | heat shock 27kDa protein family, member 7 (cardiovascular) |
C00000958
|
| 503731 |
C00000958
|
||
| 10553 | HTATIP2, CC3, SDR44U1, TIP30 | HIV-1 Tat interactive protein 2, 30kDa (EC:1.1.1.-) |
C00000958
|
| 3352 | HTR1D, 5-HT1D, HT1DA, HTR1DA, HTRL, RDC4 | 5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
C00000958
|
| 27429 | HTRA2, OMI, PARK13, PRSS25 | HtrA serine peptidase 2 (EC:3.4.21.108) |
C00000958
|
| 629 | CFB, AHUS4, BF, BFD, CFAB, FB, FBI12, GBG, H2-Bf, PBF2 | complement factor B (EC:3.4.21.47) |
C00000958
|
| 25998 | IBTK, BTBD26, BTKI, RP1-93K22.1 | inhibitor of Bruton agammaglobulinemia tyrosine kinase |
C00000958
|
| 23308 | ICOSLG, B7-H2, B7H2, B7RP-1, B7RP1, CD275, GL50, ICOS-L, ICOSL, LICOS | inducible T-cell co-stimulator ligand |
C00000958
|
| 3398 | ID2, GIG8, ID2A, ID2H, bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
C00000958
|
| 3416 | IDE, INSULYSIN | insulin-degrading enzyme (EC:3.4.24.56) |
C00000958
|
| 55853 | IDI2-AS1, C10orf110, IDI2-AS | IDI2 antisense RNA 1 |
C00000958
|
| 1070 | CETN3, CDC31, CEN3 | centrin, EF-hand protein, 3 |
C00000958
|
| 64135 | IFIH1, Hlcd, IDDM19, MDA-5, MDA5, RLR-2 | interferon induced with helicase C domain 1 (EC:3.6.4.13) |
C00000958
|
| 3434 | IFIT1, C56, G10P1, IFI-56, IFI-56K, IFI56, IFIT-1, IFNAI1, ISG56, P56, RNM561 | interferon-induced protein with tetratricopeptide repeats 1 |
C00000958
|
| 3437 | IFIT3, CIG-49, GARG-49, IFI60, IFIT4, IRG2, ISG60, P60, RIG-G | interferon-induced protein with tetratricopeptide repeats 3 |
C00000958
|
| 24138 | IFIT5, P58, RI58 | interferon-induced protein with tetratricopeptide repeats 5 |
C00000958
|
| 163702 | IFNLR1, CRF2/12, IFNLR, IL-28R1, IL28RA, LICR2 | interferon, lambda receptor 1 |
C00000958
|
| 56912 | IFT46, C11orf2, C11orf60 | intraflagellar transport 46 homolog (Chlamydomonas) |
C00000958
|
| 80173 | IFT74, CCDC2, CMG-1, CMG1 | intraflagellar transport 74 homolog (Chlamydomonas) |
C00000958
|
| 57560 | IFT80, ATD2, WDR56 | intraflagellar transport 80 homolog (Chlamydomonas) |
C00000958
|
| 28981 | IFT81, CDV-1, CDV-1R, CDV1, CDV1R | intraflagellar transport 81 homolog (Chlamydomonas) |
C00000958
|
| 8100 | IFT88, D13S1056E, DAF19, TG737, TTC10, hTg737 | intraflagellar transport 88 homolog (Chlamydomonas) |
C00000958
|
| 10390 | CEPT1 | choline/ethanolamine phosphotransferase 1 (EC:2.7.8.2 2.7.8.1) |
C00000958
|
| 80321 | CEP70, BITE | centrosomal protein 70kDa |
C00000958
|
| 95681 | CEP41, JBTS15, TSGA14 | centrosomal protein 41kDa |
C00000958
|
| 10643 | IGF2BP3, CT98, IMP-3, IMP3, KOC1, VICKZ3 | insulin-like growth factor 2 mRNA binding protein 3 |
C00000958
|
| 80184 | CEP290, 3H11Ag, BBS14, CT87, JBTS5, LCA10, MKS4, NPHP6, POC3, SLSN6, rd16 | centrosomal protein 290kDa |
C00000958
|
| 9859 | CEP170, FAM68A, KAB, KIAA0470 | centrosomal protein 170kDa |
C00000958
|
| 22807 | IKZF2, ANF1A2, HELIOS, ZNF1A2, ZNFN1A2 | IKAROS family zinc finger 2 (Helios) |
C00000958
|
| 3588 | IL10RB, CDW210B, CRF2-4, CRFB4, D21S58, D21S66, IL-10R2 | interleukin 10 receptor, beta |
C00000958
|
| 3589 | IL11, AGIF, IL-11 | interleukin 11 |
C00000958
|
| 201134 | CEP112, CCDC46, MACOCO | centrosomal protein 112kDa |
C00000958
|
| 401541 | CENPP, CENP-P | centromere protein P |
C00000958
|
| 3554 | IL1R1, CD121A, D2S1473, IL-1R-alpha, IL1R, IL1RA, P80 | interleukin 1 receptor, type I |
C00000958
|
| 2491 | CENPI, CENP-I, FSHPRH1, LRPR1, Mis6 | centromere protein I |
C00000958
|
| 9466 | IL27RA, CRL1, IL-27RA, IL27R, TCCR, WSX1, zcytor1 | interleukin 27 receptor, alpha |
C00000958
|
| 1057 | CELP, CELL, cell1, cell2, cell3 | carboxyl ester lipase pseudogene |
C00000958
|
| 3574 | IL7, IL-7 | interleukin 7 |
C00000958
|
| 50937 | CDON, CDO, CDON1, HPE11, ORCAM | cell adhesion associated, oncogene regulated |
C00000958
|
| 55602 | CDKN2AIP, CARF | CDKN2A interacting protein |
C00000958
|
| 8851 | CDK5R1, CDK5P35, CDK5R, NCK5A, p23, p25, p35, p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
C00000958
|
| 54928 | IMPAD1, GPAPP, IMP_3, IMP-3, IMPA3 | inositol monophosphatase domain containing 1 (EC:3.1.3.7 3.1.3.25) |
C00000958
|
| 50939 | IMPG2, IPM200, RP56, SPACRCAN | interphotoreceptor matrix proteoglycan 2 |
C00000958
|
| 3632 | INPP5A, 5PTASE | inositol polyphosphate-5-phosphatase, 40kDa (EC:3.1.3.56) |
C00000958
|
| 1012 | CDH13, CDHH, P105 | cadherin 13 (EC:3.6.1.3) |
C00000958
|
| 55174 | INTS10, C8orf35, INT10 | integrator complex subunit 10 |
C00000958
|
| 55656 | INTS8, C8orf52, INT8 | integrator complex subunit 8 |
C00000958
|
| 55756 | INTS9, CPSF2L, INT9, RC74 | integrator complex subunit 9 |
C00000958
|
| 27152 | INTU, INT, PDZD6, PDZK6 | inturned planar cell polarity protein |
C00000958
|
| 27130 | INVS, INV, NPH2, NPHP2 | inversin |
C00000958
|
| 51447 | IP6K2, IHPK2, PIUS | inositol hexakisphosphate kinase 2 (EC:2.7.4.21) |
C00000958
|
| 51194 | IPO11, RanBP11 | importin 11 |
C00000958
|
| 134728 | IRAK1BP1, AIP70, SIMPL | interleukin-1 receptor-associated kinase 1 binding protein 1 |
C00000958
|
| 11213 | IRAK3, ASRT5, IRAKM | interleukin-1 receptor-associated kinase 3 (EC:2.7.11.1) |
C00000958
|
| 3658 | IREB2, ACO3, IRP2, IRP2AD | iron-responsive element binding protein 2 |
C00000958
|
| 3660 | IRF2, IRF-2 | interferon regulatory factor 2 |
C00000958
|
| 8660 | IRS2, IRS-2 | insulin receptor substrate 2 |
C00000958
|
| 122961 | ISCA2, HBLD1, ISA2, c14_5557 | iron-sulfur cluster assembly 2 |
C00000958
|
| 64843 | ISL2 | ISL LIM homeobox 2 |
C00000958
|
| 83737 | ITCH, AIF4, AIP4, NAPP1, dJ468O1.1 | itchy E3 ubiquitin protein ligase |
C00000958
|
| 3676 | ITGA4, CD49D, IA4 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
C00000958
|
| 3655 | ITGA6, CD49f, ITGA6B, VLA-6 | integrin, alpha 6 |
C00000958
|
| 79577 | CDC73, C1orf28, HPTJT, HRPT2, HYX | cell division cycle 73 |
C00000958
|
| 26548 | ITGB1BP2, CHORDC3, ITGB1BP, MELUSIN | integrin beta 1 binding protein (melusin) 2 |
C00000958
|
| 56990 | CDC42SE2, SPEC2 | CDC42 small effector 2 |
C00000958
|
| 3691 | ITGB4, CD104 | integrin, beta 4 |
C00000958
|
| 3709 | ITPR2, IP3R2 | inositol 1,4,5-trisphosphate receptor, type 2 |
C00000958
|
| 85450 | ITPRIP, DANGER, KIAA1754, bA127L20, bA127L20.2 | inositol 1,4,5-trisphosphate receptor interacting protein |
C00000958
|
| 6453 | ITSN1, ITSN, SH3D1A, SH3P17 | intersectin 1 (SH3 domain protein) |
C00000958
|
| 55677 | IWS1 | IWS1 homolog (S. cerevisiae) |
C00000958
|
| 182 | JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1 | jagged 1 |
C00000958
|
| 10602 | CDC42EP3, BORG2, CEP3, UB1 | CDC42 effector protein (Rho GTPase binding) 3 |
C00000958
|
| 3720 | JARID2, JMJ | jumonji, AT rich interactive domain 2 |
C00000958
|
| 965 | CD58, LFA-3, LFA3, ag3 | CD58 molecule |
C00000958
|
| 23189 | KANK1, ANKRD15, CPSQ2, KANK | KN motif and ankyrin repeat domains 1 |
C00000958
|
| 54934 | KANSL2, C12orf41, NSL2 | KAT8 regulatory NSL complex subunit 2 |
C00000958
|
| 23522 | KAT6B, GTPTS, MORF, MOZ2, MYST4, ZC2HC6B, qkf, querkopf | K(lysine) acetyltransferase 6B (EC:2.3.1.48) |
C00000958
|
| 23607 | CD2AP, CMS | CD2-associated protein |
C00000958
|
| 79768 | KATNBL1, C15orf29 | katanin p80 subunit B-like 1 |
C00000958
|
| 23254 | KAZN, KAZ | kazrin, periplakin interacting protein |
C00000958
|
| 25948 | KBTBD2, BKLHD1 | kelch repeat and BTB (POZ) domain containing 2 |
C00000958
|
| 27345 | KCNMB4 | potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
C00000958
|
| 83892 | KCTD10, BTBD28, ULRO61, hBACURD3 | potassium channel tetramerization domain containing 10 |
C00000958
|
| 57528 | KCTD16 | potassium channel tetramerization domain containing 16 |
C00000958
|
| 11015 | KDELR3, ERD2L3 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
C00000958
|
| 23081 | KDM4C, GASC1, JHDM3C, JMJD2C, TDRD14C, bA146B14.1 | lysine (K)-specific demethylase 4C (EC:1.14.11.-) |
C00000958
|
| 54462 | CCSER2, FAM190B, Gcap14, KIAA1128, bA486O22.1 | coiled-coil serine-rich protein 2 |
C00000958
|
| 3795 | KHK | ketohexokinase (fructokinase) (EC:2.7.1.3) |
C00000958
|
| 9897 | SPG8 | KIAA0196 |
C00000958
|
| 9692 | MRPP3, PRORP | KIAA0391 |
C00000958
|
| 23247 | KIAA0556 |
C00000958
|
|
| 9786 | Talpid3 | KIAA0586 |
C00000958
|
| 22889 | BLOM7, RP11-336K24.1 | KIAA0907 |
C00000958
|
| 23240 | TMEM131L | KIAA0922 |
C00000958
|
| 23325 | SWIP | KIAA1033 |
C00000958
|
| 23285 | KIAA1107 |
C00000958
|
|
| 84162 | FSA, Tweek | KIAA1109 |
C00000958
|
| 57536 | KIAA1328 |
C00000958
|
|
| 57589 | CIP150, bA207C16.1 | KIAA1432 |
C00000958
|
| 57698 | KIAA1598 |
C00000958
|
|
| 84542 | KIAA1841 |
C00000958
|
|
| 158358 | KIAA2026 |
C00000958
|
|
| 63971 | KIF13A, RBKIN, bA500C11.2 | kinesin family member 13A |
C00000958
|
| 25819 | CCRN4L, CCR4L, NOC | CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
C00000958
|
| 56992 | KIF15, HKLP2, KNSL7, NY-BR-62 | kinesin family member 15 |
C00000958
|
| 55614 | KIF16B, C20orf23, KISC20ORF, SNX23 | kinesin family member 16B |
C00000958
|
| 219771 | CCNY, C10orf9, CBCP1, CCNX, CFP1 | cyclin Y |
C00000958
|
| 9585 | KIF20B, CT90, KRMP1, MPHOSPH1, MPP-1, MPP1 | kinesin family member 20B |
C00000958
|
| 24137 | KIF4A, KIF4, KIF4G1 | kinesin family member 4A |
C00000958
|
| 22920 | KIFAP3, FLA3, KAP-1, KAP-3, KAP3, SMAP, Smg-GDS, dJ190I16.1 | kinesin-associated protein 3 |
C00000958
|
| 3815 | KIT, C-Kit, CD117, PBT, SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (EC:2.7.10.1) |
C00000958
|
| 905 | CCNT2, CYCT2 | cyclin T2 |
C00000958
|
| 7071 | KLF10, EGR-alpha, EGRA, TIEG, TIEG1 | Kruppel-like factor 10 |
C00000958
|
| 57018 | CCNL1, ANIA6A, PRO1073, ania-6a | cyclin L1 |
C00000958
|
| 8609 | KLF7, UKLF | Kruppel-like factor 7 (ubiquitous) |
C00000958
|
| 23588 | KLHDC2, HCLP-1, HCLP1, LCP | kelch domain containing 2 |
C00000958
|
| 116138 | KLHDC3, PEAS, RP1-20C7.3, dJ20C7.3 | kelch domain containing 3 |
C00000958
|
| 59349 | KLHL12, C3IP1, DKIR, hDKIR | kelch-like family member 12 |
C00000958
|
| 80311 | KLHL15 | kelch-like family member 15 |
C00000958
|
| 23276 | KLHL18 | kelch-like family member 18 |
C00000958
|
| 27252 | KLHL20, KHLHX, KLEIP, KLHLX, RP3-383J4.3 | kelch-like family member 20 |
C00000958
|
| 9903 | KLHL21 | kelch-like family member 21 |
C00000958
|
| 55295 | KLHL26 | kelch-like family member 26 |
C00000958
|
| 55958 | KLHL9 | kelch-like family member 9 |
C00000958
|
| 16627 |
C00000958
|
||
| 58508 | KMT2C, HALR, MLL3 | lysine (K)-specific methyltransferase 2C (EC:2.1.1.43) |
C00000958
|
| 901 | CCNG2 | cyclin G2 |
C00000958
|
| 900 | CCNG1, CCNG | cyclin G1 |
C00000958
|
| 889 | KRIT1, CAM, CCM1 | KRIT1, ankyrin repeat containing |
C00000958
|
| 84456 | L3MBTL3, MBT-1, MBT1, RP11-73O6.1 | l(3)mbt-like 3 (Drosophila) |
C00000958
|
| 246269 | LACE1, AFG1 | lactation elevated 1 |
C00000958
|
| 114294 | LACTB, G24, MRPL56 | lactamase, beta |
C00000958
|
| 51110 | LACTB2 | lactamase, beta 2 |
C00000958
|
| 28956 | LAMTOR2, ENDAP, HSPC003, MAPBPIP, MAPKSP1AP, ROBLD3, Ragulator2, p14 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
C00000958
|
| 26524 | LATS2, KPM | large tumor suppressor kinase 2 (EC:2.7.11.1) |
C00000958
|
| 899 | CCNF, FBX1, FBXO1 | cyclin F |
C00000958
|
| 143458 | LDLRAD3, LRAD3 | low density lipoprotein receptor class A domain containing 3 |
C00000958
|
| 116842 | LEAP2, LEAP-2 | liver expressed antimicrobial peptide 2 |
C00000958
|
| 114823 | LENG8, pp13842 | leukocyte receptor cluster (LRC) member 8 |
C00000958
|
| 3959 | LGALS3BP, 90K, BTBD17B, MAC-2-BP, TANGO10B | lectin, galactoside-binding, soluble, 3 binding protein |
C00000958
|
| 55366 | LGR4, BNMD17, GPR48 | leucine-rich repeat containing G protein-coupled receptor 4 |
C00000958
|
| 10186 | LHFP | lipoma HMGIC fusion partner (EC:3.6.5.4) |
C00000958
|
| 3980 | LIG3, LIG2 | ligase III, DNA, ATP-dependent (EC:6.5.1.1) |
C00000958
|
| 8994 | LIMD1 | LIM domains containing 1 |
C00000958
|
| 286826 | LIN9, BARA, BARPsv, Lin-9, TGS, TGS1, TGS2 | lin-9 homolog (C. elegans) |
C00000958
|
| 284029 | LINC00324, C17orf44, NCRNA00324 | long intergenic non-protein coding RNA 324 |
C00000958
|
| 81562 | LMAN2L, VIPL | lectin, mannose-binding 2-like |
C00000958
|
| 64327 | LMBR1, ACHP, C7orf2, DIF14, PPD2, TPT, ZRS | limb development membrane protein 1 |
C00000958
|
| 55788 | LMBRD1, C6orf209, LMBD1, MAHCF, NESI | LMBR1 domain containing 1 |
C00000958
|
| 57820 | CCNB1IP1, C14orf18, HEI10 | cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
C00000958
|
| 4012 | LNPEP, CAP, IRAP, P-LAP, PLAP | leucyl/cystinyl aminopeptidase (EC:3.4.11.3) |
C00000958
|
| 4015 | LOX | lysyl oxidase (EC:1.4.3.13) |
C00000958
|
| 2846 | LPAR4, GPR23, LPA4, P2RY9, P2Y5-LIKE, P2Y9 | lysophosphatidic acid receptor 4 |
C00000958
|
| 254531 | LPCAT4, AGPAT7, AYTL3, LPAAT-eta, LPEAT2 | lysophosphatidylcholine acyltransferase 4 (EC:2.3.1.23 2.3.1.67 2.3.1.n6 2.3.1.n7 2.3.1.121) |
C00000958
|
| 987 | LRBA, BGL, CDC4L, CVID8, LAB300, LBA | LPS-responsive vesicle trafficking, beach and anchor containing |
C00000958
|
| 23143 | LRCH1, CHDC1, NP81 | leucine-rich repeats and calponin homology (CH) domain containing 1 |
C00000958
|
| 84859 | LRCH3 | leucine-rich repeats and calponin homology (CH) domain containing 3 |
C00000958
|
| 84918 | LRP11, MANSC3, bA350J20.3 | low density lipoprotein receptor-related protein 11 |
C00000958
|
| 54535 | CCHCR1, C6orf18, HCR, SBP | coiled-coil alpha-helical rod protein 1 |
C00000958
|
| 55604 | LRRC16A, CARMIL, CARMIL1, CARMIL1a, LRRC16, dJ501N12.1, dJ501N12.5 | leucine rich repeat containing 16A |
C00000958
|
| 123355 | LRRC28 | leucine rich repeat containing 28 |
C00000958
|
| 55631 | LRRC40, dJ677H15.1 | leucine rich repeat containing 40 |
C00000958
|
| 55297 | CCDC91, p56 | coiled-coil domain containing 91 |
C00000958
|
| 65999 | LRRC61, HSPC295 | leucine rich repeat containing 61 |
C00000958
|
| 57554 | LRRC7, DENSIN | leucine rich repeat containing 7 |
C00000958
|
| 56262 | LRRC8A, AGM5, LRRC8 | leucine rich repeat containing 8 family, member A |
C00000958
|
| 55144 | LRRC8D, LRRC5 | leucine rich repeat containing 8 family, member D |
C00000958
|
| 85444 | LRRCC1, CLERC, SAP2, VFL1 | leucine rich repeat and coiled-coil centrosomal protein 1 |
C00000958
|
| 79080 | CCDC86 | coiled-coil domain containing 86 |
C00000958
|
| 90678 | LRSAM1, CMT2P, RIFLE, TAL | leucine rich repeat and sterile alpha motif containing 1 |
C00000958
|
| 84967 | LSM10, MST074, MSTP074 | LSM10, U7 small nuclear RNA associated |
C00000958
|
| 11157 | LSM6, YDR378C | LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
C00000958
|
| 286006 | LSMEM1, C7orf53 | leucine-rich single-pass membrane protein 1 |
C00000958
|
| 51599 | LSR, ILDR3, LISCH7 | lipolysis stimulated lipoprotein receptor |
C00000958
|
| 56925 | LXN, ECI, TCI | latexin |
C00000958
|
| 84318 | CCDC77 | coiled-coil domain containing 77 |
C00000958
|
| 144363 | LYRM5 | LYR motif containing 5 |
C00000958
|
| 1130 | LYST, CHS, CHS1 | lysosomal trafficking regulator |
C00000958
|
| 8216 | LZTR1, BTBD29, LZTR-1 | leucine-zipper-like transcription regulator 1 |
C00000958
|
| 130951 | M1AP, C2orf65, D6Mm5e, SPATA37 | meiosis 1 associated protein |
C00000958
|
| 23499 | MACF1, ABP620, ACF7, MACF, OFC4 | microtubule-actin crosslinking factor 1 |
C00000958
|
| 23764 | MAFF, U-MAF, hMafF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
C00000958
|
| 4097 | MAFG, hMAF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
C00000958
|
| 9223 | MAGI1, AIP-3, AIP3, BAIAP1, BAP-1, BAP1, MAGI-1, TNRC19, WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 |
C00000958
|
| 114569 | MAL2 | mal, T-cell differentiation protein 2 (gene/pseudogene) |
C00000958
|
| 10892 | MALT1, IMD12, MLT, MLT1 | mucosa associated lymphoid tissue lymphoma translocation gene 1 (EC:3.4.22.-) |
C00000958
|
| 84441 | MAML2, MAM-3, MAM2, MAM3, MLL-MAML2 | mastermind-like 2 (Drosophila) |
C00000958
|
| 4122 | MAN2A2, MANA2X | mannosidase, alpha, class 2A, member 2 (EC:3.2.1.114) |
C00000958
|
| 64925 | CCDC71 | coiled-coil domain containing 71 |
C00000958
|
| 81631 | MAP1LC3B, ATG8F, LC3B, MAP1A/1BLC3, MAP1LC3B-a | microtubule-associated protein 1 light chain 3 beta |
C00000958
|
| 5608 | MAP2K6, MAPKK6, MEK6, MKK6, PRKMK6, SAPKK-3, SAPKK3 | mitogen-activated protein kinase kinase 6 (EC:2.7.12.2) |
C00000958
|
| 4214 | MAP3K1, MAPKKK1, MEKK, MEKK_1, MEKK1, SRXY6 | mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase (EC:2.7.11.25) |
C00000958
|
| 9020 | MAP3K14, FTDCR1B, HS, HSNIK, NIK | mitogen-activated protein kinase kinase kinase 14 (EC:2.7.11.25) |
C00000958
|
| 1326 | MAP3K8, COT, EST, ESTF, MEKK8, TPL2, Tpl-2, c-COT | mitogen-activated protein kinase kinase kinase 8 (EC:2.7.11.25) |
C00000958
|
| 8491 | MAP4K3, GLK, MAPKKKK3, MEKKK_3, MEKKK3, RAB8IPL1 | mitogen-activated protein kinase kinase kinase kinase 3 (EC:2.7.11.1) |
C00000958
|
| 84660 | CCDC62, CT109, ERAP75, TSP-NY | coiled-coil domain containing 62 |
C00000958
|
| 51134 | CCDC41, Cep83, NY-REN-58 | coiled-coil domain containing 41 |
C00000958
|
| 374969 | CCDC23 | coiled-coil domain containing 23 |
C00000958
|
| 343099 | CCDC18, NY-SAR-41, dJ717I23.1 | coiled-coil domain containing 18 |
C00000958
|
| 154467 | CCDC167, C6orf129, HSPC265 | coiled-coil domain containing 167 |
C00000958
|
| 8550 | MAPKAPK5, MAPKAP-K5, MK-5, MK5, PRAK | mitogen-activated protein kinase-activated protein kinase 5 (EC:2.7.11.1) |
C00000958
|
| 22924 | MAPRE3, EB3, EBF3, EBF3-S, RP3 | microtubule-associated protein, RP/EB family, member 3 |
C00000958
|
| 115123 | MARCH3, MARCH-III, RNF173 | membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase (EC:6.3.2.19) |
C00000958
|
| 165055 | CCDC138 | coiled-coil domain containing 138 |
C00000958
|
| 55777 | MBD5, MRD1 | methyl-CpG binding domain protein 5 |
C00000958
|
| 10150 | MBNL2, MBLL, MBLL39, PRO2032 | muscleblind-like splicing regulator 2 |
C00000958
|
| 154141 | MBOAT1, 1, LPEAT1, LPLAT, LPLAT_1, LPSAT, OACT1, dJ434O11.1 | membrane bound O-acyltransferase domain containing 1 (EC:2.3.1.n6) |
C00000958
|
| 55610 | CCDC132 | coiled-coil domain containing 132 |
C00000958
|
| 90693 | CCDC126 | coiled-coil domain containing 126 |
C00000958
|
| 255231 | MCOLN2, TRP-ML2, TRPML2 | mucolipin 2 |
C00000958
|
| 28985 | MCTS1, MCT-1, MCT1 | malignant T cell amplified sequence 1 |
C00000958
|
| 90550 | MCU, C10orf42, CCDC109A | mitochondrial calcium uniporter |
C00000958
|
| 400569 | MED11 | mediator complex subunit 11 |
C00000958
|
| 116931 | MED12L, NOPAR, TNRC11L, TRALP, TRALPUSH | mediator complex subunit 12-like |
C00000958
|
| 23389 | MED13L, PROSIT240, THRAP2, TRAP240L | mediator complex subunit 13-like |
C00000958
|
| 9441 | MED26, CRSP7, CRSP70 | mediator complex subunit 26 |
C00000958
|
| 9443 | MED7, ARC34, CRSP33, CRSP9 | mediator complex subunit 7 |
C00000958
|
| 55090 | MED9, MED25 | mediator complex subunit 9 |
C00000958
|
| 84935 | MEDAG, AWMS3, C13orf33, MEDA-4, MEDA4, hAWMS3 | mesenteric estrogen-dependent adipogenesis |
C00000958
|
| 4212 | MEIS2, HsT18361, MRG1 | Meis homeobox 2 |
C00000958
|
| 202243 | CCDC125, KENAE | coiled-coil domain containing 125 |
C00000958
|
| 388389 | CCDC103, CILD17, PR46b, SMH | coiled-coil domain containing 103 |
C00000958
|
| 196074 | METTL15, METT5D1 | methyltransferase like 15 (EC:2.1.1.-) |
C00000958
|
| 57545 | CC2D2A, JBTS9, MKS6 | coiled-coil and C2 domain containing 2A |
C00000958
|
| 64863 | METTL4, HsT661 | methyltransferase like 4 |
C00000958
|
| 79828 | METTL8, TIP | methyltransferase like 8 |
C00000958
|
| 51320 | MEX3C, MEX-3C, RKHD2, RNF194 | mex-3 RNA binding family member C |
C00000958
|
| 200014 | CC2D1B | coiled-coil and C2 domain containing 1B |
C00000958
|
| 25776 | CBY1, C22orf2, CBY, HS508I15A, PGEA1, PIGEA-14, PIGEA14, arb1 | chibby homolog 1 (Drosophila) |
C00000958
|
| 84709 | MGARP, C4orf49, CESP-1, HUMMR, OSAP | mitochondria-localized glutamic acid-rich protein |
C00000958
|
| 4249 | MGAT5, GNT-V, GNT-VA | mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase (EC:2.4.1.155) |
C00000958
|
| 10724 | MGEA5, MEA5, NCOAT, OGA | meningioma expressed antigen 5 (hyaluronidase) (EC:2.3.1.48 3.2.1.169) |
C00000958
|
| 8535 | CBX4, NBP16, PC2 | chromobox homolog 4 |
C00000958
|
| 286097 | MICU3, EFHA2 | mitochondrial calcium uptake family, member 3 |
C00000958
|
| 4281 | MID1, BBBG1, FXY, GBBB1, MIDIN, OGS1, OS, OSX, RNF59, TRIM18, XPRF, ZNFXY | midline 1 (Opitz/BBB syndrome) |
C00000958
|
| 54471 | MIEF1, HSU79252, MID51, SMCR7L, dJ1104E15.3 | mitochondrial elongation factor 1 |
C00000958
|
| 57708 | MIER1, ER1, MI-ER1 | mesoderm induction early response 1, transcriptional regulator |
C00000958
|
| 84864 | MINA, MDIG, MINA53, NO52, ROX | MYC induced nuclear antigen |
C00000958
|
| 145282 | MIPOL1 | mirror-image polydactyly 1 |
C00000958
|
| 54069 | MIS18A, B28, C21orf45, C21orf46, FASP1, MIS18alpha, hMis18alpha | MIS18 kinetochore protein A |
C00000958
|
| 4286 | MITF, CMM8, MI, WS2, WS2A, bHLHe32 | microphthalmia-associated transcription factor |
C00000958
|
| 57496 | MKL2, MRTF-B, NPD001 | MKL/myocardin-like 2 |
C00000958
|
| 4289 | MKLN1, TWA2 | muskelin 1, intracellular mediator containing kelch motifs |
C00000958
|
| 23608 | MKRN1, RNF61 | makorin ring finger protein 1 |
C00000958
|
| 283078 | MKX, C10orf48, IFRX, IRXL1 | mohawk homeobox |
C00000958
|
| 4292 | MLH1, COCA2, FCC2, HNPCC, HNPCC2, hMLH1 | mutL homolog 1 |
C00000958
|
| 8028 | MLLT10, AF10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
C00000958
|
| 25974 | MMACHC, RP11-291L19.3, cblC | methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
C00000958
|
| 79872 | CBLL1, HAKAI, RNF188 | Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
C00000958
|
| 868 | CBLB, Cbl-b, RNF56 | Cbl proto-oncogene B, E3 ubiquitin protein ligase |
C00000958
|
| 55329 | MNS1, SPATA40 | meiosis-specific nuclear structural 1 |
C00000958
|
| 64112 | MOAP1, MAP-1, PNMA4 | modulator of apoptosis 1 |
C00000958
|
| 55034 | MOCOS, HMCS, MCS, MOS | molybdenum cofactor sulfurase (EC:2.8.1.9) |
C00000958
|
| 23041 | MON2 | MON2 homolog (S. cerevisiae) |
C00000958
|
| 158747 | MOSPD2 | motile sperm domain containing 2 |
C00000958
|
| 4343 | MOV10, fSAP113, gb110 | Mov10, Moloney leukemia virus 10, homolog (mouse) (EC:3.6.4.13) |
C00000958
|
| 10200 | MPHOSPH6, MPP, MPP-6, MPP6 | M-phase phosphoprotein 6 |
C00000958
|
| 858 | CAV2, CAV | caveolin 2 |
C00000958
|
| 4351 | MPI, CDG1B, PMI, PMI1 | mannose phosphate isomerase (EC:5.3.1.8) |
C00000958
|
| 51678 | MPP6, PALS2, VAM-1, VAM1, p55T | membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
C00000958
|
| 64921 | CASD1, C7orf12, NBLA04196 | CAS1 domain containing 1 |
C00000958
|
| 54948 | MRPL16, L16mt, MRP-L16 | mitochondrial ribosomal protein L16 |
C00000958
|
| 9553 | MRPL33, C2orf1, L33mt, MRP-L33, RPL33L | mitochondrial ribosomal protein L33 |
C00000958
|
| 26589 | MRPL46, C15orf4, LIECG2, P2ECSL | mitochondrial ribosomal protein L46 |
C00000958
|
| 64963 | MRPS11, HCC-2 | mitochondrial ribosomal protein S11 |
C00000958
|
| 55168 | MRPS18A, HumanS18b, MRP-S18-3, MRPS18-3, S18bmt | mitochondrial ribosomal protein S18A |
C00000958
|
| 51116 | MRPS2, MRP-S2, S2mt | mitochondrial ribosomal protein S2 |
C00000958
|
| 4436 | MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2 | mutS homolog 2 |
C00000958
|
| 73390 |
C00000958
|
||
| 4501 | MT1X, MT-1l, MT1 | metallothionein 1X |
C00000958
|
| 4502 | MT2A, MT2 | metallothionein 2A |
C00000958
|
| 51001 | MTERFD1, mTERF3 | MTERF domain containing 1 |
C00000958
|
| 123263 | MTFMT, COXPD15, FMT1 | mitochondrial methionyl-tRNA formyltransferase (EC:2.1.2.9) |
C00000958
|
| 22794 | CASC3, BTZ, MLN51 | cancer susceptibility candidate 3 |
C00000958
|
| 4534 | MTM1, CNM, MTMX, XLMTM | myotubularin 1 (EC:3.1.3.64) |
C00000958
|
| 54516 | MTRF1L, HMRF1L, MRF1L, mtRF1a | mitochondrial translational release factor 1-like |
C00000958
|
| 10651 | MTX2 | metaxin 2 |
C00000958
|
| 347273 | MURC, CAVIN4, cavin-4 | muscle-related coiled-coil protein |
C00000958
|
| 65981 | CAPRIN2, C1QDC1, EEG-1, EEG1, RNG140 | caprin family member 2 |
C00000958
|
| 4599 | MX1, IFI-78K, IFI78, MX, MxA | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
C00000958
|
| 4600 | MX2, MXB | myxovirus (influenza virus) resistance 2 (mouse) |
C00000958
|
| 4603 | MYBL1, A-MYB, AMYB | v-myb avian myeloblastosis viral oncogene homolog-like 1 |
C00000958
|
| 819 | CAMLG, CAML | calcium modulating ligand |
C00000958
|
| 23077 | MYCBP2, PAM | MYC binding protein 2, E3 ubiquitin protein ligase |
C00000958
|
| 50804 | MYEF2, HsT18564, MEF-2, MST156, MSTP156 | myelin expression factor 2 |
C00000958
|
| 4628 | MYH10, NMMHC-IIB, NMMHCB | myosin, heavy chain 10, non-muscle |
C00000958
|
| 29116 | MYLIP, IDOL, MIR | myosin regulatory light chain interacting protein (EC:6.3.2.-) |
C00000958
|
| 4651 | MYO10 | myosin X |
C00000958
|
| 79823 | CAMKMT, C2orf34, CLNMT, CaM_KMT, Cam, KMT | calmodulin-lysine N-methyltransferase (EC:2.1.1.60) |
C00000958
|
| 4643 | MYO1E, FSGS6, HuncM-IC, MYO1C | myosin IE |
C00000958
|
| 4646 | MYO6, DFNA22, DFNB37 | myosin VI |
C00000958
|
| 4649 | MYO9A | myosin IXA |
C00000958
|
| 84665 | MYPN, CMD1DD, CMH22, MYOP, RCM4 | myopalladin |
C00000958
|
| 114803 | MYSM1, 2A-DUB, 2ADUB, RP4-592A1.1 | Myb-like, SWIRM and MPN domains 1 (EC:3.1.2.15) |
C00000958
|
| 817 | CAMK2D, CAMKD | calcium/calmodulin-dependent protein kinase II delta (EC:2.7.11.17) |
C00000958
|
| 8883 | NAE1, A-116A10.1, APPBP1, ula-1 | NEDD8 activating enzyme E1 subunit 1 |
C00000958
|
| 55577 | NAGK, GNK, HSA242910 | N-acetylglucosamine kinase (EC:2.7.1.59) |
C00000958
|
| 222236 | NAPEPLD, FMP30, NAPE-PLD | N-acyl phosphatidylethanolamine phospholipase D (EC:3.1.4.54) |
C00000958
|
| 8774 | NAPG, GAMMASNAP | N-ethylmaleimide-sensitive factor attachment protein, gamma |
C00000958
|
| 100195127 |
C00000958
|
||
| 79731 | NARS2, SLM5, asnRS | asparaginyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.22) |
C00000958
|
| 9 | NAT1, AAC1, MNAT, NAT-1, NATI | N-acetyltransferase 1 (arylamine N-acetyltransferase) (EC:2.3.1.5) |
C00000958
|
| 55226 | NAT10, ALP, NET43 | N-acetyltransferase 10 (GCN5-related) (EC:2.3.1.-) |
C00000958
|
| 51594 | NBAS, NAG, SOPH | neuroblastoma amplified sequence |
C00000958
|
| 65065 | NBEAL1, A530083I02Rik, ALS2CR16, ALS2CR17 | neurobeachin-like 1 |
C00000958
|
| 782 | CACNB1, CAB1, CACNLB1, CCHLB1 | calcium channel, voltage-dependent, beta 1 subunit |
C00000958
|
| 23310 | NCAPD3, CAP-D3, CAPD3, hCAP-D3, hHCP-6, hcp-6 | non-SMC condensin II complex, subunit D3 |
C00000958
|
| 138199 | C9orf41 | chromosome 9 open reading frame 41 |
C00000958
|
| 4690 | NCK1, NCK, NCKalpha, nck-1 | NCK adaptor protein 1 |
C00000958
|
| 84909 | C9orf3, AOPEP, AP-O, APO, C90RF3, ONPEP | chromosome 9 open reading frame 3 |
C00000958
|
| 84688 | C9orf24, CBE1, NYD-SP22, SMRP1, bA573M23.4 | chromosome 9 open reading frame 24 |
C00000958
|
| 138724 | C9orf131 | chromosome 9 open reading frame 131 |
C00000958
|
| 4541 | ND6, MTND6, MT-ND6 | NADH dehydrogenase, subunit 6 (complex I) |
C00000958
|
| 78996 | C7orf49, MRI | chromosome 7 open reading frame 49 |
C00000958
|
| 57827 | C6orf47, D6S53E, G4, NG34 | chromosome 6 open reading frame 47 |
C00000958
|
| 8509 | NDST2, HSST2, NST2 | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 (EC:2.8.2.8) |
C00000958
|
| 51103 | NDUFAF1, CGI65, CIA30 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
C00000958
|
| 4729 | NDUFV2, CI-24k | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa (EC:1.6.5.3 1.6.99.3) |
C00000958
|
| 121441 | NEDD1, GCP-WD, TUBGCP7 | neural precursor cell expressed, developmentally down-regulated 1 |
C00000958
|
| 4734 | NEDD4, NEDD4-1, RPF1 | neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00000958
|
| 23327 | NEDD4L, NEDD4-2, NEDD4.2, RSP5, hNEDD4-2 | neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase (EC:6.3.2.-) |
C00000958
|
| 51250 | C6orf203, PRED31, RP11-59I9.1 | chromosome 6 open reading frame 203 |
C00000958
|
| 100425666 |
C00000958
|
||
| 4750 | NEK1, NY-REN-55, SRPS2, SRPS2A | NIMA-related kinase 1 (EC:2.7.11.1) |
C00000958
|
| 4752 | NEK3, HSPK36 | NIMA-related kinase 3 (EC:2.7.11.1) |
C00000958
|
| 10783 | NEK6, SID6-1512 | NIMA-related kinase 6 (EC:2.7.11.1) |
C00000958
|
| 374987 | NEXN-AS1, C1orf118 | NEXN antisense RNA 1 |
C00000958
|
| 4763 | NF1, NFNS, VRNF, WSS | neurofibromin 1 |
C00000958
|
| 65250 | C5orf42, JBTS17 | chromosome 5 open reading frame 42 |
C00000958
|
| 4774 | NFIA, CTF, NF-I/A, NF1-A, NFI-A, NFI-L | nuclear factor I/A |
C00000958
|
| 4783 | NFIL3, E4BP4, IL3BP1, NF-IL3A, NFIL3A | nuclear factor, interleukin 3 regulated |
C00000958
|
| 64417 | C5orf28 | chromosome 5 open reading frame 28 |
C00000958
|
| 54969 | C4orf27 | chromosome 4 open reading frame 27 |
C00000958
|
| 152078 | C3orf55 | chromosome 3 open reading frame 55 |
C00000958
|
| 9054 | NFS1, IscS, NIFS | NFS1 cysteine desulfurase (EC:2.8.1.7) |
C00000958
|
| 4799 | NFX1, NFX2 | nuclear transcription factor, X-box binding 1 (EC:6.3.2.-) |
C00000958
|
| 4800 | NFYA, CBF-A, CBF-B, HAP2, NF-YA | nuclear transcription factor Y, alpha |
C00000958
|
| 4801 | NFYB, CBF-A, CBF-B, HAP3, NF-YB | nuclear transcription factor Y, beta |
C00000958
|
| 55768 | NGLY1, CDG1V, PNG1, PNGase | N-glycanase 1 (EC:3.5.1.52) |
C00000958
|
| 51335 | NGRN, DSC92 | neugrin, neurite outgrowth associated |
C00000958
|
| 60491 | NIF3L1, ALS2CR1, CALS-7 | NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
C00000958
|
| 307652 |
C00000958
|
||
| 4817 | NIT1 | nitrilase 1 |
C00000958
|
| 51701 | NLK | nemo-like kinase (EC:2.7.11.24) |
C00000958
|
| 29922 | NME7, MN23H7, NDK_7, NDK7, nm23-H7 | NME/NM23 family member 7 (EC:2.7.4.6) |
C00000958
|
| 9111 | NMI | N-myc (and STAT) interactor |
C00000958
|
| 4837 | NNMT | nicotinamide N-methyltransferase (EC:2.1.1.1) |
C00000958
|
| 28987 | NOB1, ART-4, MST158, NOB1P, PSMD8BP1 | NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
C00000958
|
| 64318 | NOC3L, AD24, C10orf117, FAD24 | nucleolar complex associated 3 homolog (S. cerevisiae) |
C00000958
|
| 9221 | NOLC1, NOPP130, NOPP140, NS5ATP13, P130 | nucleolar and coiled-body phosphoprotein 1 |
C00000958
|
| 57415 | C3orf14 | chromosome 3 open reading frame 14 |
C00000958
|
| 9520 | NPEPPS, AAP-S, MP100, PSA | aminopeptidase puromycin sensitive (EC:3.4.11.14) |
C00000958
|
| 8431 | NR0B2, SHP, SHP1 | nuclear receptor subfamily 0, group B, member 2 |
C00000958
|
| 9971 | NR1H4, BAR, FXR, HRR-1, HRR1, RIP14 | nuclear receptor subfamily 1, group H, member 4 |
C00000958
|
| 126382 | NR2C2AP, TRA16 | nuclear receptor 2C2-associated protein |
C00000958
|
| 60526 | C2orf43 | chromosome 2 open reading frame 43 |
C00000958
|
| 3164 | NR4A1, GFRP1, HMR, N10, NAK-1, NGFIB, NP10, NUR77, TR3 | nuclear receptor subfamily 4, group A, member 1 |
C00000958
|
| 4929 | NR4A2, HZF-3, NOT, NURR1, RNR1, TINUR | nuclear receptor subfamily 4, group A, member 2 |
C00000958
|
| 8013 | NR4A3, CHN, CSMF, MINOR, NOR1, TEC | nuclear receptor subfamily 4, group A, member 3 |
C00000958
|
| 9315 | NREP, C5orf13, D4S114, P311, PRO1873, PTZ17, SEZ17 | neuronal regeneration related protein |
C00000958
|
| 9847 | C2CD5, CDP138, KIAA0528 | C2 calcium-dependent domain containing 5 |
C00000958
|
| 8204 | NRIP1, RIP140 | nuclear receptor interacting protein 1 |
C00000958
|
| 4905 | NSF, SKD2 | N-ethylmaleimide-sensitive factor (EC:3.6.4.6) |
C00000958
|
| 25936 | NSL1, C1orf48, MIS14 | NSL1, MIS12 kinetochore complex component |
C00000958
|
| 286053 | NSMCE2, C8orf36, MMS21, NSE2, ZMIZ7 | non-SMC element 2, MMS21 homolog (S. cerevisiae) |
C00000958
|
| 54780 | NSMCE4A, C10orf86, NS4EA, NSE4A | non-SMC element 4 homolog A (S. cerevisiae) |
C00000958
|
| 221078 | NSUN6, 4933414E04Rik, NOPD1 | NOP2/Sun domain family, member 6 |
C00000958
|
| 9891 | NUAK1, ARK5 | NUAK family, SNF1-like kinase, 1 (EC:2.7.11.1) |
C00000958
|
| 4682 | NUBP1, NBP, NBP1 | nucleotide binding protein 1 |
C00000958
|
| 84955 | NUDCD1, CML66, OVA66 | NudC domain containing 1 |
C00000958
|
| 83594 | NUDT12 | nudix (nucleoside diphosphate linked moiety X)-type motif 12 (EC:3.6.1.22) |
C00000958
|
| 11163 | NUDT4, DIPP2, DIPP2alpha, DIPP2beta | nudix (nucleoside diphosphate linked moiety X)-type motif 4 (EC:3.6.1.52) |
C00000958
|
| 83540 | NUF2, CDCA1, CT106, NUF2R | NUF2, NDC80 kinetochore complex component |
C00000958
|
| 8650 | NUMB, C14orf41, S171, c14_5527 | numb homolog (Drosophila) |
C00000958
|
| 26005 | C2CD3 | C2 calcium-dependent domain containing 3 |
C00000958
|
| 55746 | NUP133, hNUP133 | nucleoporin 133kDa |
C00000958
|
| 23279 | NUP160 | nucleoporin 160kDa |
C00000958
|
| 79023 | NUP37, p37 | nucleoporin 37kDa |
C00000958
|
| 79902 | NUP85, Nup75 | nucleoporin 85kDa |
C00000958
|
| 4928 | NUP98, ADIR2, NUP196, NUP96 | nucleoporin 98kDa |
C00000958
|
| 10482 | NXF1, MEX67, TAP | nuclear RNA export factor 1 |
C00000958
|
| 11247 | NXPH4, NPH4 | neurexophilin 4 |
C00000958
|
| 4938 | OAS1, IFI-4, OIAS, OIASI | 2'-5'-oligoadenylate synthetase 1, 40/46kDa (EC:2.7.7.84) |
C00000958
|
| 9854 | C2CD2L, TMEM24 | C2CD2-like |
C00000958
|
| 57489 | ODF2L, RP5-977L11.1, dJ977L11.1 | outer dense fiber of sperm tails 2-like |
C00000958
|
| 4967 | OGDH, AKGDH, E1k, OGDC | oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (EC:1.2.4.2) |
C00000958
|
| 8473 | OGT, HRNT1, O-GLCNAC | O-linked N-acetylglucosamine (GlcNAc) transferase (EC:2.4.1.255) |
C00000958
|
| 148423 | C1orf52, RP11-234D19.1, gm117 | chromosome 1 open reading frame 52 |
C00000958
|
| 25903 | OLFML2B, RP11-227F8.1 | olfactomedin-like 2B |
C00000958
|
| 148523 | C1orf51 | chromosome 1 open reading frame 51 |
C00000958
|
| 9480 | ONECUT2, OC-2, OC2 | one cut homeobox 2 |
C00000958
|
| 4983 | OPHN1, ARHGAP41, MRX60, OPN1 | oligophrenin 1 |
C00000958
|
| 80228 | ORAI2, C7orf19, CBCIP2, MEM142B, TMEM142B | ORAI calcium release-activated calcium modulator 2 |
C00000958
|
| 29095 | ORMDL2, MST095, MSTP095, adoplin-2 | ORM1-like 2 (S. cerevisiae) |
C00000958
|
| 26031 | OSBPL3, ORP-3, ORP3, OSBP3 | oxysterol binding protein-like 3 |
C00000958
|
| 51526 | OSER1, C20orf111, HSPC207, Osr1, Perit1, dJ1183I21.1 | oxidative stress responsive serine-rich 1 |
C00000958
|
| 55644 | OSGEP, GCPL1, KAE1, OSGEP1, PRSMG1 | O-sialoglycoprotein endopeptidase (EC:3.4.24.57) |
C00000958
|
| 64172 | OSGEPL1, Qri7 | O-sialoglycoprotein endopeptidase-like 1 (EC:3.4.24.57) |
C00000958
|
| 5016 | OVGP1, CHIT5, EGP, MUC9, OGP | oviductal glycoprotein 1, 120kDa |
C00000958
|
| 54995 | OXSM, FASN2D, KASI, KS | 3-oxoacyl-ACP synthase, mitochondrial (EC:2.3.1.41) |
C00000958
|
| 655678 |
C00000958
|
||
| 80227 | PAAF1, PAAF, Rpn14, WDR71 | proteasomal ATPase-associated factor 1 |
C00000958
|
| 26986 | PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1 | poly(A) binding protein, cytoplasmic 1 |
C00000958
|
| 5042 | PABPC3, PABP3, PABPL3, tPABP | poly(A) binding protein, cytoplasmic 3 |
C00000958
|
| 8761 | PABPC4, APP-1, APP1, PABP4, iPABP | poly(A) binding protein, cytoplasmic 4 (inducible form) |
C00000958
|
| 79447 | PAGR1, C16orf53, GAS, PA1 | PAXIP1 associated glutamate-rich protein 1 |
C00000958
|
| 8505 | PARG, PARG99 | poly (ADP-ribose) glycohydrolase (EC:3.2.1.143) |
C00000958
|
| 81563 | C1orf21, PIG13 | chromosome 1 open reading frame 21 |
C00000958
|
| 64761 | PARP12, ARTD12, MST109, MSTP109, ZC3H1, ZC3HDC1 | poly (ADP-ribose) polymerase family, member 12 (EC:2.4.2.30) |
C00000958
|
| 143 | PARP4, ADPRTL1, ARTD4, PARP-4, PARPL, PH5P, VAULT3, VPARP, VWA5C, p193 | poly (ADP-ribose) polymerase family, member 4 (EC:2.4.2.30) |
C00000958
|
| 56965 | PARP6, ARTD17, PARP-6-B1, PARP-6-C, pART17 | poly (ADP-ribose) polymerase family, member 6 (EC:2.4.2.30) |
C00000958
|
| 79668 | PARP8, ARTD16, pART16 | poly (ADP-ribose) polymerase family, member 8 (EC:2.4.2.30) |
C00000958
|
| 83666 | PARP9, ARTD9, BAL, BAL1, MGC:7868 | poly (ADP-ribose) polymerase family, member 9 (EC:2.4.2.30) |
C00000958
|
| 64098 | PARVG | parvin, gamma |
C00000958
|
| 55193 | PBRM1, BAF180, PB1 | polybromo 1 |
C00000958
|
| 5089 | PBX2, G17, HOX12, PBX2MHC | pre-B-cell leukemia homeobox 2 |
C00000958
|
| 128346 | C1orf162 | chromosome 1 open reading frame 162 |
C00000958
|
| 5095 | PCCA | propionyl CoA carboxylase, alpha polypeptide (EC:6.4.1.3) |
C00000958
|
| 57575 | PCDH10, OL-PCDH, PCDH19 | protocadherin 10 |
C00000958
|
| 54510 | PCDH18, PCDH68L | protocadherin 18 |
C00000958
|
| 51585 | PCF11 | PCF11 cleavage and polyadenylation factor subunit |
C00000958
|
| 7703 | PCGF2, MEL-18, RNF110, ZNF144 | polycomb group ring finger 2 |
C00000958
|
| 55795 | PCID2, F10 | PCI domain containing 2 |
C00000958
|
| 100380451 |
C00000958
|
||
| 11235 | PDCD10, CCM3, TFAR15 | programmed cell death 10 |
C00000958
|
| 9141 | PDCD5, TFAR19 | programmed cell death 5 |
C00000958
|
| 27115 | PDE7B, bA472E5.1 | phosphodiesterase 7B (EC:3.1.4.17) |
C00000958
|
| 56034 | PDGFC, FALLOTEIN, SCDGF | platelet derived growth factor C |
C00000958
|
| 56913 | C1GALT1, C1GALT, T-synthase | core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 (EC:2.4.1.122) |
C00000958
|
| 5162 | PDHB, PDHBD, PDHE1-B, PHE1B | pyruvate dehydrogenase (lipoamide) beta (EC:1.2.4.1) |
C00000958
|
| 5163 | PDK1 | pyruvate dehydrogenase kinase, isozyme 1 (EC:2.7.11.2) |
C00000958
|
| 10611 | PDLIM5, ENH, ENH1, LIM | PDZ and LIM domain 5 |
C00000958
|
| 81572 | PDRG1, C20orf126, PDRG | p53 and DNA-damage regulated 1 |
C00000958
|
| 23047 | PDS5B, APRIN, AS3, CG008 | PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
C00000958
|
| 57026 | PDXP, CIN, PLP, dJ37E16.5 | pyridoxal (pyridoxine, vitamin B6) phosphatase (EC:3.1.3.3 3.1.3.74) |
C00000958
|
| 100196721 |
C00000958
|
||
| 57162 | PELI1 | pellino E3 ubiquitin protein ligase 1 |
C00000958
|
| 8864 | PER2, FASPS, FASPS1 | period circadian clock 2 |
C00000958
|
| 5188 | PET112, PET112L | PET112 homolog (yeast) |
C00000958
|
| 8799 | PEX11B, PEX11-BETA, PEX14B | peroxisomal biogenesis factor 11 beta |
C00000958
|
| 5828 | PEX2, PAF1, PBD5A, PBD5B, PMP3, PMP35, PXMP3, RNF72, ZWS3 | peroxisomal biogenesis factor 2 |
C00000958
|
| 8504 | PEX3, PBD10A, TRG18 | peroxisomal biogenesis factor 3 |
C00000958
|
| 5209 | PFKFB3, IPFK2, PFK2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (EC:2.7.1.105 3.1.3.46) |
C00000958
|
| 125144 | C17orf76-AS1, C17orf45, NCRNA00188, TSAP19, FAM211A-AS1 | C17orf76 antisense RNA 1 |
C00000958
|
| 9749 | PHACTR2, C6orf56 | phosphatase and actin regulator 2 |
C00000958
|
| 65979 | PHACTR4 | phosphatase and actin regulator 4 |
C00000958
|
| 51131 | PHF11, APY, BCAP, IGEL, IGER, IGHER, NY-REN-34, NYREN34 | PHD finger protein 11 |
C00000958
|
| 29035 | C16orf72, PRO0149 | chromosome 16 open reading frame 72 |
C00000958
|
| 55023 | PHIP, BRWD2, DCAF14, WDR11, ndrp | pleckstrin homology domain interacting protein |
C00000958
|
| 5260 | PHKG1, PHKG | phosphorylase kinase, gamma 1 (muscle) (EC:2.7.11.1 2.7.11.19 2.7.11.26) |
C00000958
|
| 7262 | PHLDA2, BRW1C, BWR1C, HLDA2, IPL, TSSC3 | pleckstrin homology-like domain, family A, member 2 |
C00000958
|
| 23187 | PHLDB1, LL5A | pleckstrin homology-like domain, family B, member 1 |
C00000958
|
| 8554 | PIAS1, DDXBP1, GBP, GU/RH-II, ZMIZ3 | protein inhibitor of activated STAT, 1 |
C00000958
|
| 10464 | PIBF1, C13orf24, CEP90, PIBF, RP11-505F3.1 | progesterone immunomodulatory binding factor 1 |
C00000958
|
| 8301 | PICALM, CALM, CLTH, LAP | phosphatidylinositol binding clathrin assembly protein |
C00000958
|
| 9463 | PICK1, PICK, PRKCABP | protein interacting with PRKCA 1 |
C00000958
|
| 55022 | PID1, NYGGF4, P-CLI1, PCLI1 | phosphotyrosine interaction domain containing 1 |
C00000958
|
| 5277 | PIGA, GPI3, MCAHS2, PIG-A | phosphatidylinositol glycan anchor biosynthesis, class A (EC:2.4.1.198) |
C00000958
|
| 5281 | PIGF | phosphatidylinositol glycan anchor biosynthesis, class F (EC:2.7.8.1) |
C00000958
|
| 51227 | PIGP, DCRC, DCRC-S, DSCR5, DSRC | phosphatidylinositol glycan anchor biosynthesis, class P (EC:2.4.1.198) |
C00000958
|
| 55650 | PIGV, GPI-MT-II, HPMRS1, PIG-V | phosphatidylinositol glycan anchor biosynthesis, class V |
C00000958
|
| 84529 | C15orf41, HH114 | chromosome 15 open reading frame 41 |
C00000958
|
| 5293 | PIK3CD, P110DELTA, PI3K, p110D | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta (EC:2.7.1.153) |
C00000958
|
| 11040 | PIM2 | pim-2 oncogene (EC:2.7.11.1) |
C00000958
|
| 5305 | PIP4K2A, PI5P4KA, PIP5K2A, PIP5KII-alpha, PIP5KIIA, PIPK | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha (EC:2.7.1.149) |
C00000958
|
| 5311 | PKD2, APKD2, PC2, PKD4, Pc-2, TRPP2 | polycystic kidney disease 2 (autosomal dominant) |
C00000958
|
| 56905 | C15orf39 | chromosome 15 open reading frame 39 |
C00000958
|
| 5318 | PKP2, ARVD9 | plakophilin 2 |
C00000958
|
| 5325 | PLAGL1, LOT1, ZAC, ZAC1 | pleiomorphic adenoma gene-like 1 |
C00000958
|
| 55017 | C14orf119 | chromosome 14 open reading frame 119 |
C00000958
|
| 51501 | C11orf73, HSPC179, Hikeshi, L7RN6, OPI10 | chromosome 11 open reading frame 73 |
C00000958
|
| 51196 | PLCE1, NPHS3, PLCE, PPLC | phospholipase C, epsilon 1 (EC:3.1.4.11) |
C00000958
|
| 56652 | C10orf2, ATXN8, IOSCA, MTDPS7, PEO, PEO1, PEOA3, SANDO, SCA8, TWINL | chromosome 10 open reading frame 2 (EC:3.6.4.12) |
C00000958
|
| 5338 | PLD2 | phospholipase D2 (EC:3.1.4.4) |
C00000958
|
| 26499 | PLEK2 | pleckstrin 2 |
C00000958
|
| 8896 | BUD31, Cwc14, EDG-2, EDG2, G10, YCR063W, fSAP17 | BUD31 homolog (S. cerevisiae) |
C00000958
|
| 144100 | PLEKHA7 | pleckstrin homology domain containing, family A member 7 |
C00000958
|
| 55848 | PLGRKT, C9orf46, MDS030, PLG-RKT, Plg-R(KT) | plasminogen receptor, C-terminal lysine transmembrane protein |
C00000958
|
| 10769 | PLK2, SNK, hPlk2, hSNK | polo-like kinase 2 (EC:2.7.11.21) |
C00000958
|
| 8945 | BTRC, BETA-TRCP, FBW1A, FBXW1, FBXW1A, FWD1, bTrCP, bTrCP1, betaTrCP | beta-transducin repeat containing E3 ubiquitin protein ligase |
C00000958
|
| 5357 | PLS1 | plastin 1 |
C00000958
|
| 5362 | PLXNA2, OCT, PLXN2 | plexin A2 |
C00000958
|
| 10384 | BTN3A3, BTF3 | butyrophilin, subfamily 3, member A3 |
C00000958
|
| 11118 | BTN3A2, BT3.2, BTF4, CD277 | butyrophilin, subfamily 3, member A2 |
C00000958
|
| 11119 | BTN3A1, BT3.1, BTF5, CD277 | butyrophilin, subfamily 3, member A1 |
C00000958
|
| 741857 |
C00000958
|
||
| 5411 | PNN, DRS, DRSP, SDK3, memA | pinin, desmosome associated protein |
C00000958
|
| 91408 | BTF3L4 | basic transcription factor 3-like 4 |
C00000958
|
| 57104 | PNPLA2, 1110001C14Rik, ATGL, PEDF-R, TTS-2.2, TTS2, iPLA2zeta | patatin-like phospholipase domain containing 2 (EC:3.1.1.3) |
C00000958
|
| 87178 | PNPT1, COXPD13, DFNB70, OLD35, PNPASE, old-35 | polyribonucleotide nucleotidyltransferase 1 (EC:2.7.7.8) |
C00000958
|
| 25886 | POC1A, PIX2, SOFT, WDR51A | POC1 centriolar protein A |
C00000958
|
| 134359 | POC5, C5orf37 | POC5 centriolar protein |
C00000958
|
| 689 | BTF3, BETA-NAC, BTF3a, BTF3b, NACB | basic transcription factor 3 |
C00000958
|
| 254065 | BRWD3, BRODL, MRX93 | bromodomain and WD repeat domain containing 3 |
C00000958
|
| 54107 | POLE3, CHARAC17, CHRAC17, YBL1, p17 | polymerase (DNA directed), epsilon 3, accessory subunit (EC:2.7.7.7) |
C00000958
|
| 27154 | BRPF3 | bromodomain and PHD finger containing, 3 |
C00000958
|
| 51082 | POLR1D, AC19, POLR1C, RPA16, RPA9, RPAC2, RPC16, RPO1-3, TCS2 | polymerase (RNA) I polypeptide D, 16kDa |
C00000958
|
| 55703 | POLR3B, C128, HLD8, RPC2 | polymerase (RNA) III (DNA directed) polypeptide B (EC:2.7.7.6) |
C00000958
|
| 171568 | POLR3H, RPC22.9, RPC8 | polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) (EC:2.7.7.6) |
C00000958
|
| 5443 | POMC, ACTH, CLIP, LPH, MSH, NPP, POC | proopiomelanocortin |
C00000958
|
| 10940 | POP1 | processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (EC:3.1.26.5) |
C00000958
|
| 51367 | POP5, HSPC004, RPP2, RPP20, hPop5 | processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) (EC:3.1.26.5) |
C00000958
|
| 5447 | POR, CPR, CYPOR, P450R | P450 (cytochrome) oxidoreductase (EC:1.6.2.4) |
C00000958
|
| 25913 | POT1, HPOT1 | protection of telomeres 1 |
C00000958
|
| 5462 | POU5F1B, OCT4-PG1, OTF3C, OTF3P1, POU5F1P1, POU5F1P4, POU5FLC20, POU5FLC8 | POU class 5 homeobox 1B |
C00000958
|
| 8611 | PPAP2A, LLP1a, LPP1, PAP-2a, PAP2 | phosphatidic acid phosphatase type 2A (EC:3.1.3.4) |
C00000958
|
| 8613 | PPAP2B, Dri42, LPP3, PAP2B, VCIP | phosphatidic acid phosphatase type 2B (EC:3.1.3.4) |
C00000958
|
| 83990 | BRIP1, BACH1, FANCJ, OF | BRCA1 interacting protein C-terminal helicase 1 (EC:3.6.4.13) |
C00000958
|
| 79144 | PPDPF, C20orf149, dJ697K14.9, exdpf | pancreatic progenitor cell differentiation and proliferation factor |
C00000958
|
| 8496 | PPFIBP1, L2, SGT2, hSGT2, hSgt2p | PTPRF interacting protein, binding protein 1 (liprin beta 1) |
C00000958
|
| 23759 | PPIL2, CYC4, CYP60, Cyp-60, UBOX7, hCyP-60 | peptidylprolyl isomerase (cyclophilin)-like 2 (EC:5.2.1.8) |
C00000958
|
| 23262 | PPIP5K2, HISPPD1, IP7K2, VIP2 | diphosphoinositol pentakisphosphate kinase 2 (EC:2.7.4.21 2.7.4.24) |
C00000958
|
| 51400 | PPME1, PME-1 | protein phosphatase methylesterase 1 (EC:3.1.1.89) |
C00000958
|
| 5499 | PPP1CA, PP-1A, PP1A, PP1alpha, PPP1A | protein phosphatase 1, catalytic subunit, alpha isozyme (EC:3.1.3.16) |
C00000958
|
| 4659 | PPP1R12A, M130, MBS, MYPT1 | protein phosphatase 1, regulatory subunit 12A |
C00000958
|
| 23645 | PPP1R15A, GADD34 | protein phosphatase 1, regulatory subunit 15A |
C00000958
|
| 170954 | PPP1R18, HKMT1098, KIAA1949 | protein phosphatase 1, regulatory subunit 18 |
C00000958
|
| 5523 | PPP2R3A, PPP2R3, PR130, PR72 | protein phosphatase 2, regulatory subunit B'', alpha (EC:3.1.3.16) |
C00000958
|
| 55012 | PPP2R3C, C14orf10, G4-1, G5pr | protein phosphatase 2, regulatory subunit B'', gamma |
C00000958
|
| 5533 | PPP3CC, CALNA3, CNA3, PP2Bgamma | protein phosphatase 3, catalytic subunit, gamma isozyme (EC:3.1.3.16) |
C00000958
|
| 57718 | PPP4R4, KIAA1622, PP4R4 | protein phosphatase 4, regulatory subunit 4 |
C00000958
|
| 55291 | PPP6R3, C11orf23, PP6R3, SAP190, SAPL, SAPLa, SAPS3 | protein phosphatase 6, regulatory subunit 3 |
C00000958
|
| 23082 | PPRC1, PRC, RP11-302K17.6 | peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
C00000958
|
| 23398 | PPWD1 | peptidylprolyl isomerase domain and WD repeat containing 1 (EC:5.2.1.8) |
C00000958
|
| 10113 | PREB, SEC12 | prolactin regulatory element binding |
C00000958
|
| 153768 | PRELID2 | PRELI domain containing 2 |
C00000958
|
| 166336 | PRICKLE2, EPM5 | prickle homolog 2 (Drosophila) |
C00000958
|
| 23774 | BRD1, BRL, BRPF1, BRPF2 | bromodomain containing 1 |
C00000958
|
| 669 | BPGM, DPGM | 2,3-bisphosphoglycerate mutase (EC:3.1.3.13 5.4.2.4 5.4.2.11) |
C00000958
|
| 201973 | PRIMPOL, CCDC111, MYP22 | primase and polymerase (DNA-directed) |
C00000958
|
| 5583 | PRKCH, PKC-L, PKCL, PRKCL, nPKC-eta | protein kinase C, eta (EC:2.7.11.13) |
C00000958
|
| 5589 | PRKCSH, AGE-R2, G19P1, PCLD, PKCSH, PLD1 | protein kinase C substrate 80K-H (EC:2.7.11.1) |
C00000958
|
| 51027 | BOLA1 | bolA family member 1 |
C00000958
|
| 5618 | PRLR, hPRLrI | prolactin receptor |
C00000958
|
| 10196 | PRMT3, HRMT1L3 | protein arginine methyltransferase 3 |
C00000958
|
| 29053 |
C00000958
|
||
| 55478 |
C00000958
|
||
| 55390 |
C00000958
|
||
| 55119 | PRPF38B, NET1, RP11-293A10.1 | pre-mRNA processing factor 38B |
C00000958
|
| 5636 | PRPSAP2, PAP41 | phosphoribosyl pyrophosphate synthetase-associated protein 2 |
C00000958
|
| 657 | BMPR1A, 10q23del, ACVRLK3, ALK3, CD292, SKR5 | bone morphogenetic protein receptor, type IA (EC:2.7.11.30) |
C00000958
|
| 58497 | PRUNE, DRES-17, DRES17, HTCD37 | prune exopolyphosphatase (EC:3.6.1.1) |
C00000958
|
| 5671 | PSG3 | pregnancy specific beta-1-glycoprotein 3 |
C00000958
|
| 168667 | BMPER, CRIM3, CV-2, CV2 | BMP binding endothelial regulator |
C00000958
|
| 652 | BMP4, BMP2B, BMP2B1, MCOPS6, OFC11, ZYME | bone morphogenetic protein 4 |
C00000958
|
| 5724 | PTAFR, PAFR | platelet-activating factor receptor |
C00000958
|
| 9317 | PTER, HPHRP, RPR-1 | phosphotriesterase related |
C00000958
|
| 5730 | PTGDS, L-PGDS, LPGDS, PDS, PGD2, PGDS, PGDS2 | prostaglandin D2 synthase 21kDa (brain) (EC:5.3.99.2) |
C00000958
|
| 55589 | BMP2K, BIKE | BMP2 inducible kinase (EC:2.7.11.1) |
C00000958
|
| 644 | BLVRA, BLVR, BVR, BVRA | biliverdin reductase A (EC:1.3.1.24) |
C00000958
|
| 414899 | BLID, BRCC2 | BH3-like motif containing, cell death inducer |
C00000958
|
| 55909 | BIN3 | bridging integrator 3 |
C00000958
|
| 7803 | PTP4A1, HH72, PRL-1, PRL1, PTP(CAAX1), PTPCAAX1 | protein tyrosine phosphatase type IVA, member 1 (EC:3.1.3.48) |
C00000958
|
| 401494 | PTPLAD2, HACD4 | protein tyrosine phosphatase-like A domain containing 2 (EC:4.2.1.134) |
C00000958
|
| 23299 | BICD2, SMALED2, bA526D8.1 | bicaudal D homolog 2 (Drosophila) |
C00000958
|
| 5783 | PTPN13, FAP-1, PNP1, PTP-BAS, PTP-BL, PTP1E, PTPL1, PTPLE, hPTP1E | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) (EC:3.1.3.48) |
C00000958
|
| 5774 | PTPN3, PTP-H1, PTPH1 | protein tyrosine phosphatase, non-receptor type 3 (EC:3.1.3.48) |
C00000958
|
| 5775 | PTPN4, MEG, PTPMEG, PTPMEG1 | protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) (EC:3.1.3.48) |
C00000958
|
| 5791 | PTPRE, HPTPE, PTPE, R-PTP-EPSILON | protein tyrosine phosphatase, receptor type, E (EC:3.1.3.48) |
C00000958
|
| 5795 | PTPRJ, CD148, DEP1, HPTPeta, R-PTP-ETA, SCC1 | protein tyrosine phosphatase, receptor type, J (EC:3.1.3.48) |
C00000958
|
| 138428 | PTRH1, C9orf115, PTH1 | peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) (EC:3.1.1.29) |
C00000958
|
| 5806 | PTX3, TNFAIP5, TSG-14 | pentraxin 3, long |
C00000958
|
| 150962 | PUS10, CCDC139, DOBI | pseudouridylate synthase 10 |
C00000958
|
| 54517 | PUS7 | pseudouridylate synthase 7 homolog (S. cerevisiae) (EC:4.2.1.70) |
C00000958
|
| 83448 | PUS7L | pseudouridylate synthase 7 homolog (S. cerevisiae)-like (EC:5.4.99.-) |
C00000958
|
| 11137 | PWP1, IEF-SSP-9502 | PWP1 homolog (S. cerevisiae) |
C00000958
|
| 11264 | PXMP4, PMP24 | peroxisomal membrane protein 4, 24kDa |
C00000958
|
| 5837 | PYGM | phosphorylase, glycogen, muscle (EC:2.4.1.1) |
C00000958
|
| 9230 | RAB11B, H-YPT3 | RAB11B, member RAS oncogene family |
C00000958
|
| 26056 | RAB11FIP5, GAF1, RIP11, pp75 | RAB11 family interacting protein 5 (class I) |
C00000958
|
| 201475 | RAB12 | RAB12, member RAS oncogene family |
C00000958
|
| 5861 | RAB1A, RAB1, YPT1 | RAB1A, member RAS oncogene family |
C00000958
|
| 441400 |
C00000958
|
||
| 5874 | RAB27B, C25KG | RAB27B, member RAS oncogene family |
C00000958
|
| 9364 | RAB28, CORD18 | RAB28, member RAS oncogene family |
C00000958
|
| 623 | BDKRB1, B1BKR, B1R, BKB1R, BKR1, BRADYB1 | bradykinin receptor B1 |
C00000958
|
| 83452 | RAB33B, SMC2 | RAB33B, member RAS oncogene family |
C00000958
|
| 25782 | RAB3GAP2, RAB3-GAP150, RAB3GAP150, WARBM2, p150 | RAB3 GTPase activating protein subunit 2 (non-catalytic) |
C00000958
|
| 117177 | RAB3IP, RABIN3 | RAB3A interacting protein |
C00000958
|
| 10244 | RABEPK, RAB9P40, bA65N13.1, p40 | Rab9 effector protein with kelch motifs |
C00000958
|
| 23637 | RABGAP1, GAPCENA, RP11-123N4.2, TBC1D11 | RAB GTPase activating protein 1 |
C00000958
|
| 9910 | RABGAP1L, HHL, TBC1D18 | RAB GTPase activating protein 1-like |
C00000958
|
| 605 | BCL7A, BCL7 | B-cell CLL/lymphoma 7A |
C00000958
|
| 5886 | RAD23A, HHR23A, HR23A | RAD23 homolog A (S. cerevisiae) |
C00000958
|
| 56987 | BBX, ARTC1, HBP2, HSPC339, MDS001 | bobby sox homolog (Drosophila) |
C00000958
|
| 585 | BBS4 | Bardet-Biedl syndrome 4 |
C00000958
|
| 5890 | RAD51B, R51H2, RAD51L1, REC2 | RAD51 paralog B |
C00000958
|
| 583 | BBS2, BBS | Bardet-Biedl syndrome 2 |
C00000958
|
| 353091 | RAET1G, ULBP5 | retinoic acid early transcript 1G |
C00000958
|
| 100313526 |
C00000958
|
||
| 57186 | RALGAPA2, AS250, C20orf74, bA287B20.1, dJ1049G11, dJ1049G11.4, p220 | Ral GTPase activating protein, alpha subunit 2 (catalytic) |
C00000958
|
| 55103 | RALGPS2, dJ595C2.1 | Ral GEF with PH domain and SH3 binding motif 2 |
C00000958
|
| 9693 | RAPGEF2, CNrasGEF, NRAPGEP, PDZ-GEF1, PDZGEF1, RA-GEF, RA-GEF-1, Rap-GEP, nRap_GEP | Rap guanine nucleotide exchange factor (GEF) 2 |
C00000958
|
| 51735 | RAPGEF6, KIA001LB, PDZ-GEF2, PDZGEF2, RA-GEF-2, RAGEF2 | Rap guanine nucleotide exchange factor (GEF) 6 |
C00000958
|
| 79738 | BBS10, C12orf58 | Bardet-Biedl syndrome 10 |
C00000958
|
| 29994 | BAZ2B, WALp4 | bromodomain adjacent to zinc finger domain, 2B |
C00000958
|
| 5918 | RARRES1, LXNL, TIG1 | retinoic acid receptor responder (tazarotene induced) 1 |
C00000958
|
| 5917 | RARS, ArgRS, DALRD1 | arginyl-tRNA synthetase (EC:6.1.1.19) |
C00000958
|
| 9462 | RASAL2, NGAP | RAS protein activator like 2 |
C00000958
|
| 25780 | RASGRP3, GRP3 | RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
C00000958
|
| 387496 | RASL11A | RAS-like, family 11, member A |
C00000958
|
| 11228 | RASSF8, C12orf2, HOJ1 | Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
C00000958
|
| 11177 | BAZ1A, ACF1, WALp1, WCRF180, hACF1 | bromodomain adjacent to zinc finger domain, 1A |
C00000958
|
| 5928 | RBBP4, NURF55, RBAP48 | retinoblastoma binding protein 4 |
C00000958
|
| 23543 | RBFOX2, FOX2, Fox-2, HNRBP2, HRNBP2, RBM9, RTA, dJ106I20.3, fxh | RNA binding protein, fox-1 homolog (C. elegans) 2 |
C00000958
|
| 29890 | RBM15B, HUMAGCGB, OTT3 | RNA binding motif protein 15B |
C00000958
|
| 55147 | RBM23, CAPERbeta, RNPC4 | RNA binding motif protein 23 |
C00000958
|
| 129831 | RBM45, DRB1 | RNA binding motif protein 45 |
C00000958
|
| 7917 | BAG6, BAG-6, BAT3, D6S52E, G3 | BCL2-associated athanogene 6 |
C00000958
|
| 9978 | RBX1, BA554C12.1, RNF75, ROC1 | ring-box 1, E3 ubiquitin protein ligase |
C00000958
|
| 11123 | RCAN3, DSCR1L2, MCIP3, RCN3, hRCN3 | RCAN family member 3 |
C00000958
|
| 10171 | RCL1, RNAC, RPCL1 | RNA terminal phosphate cyclase-like 1 |
C00000958
|
| 9530 | BAG4, BAG-4, SODD | BCL2-associated athanogene 4 |
C00000958
|
| 9531 | BAG3, BAG-3, BIS, CAIR-1, MFM6 | BCL2-associated athanogene 3 |
C00000958
|
| 5980 | REV3L, POLZ, REV3 | REV3-like, polymerase (DNA directed), zeta, catalytic subunit (EC:2.7.7.7) |
C00000958
|
| 91869 | RFT1, CDG1N | RFT1 homolog (S. cerevisiae) |
C00000958
|
| 64326 | RFWD2, COP1, RNF200 | ring finger and WD repeat domain 2, E3 ubiquitin protein ligase |
C00000958
|
| 5991 | RFX3 | regulatory factor X, 3 (influences HLA class II expression) |
C00000958
|
| 8625 | RFXANK, ANKRA1, BLS, F14150_1, RFX-B | regulatory factor X-associated ankyrin-containing protein |
C00000958
|
| 28984 | RGCC, C13orf15, RGC-32, RGC32, bA157L14.2 | regulator of cell cycle |
C00000958
|
| 23179 | RGL1, RGL | ral guanine nucleotide dissociation stimulator-like 1 |
C00000958
|
| 5997 | RGS2, G0S8 | regulator of G-protein signaling 2, 24kDa |
C00000958
|
| 5999 | RGS4, RGP4, SCZD9 | regulator of G-protein signaling 4 |
C00000958
|
| 57414 | RHBDD2, NPD007, RHBDL7 | rhomboid domain containing 2 |
C00000958
|
| 54933 | RHBDL2, RRP2 | rhomboid, veinlet-like 2 (Drosophila) (EC:3.4.21.105) |
C00000958
|
| 573 | BAG1, BAG-1, HAP, RAP46 | BCL2-associated athanogene |
C00000958
|
| 29984 | RHOD, ARHD, RHOHP1, RHOM, Rho | ras homolog family member D |
C00000958
|
| 23621 | BACE1, ASP2, BACE, HSPC104 | beta-site APP-cleaving enzyme 1 (EC:3.4.23.46) |
C00000958
|
| 6018 | RLF, ZN-15L, ZNF292L | rearranged L-myc fusion |
C00000958
|
| 51115 | RMDN1, FAM82B, RMD-1, RMD1 | regulator of microtubule dynamics 1 |
C00000958
|
| 64795 | RMND5A, CTLH, GID2, GID2A, RMD5, p44CTLH | required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
C00000958
|
| 64777 | RMND5B, GID2, GID2B | required for meiotic nuclear division 5 homolog B (S. cerevisiae) |
C00000958
|
| 570 | BAAT, BACAT, BAT | bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) (EC:2.3.1.65 3.1.2.2) |
C00000958
|
| 567505 |
C00000958
|
||
| 27289 | RND1, ARHS, RHO6, RHOS | Rho family GTPase 1 |
C00000958
|
| 27246 | RNF115, BCA2, ZNF364 | ring finger protein 115 |
C00000958
|
| 11342 | RNF13, RZF | ring finger protein 13 |
C00000958
|
| 255488 | RNF144B, IBRDC2, PIR2, bA528A10.3, p53RFP | ring finger protein 144B |
C00000958
|
| 81790 | RNF170, SNAX1 | ring finger protein 170 |
C00000958
|
| 285671 | RNF180 | ring finger protein 180 |
C00000958
|
| 221687 | RNF182 | ring finger protein 182 |
C00000958
|
| 25897 | RNF19A, RNF19 | ring finger protein 19A, RBR E3 ubiquitin protein ligase |
C00000958
|
| 57674 | RNF213, ALO17, C17orf27, KIAA1618, MYMY2, MYSTR, NET57 | ring finger protein 213 |
C00000958
|
| 54476 | RNF216, CAHH, TRIAD3, U7I1, UBCE7IP1, ZIN | ring finger protein 216 |
C00000958
|
| 154214 | RNF217, C6orf172, IBRDC1, dJ84N20.1 | ring finger protein 217 |
C00000958
|
| 146712 | B3GNTL1, B3GNT8 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (EC:2.4.1.-) |
C00000958
|
| 6049 | RNF6 | ring finger protein (C3H2C3 type) 6 |
C00000958
|
| 51136 | RNFT1 | ring finger protein, transmembrane 1 |
C00000958
|
| 8732 | RNGTT, CAP1A, HCE, HCE1, hCAP | RNA guanylyltransferase and 5'-phosphatase (EC:2.7.7.50 3.1.3.33) |
C00000958
|
| 9475 | ROCK2, ROCK-II | Rho-associated, coiled-coil containing protein kinase 2 (EC:2.7.11.1) |
C00000958
|
| 4920 | ROR2, BDB, BDB1, NTRKR2 | receptor tyrosine kinase-like orphan receptor 2 (EC:2.7.10.1) |
C00000958
|
| 6100 | RP9, PAP-1 | retinitis pigmentosa 9 (autosomal dominant) |
C00000958
|
| 23322 | RPGRIP1L, CORS3, FTM, JBTS7, MKS5, NPHP8 | RPGRIP1-like |
C00000958
|
| 6134 | RPL10, AUTSX5, DXS648, DXS648E, L10, NOV, QM | ribosomal protein L10 |
C00000958
|
| 6137 | RPL13, BBC1, D16S444E, D16S44E, L13 | ribosomal protein L13 (EC:3.6.5.3) |
C00000958
|
| 9045 | RPL14, CAG-ISL-7, CTG-B33, L14, RL14, hRL14 | ribosomal protein L14 |
C00000958
|
| 6142 | RPL18A, L18A | ribosomal protein L18a |
C00000958
|
| 6157 | RPL27A, L27A | ribosomal protein L27a |
C00000958
|
| 389435 |
C00000958
|
||
| 6158 | RPL28, L28 | ribosomal protein L28 (EC:3.6.5.3) |
C00000958
|
| 6164 | RPL34, L34 | ribosomal protein L34 |
C00000958
|
| 6167 | RPL37, L37 | ribosomal protein L37 |
C00000958
|
| 6168 | RPL37A, L37A | ribosomal protein L37a |
C00000958
|
| 116832 | RPL39L, RPL39L1 | ribosomal protein L39-like |
C00000958
|
| 6125 | RPL5, DBA6, L5 | ribosomal protein L5 |
C00000958
|
| 54913 | RPP25 | ribonuclease P/MRP 25kDa subunit (EC:3.1.26.5) |
C00000958
|
| 145173 | B3GALTL, B3GLCT, B3GTL, B3Glc-T, Gal-T, beta3Glc-T | beta 1,3-galactosyltransferase-like |
C00000958
|
| 6197 | RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RSK2, S6K-alpha3, p90-RSK2, pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1) |
C00000958
|
| 51582 | AZIN1, OAZI, OAZIN, ODC1L | antizyme inhibitor 1 |
C00000958
|
| 26750 | RPS6KC1, RPK118, humS6PKh1 | ribosomal protein S6 kinase, 52kDa, polypeptide 1 (EC:2.7.11.1) |
C00000958
|
| 220885 |
C00000958
|
||
| 57521 | RPTOR, KOG1, Mip1 | regulatory associated protein of MTOR, complex 1 |
C00000958
|
| 285367 | RPUSD3 | RNA pseudouridylate synthase domain containing 3 |
C00000958
|
| 84881 | RPUSD4 | RNA pseudouridylate synthase domain containing 4 (EC:5.4.99.12) |
C00000958
|
| 10692 | RRH | retinal pigment epithelium-derived rhodopsin homolog |
C00000958
|
| 23378 | RRP8, KIAA0409 | ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
C00000958
|
| 65117 | RSRC2 | arginine/serine-rich coiled-coil 2 |
C00000958
|
| 219790 | RTKN2, PLEKHK1, bA531F24.1 | rhotekin 2 |
C00000958
|
| 84816 | RTN4IP1, NIMP | reticulon 4 interacting protein 1 |
C00000958
|
| 25914 | RTTN | rotatin |
C00000958
|
| 80230 | RUFY1, RABIP4, ZFYVE12 | RUN and FYVE domain containing 1 |
C00000958
|
| 33419 |
C00000958
|
||
| 112611 | RWDD2A, RWDD2, dJ747H23.2 | RWD domain containing 2A |
C00000958
|
| 222255 | ATXN7L1, ATXN7L4 | ataxin 7-like 1 |
C00000958
|
| 6274 | S100A3, S100E | S100 calcium binding protein A3 |
C00000958
|
| 64766 | S100PBP, S100PBPR | S100P binding protein |
C00000958
|
| 22908 | SACM1L, SAC1 | SAC1 suppressor of actin mutations 1-like (yeast) |
C00000958
|
| 9667 | SAFB2 | scaffold attachment factor B2 |
C00000958
|
| 54809 | SAMD9, C7orf5, DRIF1, NFTC, OEF1, OEF2 | sterile alpha motif domain containing 9 |
C00000958
|
| 219285 | SAMD9L, C7orf6, DRIF2, UEF1 | sterile alpha motif domain containing 9-like |
C00000958
|
| 10284 | SAP18, 2HOR0202, SAP18P | Sin3A-associated protein, 18kDa |
C00000958
|
| 23328 | SASH1, SH3D6A, dJ323M4, dJ323M4.1 | SAM and SH3 domain containing 1 |
C00000958
|
| 163786 | SASS6, SAS-6, SAS6 | spindle assembly 6 homolog (C. elegans) |
C00000958
|
| 6303 | SAT1, DC21, KFSD, KFSDX, SAT, SSAT, SSAT-1 | spermidine/spermine N1-acetyltransferase 1 (EC:2.3.1.57) |
C00000958
|
| 112483 | SAT2, SSAT2 | spermidine/spermine N1-acetyltransferase family member 2 (EC:2.3.1.57) |
C00000958
|
| 55776 | SAYSD1, C6orf64 | SAYSVFN motif domain containing 1 |
C00000958
|
| 66234 |
C00000958
|
||
| 286205 | SCAI, C9orf126, NET40 | suppressor of cancer cell invasion |
C00000958
|
| 114821 | SCAND3, Buster4, ZBED9, ZFP38-L, ZNF305P2, ZNF452, dJ1186N24.3 | SCAN domain containing 3 |
C00000958
|
| 49855 | SCAPER, ZNF291, Zfp291 | S-phase cyclin A-associated protein in the ER |
C00000958
|
| 11273 | ATXN2L, A2D, A2LG, A2LP, A2RP | ataxin 2-like |
C00000958
|
| 51540 | SCLY, SCL, hSCL | selenocysteine lyase (EC:4.4.1.16) |
C00000958
|
| 6322 | SCML1 | sex comb on midleg-like 1 (Drosophila) |
C00000958
|
| 79634 | SCRN3, SES3 | secernin 3 |
C00000958
|
| 83482 | SCRT1, SCRT, ZNF898 | scratch family zinc finger 1 |
C00000958
|
| 57147 | SCYL3, PACE-1, PACE1 | SCY1-like 3 (S. cerevisiae) |
C00000958
|
| 54949 | SDHAF2, C11orf79, PGL2, SDH5 | succinate dehydrogenase complex assembly factor 2 |
C00000958
|
| 8436 | SDPR, CAVIN2, PS-p68, SDR, cavin-2 | serum deprivation response |
C00000958
|
| 6397 | SEC14L1, PRELID4A, SEC14L | SEC14-like 1 (S. cerevisiae) |
C00000958
|
| 26984 | SEC22A, SEC22L2 | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) |
C00000958
|
| 6311 | ATXN2, ASL13, ATX2, SCA2, TNRC13 | ataxin 2 |
C00000958
|
| 6404 | SELPLG, CD162, CLA, PSGL-1, PSGL1 | selectin P ligand |
C00000958
|
| 80031 | SEMA6D | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
C00000958
|
| 57337 | SENP7 | SUMO1/sentrin specific peptidase 7 (EC:3.4.22.68) |
C00000958
|
| 57190 | SEPN1, CFTD, MDRS1, RSMD1, RSS, SELN | selenoprotein N, 1 |
C00000958
|
| 84947 | SERAC1 | serine active site containing 1 |
C00000958
|
| 1992 | SERPINB1, EI, ELANH2, LEI, M/NEI, MNEI, PI-2, PI2 | serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
C00000958
|
| 5272 | SERPINB9, CAP-3, CAP3, PI-9, PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
C00000958
|
| 29950 | SERTAD1, SEI1, TRIP-Br1 | SERTA domain containing 1 |
C00000958
|
| 9792 | SERTAD2, Sei-2, TRIP-Br2 | SERTA domain containing 2 |
C00000958
|
| 29946 | SERTAD3, RBT1 | SERTA domain containing 3 |
C00000958
|
| 91404 | SESTD1, SOLO | SEC14 and spectrin domains 1 |
C00000958
|
| 5205 | ATP8B1, ATPIC, BRIC, FIC1, ICP1, PFIC, PFIC1 | ATPase, aminophospholipid transporter, class I, type 8B, member 1 (EC:3.6.3.1) |
C00000958
|
| 10262 | SF3B4, AFD1, Hsh49, SAP49, SF3b49 | splicing factor 3b, subunit 4, 49kDa |
C00000958
|
| 51460 | SFMBT1, RU1, SFMBT, hSFMBT | Scm-like with four mbt domains 1 |
C00000958
|
| 100169978 |
C00000958
|
||
| 733142 |
C00000958
|
||
| 6446 | SGK1, SGK | serum/glucocorticoid regulated kinase 1 (EC:2.7.11.1) |
C00000958
|
| 100174271 |
C00000958
|
||
| 259230 | SGMS1, MOB, MOB1, SMS1, TMEM23, hmob33 | sphingomyelin synthase 1 (EC:2.7.8.27) |
C00000958
|
| 540 | ATP7B, PWD, WC1, WD, WND | ATPase, Cu++ transporting, beta polypeptide (EC:3.6.3.54) |
C00000958
|
| 63898 | SH2D4A, PPP1R38, SH2A | SH2 domain containing 4A |
C00000958
|
| 285590 | SH3PXD2B, FAD49, FTHS, HOFI, KIAA1295, TKS4, TSK4 | SH3 and PX domains 2B |
C00000958
|
| 527 | ATP6V0C, ATP6C, ATP6L, ATPL, VATL, VPPC, Vma3 | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c (EC:3.6.3.14) |
C00000958
|
| 8036 | SHOC2, SIAA0862, SOC2, SUR8 | soc-2 suppressor of clear homolog (C. elegans) |
C00000958
|
| 51092 | SIDT2 | SID1 transmembrane family, member 2 |
C00000958
|
| 59307 | SIGIRR, TIR8 | single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
C00000958
|
| 150094 | SIK1, MSK, SIK, SNF1LK | salt-inducible kinase 1 (EC:2.7.11.1) |
C00000958
|
| 6492 | SIM1, bHLHe14 | single-minded family bHLH transcription factor 1 |
C00000958
|
| 26037 | SIPA1L1, E6TP1 | signal-induced proliferation-associated 1 like 1 |
C00000958
|
| 57568 | SIPA1L2, SPAL2 | signal-induced proliferation-associated 1 like 2 |
C00000958
|
| 51804 | SIX4, AREC3 | SIX homeobox 4 |
C00000958
|
| 267020 | ATP5L2, ATP5K2 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
C00000958
|
| 84068 | SLC10A7, C4orf13, P7 | solute carrier family 10, member 7 |
C00000958
|
| 201232 | SLC16A13, MCT13 | solute carrier family 16, member 13 |
C00000958
|
| 9123 | SLC16A3, MCT_3, MCT_4, MCT-3, MCT-4, MCT3, MCT4 | solute carrier family 16 (monocarboxylate transporter), member 3 |
C00000958
|
| 493 | ATP2B4, ATP2B2, MXRA1, PMCA4, PMCA4b, PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 (EC:3.6.3.8) |
C00000958
|
| 8604 | SLC25A12, AGC1, ARALAR | solute carrier family 25 (aspartate/glutamate carrier), member 12 |
C00000958
|
| 10165 | SLC25A13, ARALAR2, CITRIN, CTLN2 | solute carrier family 25 (aspartate/glutamate carrier), member 13 |
C00000958
|
| 83884 | SLC25A2, ORC2, ORNT2 | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 2 |
C00000958
|
| 788 | SLC25A20, CAC, CACT | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
C00000958
|
| 55186 | SLC25A36, PNC2 | solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
C00000958
|
| 91137 | SLC25A46 | solute carrier family 25, member 46 |
C00000958
|
| 490 | ATP2B1, PMCA1, PMCA1kb | ATPase, Ca++ transporting, plasma membrane 1 (EC:3.6.3.8) |
C00000958
|
| 488 | ATP2A2, ATP2B, DAR, DD, SERCA2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (EC:3.6.3.8) |
C00000958
|
| 10999 | SLC27A4, ACSVL4, FATP4, IPS | solute carrier family 27 (fatty acid transporter), member 4 (EC:6.2.1.-) |
C00000958
|
| 6513 | SLC2A1, DYT17, DYT18, DYT9, EIG12, GLUT, GLUT1, GLUT1DS, HTLVR, PED | solute carrier family 2 (facilitated glucose transporter), member 1 |
C00000958
|
| 6515 | SLC2A3, GLUT3 | solute carrier family 2 (facilitated glucose transporter), member 3 |
C00000958
|
| 7779 | SLC30A1, ZNT1, ZRC1 | solute carrier family 30 (zinc transporter), member 1 |
C00000958
|
| 7782 | SLC30A4, ZNT4 | solute carrier family 30 (zinc transporter), member 4 |
C00000958
|
| 9197 | SLC33A1, ACATN, AT-1, AT1, CCHLND, SPG42 | solute carrier family 33 (acetyl-CoA transporter), member 1 |
C00000958
|
| 10559 | SLC35A1, CDG2F, CMPST, CST, hCST | solute carrier family 35 (CMP-sialic acid transporter), member A1 |
C00000958
|
| 55032 | SLC35A5 | solute carrier family 35, member A5 |
C00000958
|
| 347734 | SLC35B2, PAPST1, SLL, UGTrel4 | solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
C00000958
|
| 11046 | SLC35D2, HFRC1, SQV7L, UGTrel8, hfrc | solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
C00000958
|
| 55508 | SLC35E3, BLOV1 | solute carrier family 35, member E3 |
C00000958
|
| 80255 | SLC35F5 | solute carrier family 35, member F5 |
C00000958
|
| 159371 | SLC35G1, C10orf60, TMEM20 | solute carrier family 35, member G1 |
C00000958
|
| 55089 | SLC38A4, ATA3, NAT3, PAAT | solute carrier family 38, member 4 |
C00000958
|
| 145389 | SLC38A6, NAT-1 | solute carrier family 38, member 6 |
C00000958
|
| 153129 | SLC38A9 | solute carrier family 38, member 9 |
C00000958
|
| 27173 | SLC39A1, ZIP1, ZIRTL | solute carrier family 39 (zinc transporter), member 1 |
C00000958
|
| 57181 | SLC39A10, LZT-Hs2 | solute carrier family 39 (zinc transporter), member 10 |
C00000958
|
| 23516 | SLC39A14, LZT-Hs4, NET34, ZIP14, cig19 | solute carrier family 39 (zinc transporter), member 14 |
C00000958
|
| 55630 | SLC39A4, AEZ, AWMS2, ZIP4 | solute carrier family 39 (zinc transporter), member 4 |
C00000958
|
| 30061 | SLC40A1, FPN1, HFE4, IREG1, MST079, MTP1, SLC11A3 | solute carrier family 40 (iron-regulated transporter), member 1 |
C00000958
|
| 84102 | SLC41A2, SLC41A1-L1 | solute carrier family 41 (magnesium transporter), member 2 |
C00000958
|
| 29015 | SLC43A3, EEG1, FOAP-13, PRO1659, SEEEG-1 | solute carrier family 43, member 3 |
C00000958
|
| 283537 | SLC46A3, FKSG16 | solute carrier family 46, member 3 |
C00000958
|
| 9497 | SLC4A7, NBC2, NBC3, NBCN1, SBC2, SLC4A6 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
C00000958
|
| 200931 | SLC51A, OSTA, OSTalpha | solute carrier family 51, alpha subunit |
C00000958
|
| 123264 | SLC51B, OSTB, OSTBETA | solute carrier family 51, beta subunit |
C00000958
|
| 6526 | SLC5A3, SMIT, SMIT1, SMIT2 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
C00000958
|
| 8884 | SLC5A6, SMVT | solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
C00000958
|
| 6540 | SLC6A13, GAT-2, GAT2, GAT3 | solute carrier family 6 (neurotransmitter transporter), member 13 |
C00000958
|
| 6533 | SLC6A6, TAUT | solute carrier family 6 (neurotransmitter transporter), member 6 |
C00000958
|
| 6535 | SLC6A8, CCDS1, CRT, CRTR, CT1 | solute carrier family 6 (neurotransmitter transporter), member 8 |
C00000958
|
| 84138 | SLC7A6OS | solute carrier family 7, member 6 opposite strand |
C00000958
|
| 28232 | SLCO3A1, OATP-D, OATP3A1, OATPD, SLC21A11 | solute carrier organic anion transporter family, member 3A1 |
C00000958
|
| 7871 | SLMAP, SLAP | sarcolemma associated protein |
C00000958
|
| 128710 | SLX4IP, C20orf94, bA204H22.1, bA254M13.1, dJ1099D15.3 | SLX4 interacting protein |
C00000958
|
| 481 | ATP1B1, ATP1B | ATPase, Na+/K+ transporting, beta 1 polypeptide |
C00000958
|
| 57205 | ATP10D, ATPVD | ATPase, class V, type 10D (EC:3.6.3.1) |
C00000958
|
| 4091 | SMAD6, AOVD2, HsT17432, MADH6, MADH7 | SMAD family member 6 |
C00000958
|
| 6594 | SMARCA1, ISWI, NURF140, SNF2L, SNF2L1, SNF2LB, SNF2LT, SWI, SWI2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
C00000958
|
| 6595 | SMARCA2, BAF190, BRM, NCBRS, SNF2, SNF2L2, SNF2LA, SWI2, Sth1p, hBRM, hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
C00000958
|
| 56916 | SMARCAD1, ADERM, ETL1, HEL1 | SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 (EC:3.6.4.12) |
C00000958
|
| 50485 | SMARCAL1, HARP, HHARP | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
C00000958
|
| 6598 | SMARCB1, BAF47, INI1, MRD15, RDT, RTPS1, SNF5, SNF5L1, Sfh1p, Snr1, hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
C00000958
|
| 1822 | ATN1, B37, D12S755E, DRPLA, HRS, NOD | atrophin 1 |
C00000958
|
| 79677 | SMC6, SMC-6, SMC6L1, hSMC6 | structural maintenance of chromosomes 6 |
C00000958
|
| 56935 | SMCO4, C11orf75, FN5 | single-pass membrane protein with coiled-coil domains 4 |
C00000958
|
| 57150 | SMIM8, C6orf162, dJ102H19.2 | small integral membrane protein 8 |
C00000958
|
| 51062 | ATL1, AD-FSP, FSP1, GBP3, HSN1D, SPG3, SPG3A, atlastin1 | atlastin GTPase 1 |
C00000958
|
| 54498 | SMOX, C20orf16, PAO, PAO-1, PAOH1, SMO | spermine oxidase (EC:1.5.3.16) |
C00000958
|
| 23583 | SMUG1, FDG, HMUDG, UNG3 | single-strand-selective monofunctional uracil-DNA glycosylase 1 |
C00000958
|
| 84971 | ATG4D, APG4-D, APG4D, AUTL4 | autophagy related 4D, cysteine peptidase |
C00000958
|
| 23557 | SNAPIN, BLOC1S7, BLOS7, SNAPAP | SNAP-associated protein |
C00000958
|
| 85028 | SNHG12, C1orf79, LINC00100, NCRNA00100, RP4-669K10.5 | small nucleolar RNA host gene 12 (non-protein coding) |
C00000958
|
| 79753 | SNIP1, PMRED | Smad nuclear interacting protein 1 |
C00000958
|
| 677792 | SNORA1, ACA1 | small nucleolar RNA, H/ACA box 1 |
C00000958
|
| 677802 | SNORA14B, ACA14b | small nucleolar RNA, H/ACA box 14B |
C00000958
|
| 677811 | SNORA28, ACA28 | small nucleolar RNA, H/ACA box 28 |
C00000958
|
| 677812 | SNORA29, ACA29 | small nucleolar RNA, H/ACA box 29 |
C00000958
|
| 677793 | SNORA2A, ACA2A | small nucleolar RNA, H/ACA box 2A |
C00000958
|
| 594839 | SNORA33, ACA33 | small nucleolar RNA, H/ACA box 33 |
C00000958
|
| 619569 | SNORA41, ACA41 | small nucleolar RNA, H/ACA box 41 |
C00000958
|
| 677826 | SNORA45, ACA3-2 | small nucleolar RNA, H/ACA box 45 |
C00000958
|
| 677833 | SNORA54, ACA54 | small nucleolar RNA, H/ACA box 54 |
C00000958
|
| 677834 | SNORA55, ACA55 | small nucleolar RNA, H/ACA box 55 |
C00000958
|
| 654319 | SNORA5A, ACA5, ACA5A/B/C | small nucleolar RNA, H/ACA box 5A |
C00000958
|
| 677796 | SNORA5C, ACA5c | small nucleolar RNA, H/ACA box 5C |
C00000958
|
| 574040 | SNORA6, ACA6 | small nucleolar RNA, H/ACA box 6 |
C00000958
|
| 677837 | SNORA60, ACA60 | small nucleolar RNA, H/ACA box 60 |
C00000958
|
| 26779 | SNORA69, RNU69, U69, U69A | small nucleolar RNA, H/ACA box 69 |
C00000958
|
| 26821 | SNORA74A, RNU19, U19 | small nucleolar RNA, H/ACA box 74A |
C00000958
|
| 654321 | SNORA75, U23 | small nucleolar RNA, H/ACA box 75 |
C00000958
|
| 26765 | SNORD12C, E2, E2-1, E3, RNU106, SNORD106, U106 | small nucleolar RNA, C/D box 12C |
C00000958
|
| 6077 |
C00000958
|
||
| 6079 | SNORD15A, RNU15A, SNORNA, U15A | small nucleolar RNA, C/D box 15A |
C00000958
|
| 677848 | SNORD1A, R38A, snR38A | small nucleolar RNA, C/D box 1A |
C00000958
|
| 6082 | SNORD20, RNU20, U20 | small nucleolar RNA, C/D box 20 |
C00000958
|
| 6083 | SNORD21, RNU21, U21 | small nucleolar RNA, C/D box 21 |
C00000958
|
| 9300 | SNORD28, RNU28, U28 | small nucleolar RNA, C/D box 28 |
C00000958
|
| 9299 | SNORD30, RNU30, U30 | small nucleolar RNA, C/D box 30 |
C00000958
|
| 94162 | SNORD38A, RNU38A, U38A | small nucleolar RNA, C/D box 38A |
C00000958
|
| 94163 | SNORD38B, RNU38B, U38B | small nucleolar RNA, C/D box 38B |
C00000958
|
| 26809 | SNORD42A, RNU42A, U42, U42A | small nucleolar RNA, C/D box 42A |
C00000958
|
| 26808 | SNORD42B, RNU42B, U42B | small nucleolar RNA, C/D box 42B |
C00000958
|
| 26807 | SNORD43, RNU43, U43 | small nucleolar RNA, C/D box 43 |
C00000958
|
| 26806 | SNORD44, RNU44, U44 | small nucleolar RNA, C/D box 44 |
C00000958
|
| 692085 | SNORD45C, U45C | small nucleolar RNA, C/D box 45C |
C00000958
|
| 26800 | SNORD49A, RNU49, U49, U49A | small nucleolar RNA, C/D box 49A |
C00000958
|
| 692087 | SNORD49B, U49B | small nucleolar RNA, C/D box 49B |
C00000958
|
| 26773 | SNORD4A, RNU101A, Z17A, mgh18S-121 | small nucleolar RNA, C/D box 4A |
C00000958
|
| 26772 | SNORD4B, RNU101B, Z17B | small nucleolar RNA, C/D box 4B |
C00000958
|
| 26799 | SNORD50A, RNU50, U50 | small nucleolar RNA, C/D box 50A |
C00000958
|
| 692088 | SNORD50B, U50', U50B | small nucleolar RNA, C/D box 50B |
C00000958
|
| 26798 | SNORD51, RNU51, U51 | small nucleolar RNA, C/D box 51 |
C00000958
|
| 26796 | SNORD53, RNU53, U53 | small nucleolar RNA, C/D box 53 |
C00000958
|
| 26795 | SNORD54, RNU54, U54 | small nucleolar RNA, C/D box 54 |
C00000958
|
| 26811 | SNORD55, RNU39, RNU55, SNORD39, U39, U55 | small nucleolar RNA, C/D box 55 |
C00000958
|
| 26791 | SNORD58A, RNU58A, U58a | small nucleolar RNA, C/D box 58A |
C00000958
|
| 692075 | SNORD6, mgh28S-2412 | small nucleolar RNA, C/D box 6 |
C00000958
|
| 26785 | SNORD63, RNU63, U63 | small nucleolar RNA, C/D box 63 |
C00000958
|
| 8944 | SNORD73A, RNU73, RNU73A, U73, U73a | small nucleolar RNA, C/D box 73A |
C00000958
|
| 619498 | SNORD74, U74, Z18 | small nucleolar RNA, C/D box 74 |
C00000958
|
| 692195 | SNORD75, U75 | small nucleolar RNA, C/D box 75 |
C00000958
|
| 692197 | SNORD77, U77 | small nucleolar RNA, C/D box 77 |
C00000958
|
| 692198 | SNORD78, U78 | small nucleolar RNA, C/D box 78 |
C00000958
|
| 26770 | SNORD79, RNU103, U79, Z22 | small nucleolar RNA, C/D box 79 |
C00000958
|
| 25826 | SNORD82, RNU82, U82, Z25 | small nucleolar RNA, C/D box 82 |
C00000958
|
| 116938 | SNORD83B, RNU83B, U83B | small nucleolar RNA, C/D box 83B |
C00000958
|
| 619570 | SNORD95, U95 | small nucleolar RNA, C/D box 95 |
C00000958
|
| 619571 | SNORD96A, U96A | small nucleolar RNA, C/D box 96A |
C00000958
|
| 9410 | SNRNP40, 40K, HPRP8BP, PRP8BP, PRPF8BP, RP11-490K7.3, SPF38, WDR57 | small nuclear ribonucleoprotein 40kDa (U5) |
C00000958
|
| 10073 | SNUPN, KPNBL, RNUT1, Snurportin1 | snurportin 1 |
C00000958
|
| 23161 | SNX13, RGS-PX1 | sorting nexin 13 |
C00000958
|
| 28966 | SNX24, PRO1284 | sorting nexin 24 |
C00000958
|
| 83891 | SNX25, SBBI31 | sorting nexin 25 |
C00000958
|
| 92017 | SNX29, A-388D4.1, RUNDC2A | sorting nexin 29 |
C00000958
|
| 8723 | SNX4, ATG24B | sorting nexin 4 |
C00000958
|
| 29886 | SNX8, Mvp1 | sorting nexin 8 |
C00000958
|
| 51429 | SNX9, SDP1, SH3PX1, SH3PXD3A, WISP | sorting nexin 9 |
C00000958
|
| 84938 | ATG4C, APG4-C, APG4C, AUTL1, AUTL3 | autophagy related 4C, cysteine peptidase |
C00000958
|
| 115201 | ATG4A, APG4A, AUTL2 | autophagy related 4A, cysteine peptidase |
C00000958
|
| 6655 | SOS2 | son of sevenless homolog 2 (Drosophila) |
C00000958
|
| 65124 | SOWAHC, ANKRD57, C2orf26 | sosondowah ankyrin repeat domain family member C |
C00000958
|
| 83734 | ATG10, APG10, APG10L, pp12616 | autophagy related 10 |
C00000958
|
| 79582 | SPAG16, PF20, WDR29 | sperm associated antigen 16 |
C00000958
|
| 619455 | SPANXA2-OT1, CXorf18 | SPANXA2 overlapping transcript 1 (non-protein coding) |
C00000958
|
| 6683 | SPAST, ADPSP, FSP2, SPG4 | spastin (EC:3.6.4.3) |
C00000958
|
| 11101 | ATE1 | arginyltransferase 1 (EC:2.3.2.8) |
C00000958
|
| 60559 | SPCS3, PRO3567, SPC22/23, SPC3, YLR066W | signal peptidase complex subunit 3 homolog (S. cerevisiae) |
C00000958
|
| 80208 | SPG11, KIAA1840 | spastic paraplegia 11 (autosomal recessive) |
C00000958
|
| 56907 | SPIRE1, Spir-1 | spire-type actin nucleation factor 1 |
C00000958
|
| 200734 | SPRED2, Spred-2 | sprouty-related, EVH1 domain containing 2 |
C00000958
|
| 81848 | SPRY4, HH17 | sprouty homolog 4 (Drosophila) |
C00000958
|
| 84926 | SPRYD3 | SPRY domain containing 3 (EC:2.7.1.112) |
C00000958
|
| 9517 | SPTLC2, HSN1C, LCB2, LCB2A, NSAN1C, SPT2, hLCB2a | serine palmitoyltransferase, long chain base subunit 2 (EC:2.3.1.50) |
C00000958
|
| 144108 | SPTY2D1 | SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
C00000958
|
| 54454 | ATAD2B | ATPase family, AAA domain containing 2B |
C00000958
|
| 9070 | ASH2L, ASH2, ASH2L1, ASH2L2, Bre2 | ash2 (absent, small, or homeotic)-like (Drosophila) |
C00000958
|
| 8878 | SQSTM1, A170, OSIL, PDB3, ZIP3, p60, p62, p62B | sequestosome 1 |
C00000958
|
| 55133 | SRBD1 | S1 RNA binding domain 1 |
C00000958
|
| 25842 | ASF1A, CGI-98, CIA, HSPC146 | anti-silencing function 1A histone chaperone |
C00000958
|
| 10973 | ASCC3, ASC1p200, HELIC1, RNAH | activating signal cointegrator 1 complex subunit 3 (EC:3.6.4.12) |
C00000958
|
| 57522 | SRGAP1, ARHGAP13 | SLIT-ROBO Rho GTPase activating protein 1 |
C00000958
|
| 23380 | SRGAP2, ARHGAP34, FNBP2, SRGAP2A, SRGAP3 | SLIT-ROBO Rho GTPase activating protein 2 |
C00000958
|
| 6717 | SRI, CP-22, CP22, SCN, V19 | sorcin |
C00000958
|
| 140459 | ASB6 | ankyrin repeat and SOCS box containing 6 |
C00000958
|
| 10250 | SRRM1, 160-KD, POP101, SRM160 | serine/arginine repetitive matrix 1 |
C00000958
|
| 23635 | SSBP2, HSPC116, SOSS-B2 | single-stranded DNA binding protein 2 |
C00000958
|
| 23648 | SSBP3, CSDP, SSDP, SSDP1 | single stranded DNA binding protein 3 |
C00000958
|
| 619518 | SSBP3-AS1, C1orf191, MST128 | SSBP3 antisense RNA 1 |
C00000958
|
| 54434 | SSH1, SSH1L | slingshot protein phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00000958
|
| 85464 | SSH2, SSH-2, SSH-2L | slingshot protein phosphatase 2 (EC:3.1.3.16 3.1.3.48) |
C00000958
|
| 50807 | ASAP1, AMAP1, CENTB4, DDEF1, PAG2, PAP, ZG14P | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
C00000958
|
| 6764 | ST5, DENND2B, HTS1, p126 | suppression of tumorigenicity 5 |
C00000958
|
| 10610 | ST6GALNAC2, SAITL1, SIAT7, SIAT7B, SIATL1, ST6GalNAII, STHM | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (EC:2.4.99.7) |
C00000958
|
| 81849 | ST6GALNAC5, SIAT7E, ST6GalNAcV | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
C00000958
|
| 10274 | STAG1, SA1, SCC3A | stromal antigen 1 |
C00000958
|
| 10735 | STAG2, SA-2, SA2, SCC3B, bA517O1.1 | stromal antigen 2 |
C00000958
|
| 10254 | STAM2, Hbp, STAM2A, STAM2B | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
C00000958
|
| 57559 | STAMBPL1, ALMalpha, AMSH-FP, AMSH-LP, bA399O19.2 | STAM binding protein-like 1 (EC:3.1.2.15) |
C00000958
|
| 90627 | STARD13, ARHGAP37, DLC2, GT650, LINC00464 | StAR-related lipid transfer (START) domain containing 13 |
C00000958
|
| 64801 | ARV1 | ARV1 homolog (S. cerevisiae) |
C00000958
|
| 6773 | STAT2, ISGF-3, P113, STAT113 | signal transducer and activator of transcription 2, 113kDa |
C00000958
|
| 420 | ART4, ARTC4, CD297, DO, DOK1 | ADP-ribosyltransferase 4 (EC:2.4.2.31) |
C00000958
|
| 27067 | STAU2, 39K2, 39K3 | staufen double-stranded RNA binding protein 2 |
C00000958
|
| 8614 | STC2, STC-2, STCRP | stanniocalcin 2 |
C00000958
|
| 6793 | STK10, LOK, PRO2729 | serine/threonine kinase 10 (EC:2.7.11.1) |
C00000958
|
| 79642 | ARSJ | arylsulfatase family, member J (EC:3.1.6.12) |
C00000958
|
| 6788 | STK3, KRS1, MST2 | serine/threonine kinase 3 (EC:2.7.11.1) |
C00000958
|
| 27148 | STK36, FU | serine/threonine kinase 36 (EC:2.7.11.1) |
C00000958
|
| 11329 | STK38, NDR, NDR1 | serine/threonine kinase 38 (EC:2.7.11.1) |
C00000958
|
| 340075 | ARSI | arylsulfatase family, member I |
C00000958
|
| 6789 | STK4, KRS2, MST1, TIIAC, YSK3 | serine/threonine kinase 4 (EC:2.7.11.1) |
C00000958
|
| 83931 | STK40, SHIK, SgK495 | serine/threonine kinase 40 (EC:2.7.11.1) |
C00000958
|
| 90529 | STPG1, C1orf201, MAPO2 | sperm-tail PG-rich repeat containing 1 |
C00000958
|
| 92335 | STRADA, LYK5, NY-BR-96, PMSE, STRAD, Stlk | STE20-related kinase adaptor alpha |
C00000958
|
| 55437 | STRADB, ALS2CR2, CALS-21, ILPIP, ILPIPA, PAPK | STE20-related kinase adaptor beta |
C00000958
|
| 6801 | STRN, SG2NA | striatin, calmodulin binding protein |
C00000958
|
| 8676 | STX11, FHL4, HLH4, HPLH4 | syntaxin 11 |
C00000958
|
| 55014 | STX17 | syntaxin 17 |
C00000958
|
| 6810 | STX4, STX4A, p35-2 | syntaxin 4 |
C00000958
|
| 8417 | STX7 | syntaxin 7 |
C00000958
|
| 6814 | STXBP3, MUNC18-3, MUNC18C, PSP, UNC-18C | syntaxin binding protein 3 |
C00000958
|
| 134957 | STXBP5, LGL3, LLGL3, Nbla04300 | syntaxin binding protein 5 (tomosyn) |
C00000958
|
| 29091 | STXBP6, amisyn | syntaxin binding protein 6 (amisyn) |
C00000958
|
| 387082 | SUMO4, IDDM5, SMT3H4, SUMO-4, dJ281H8.4 | small ubiquitin-like modifier 4 |
C00000958
|
| 6821 | SUOX | sulfite oxidase (EC:1.8.3.1) |
C00000958
|
| 8464 | SUPT3H, SPT3, SPT3L | suppressor of Ty 3 homolog (S. cerevisiae) |
C00000958
|
| 440423 | SUZ12P1, SUZ12P | suppressor of zeste 12 homolog pseudogene 1 |
C00000958
|
| 54823 | SWT1, C1orf26, HsSwt1 | SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
C00000958
|
| 256126 | SYCE2, CESC1 | synaptonemal complex central element protein 2 |
C00000958
|
| 25949 | SYF2, CBPIN, NTC31, P29, fSAP29 | SYF2 pre-mRNA-splicing factor |
C00000958
|
| 81493 | SYNC, SYNC1, SYNCOILIN | syncoilin, intermediate filament protein |
C00000958
|
| 23224 | SYNE2, EDMD5, NUA, NUANCE, Nesp2, Nesprin-2, SYNE-2, TROPH | spectrin repeat containing, nuclear envelope 2 |
C00000958
|
| 8871 | SYNJ2, INPP5H | synaptojanin 2 (EC:3.1.3.36) |
C00000958
|
| 255928 | SYT14, SCAR11, sytXIV | synaptotagmin XIV |
C00000958
|
| 57561 | ARRDC3, TLIMP | arrestin domain containing 3 |
C00000958
|
| 51204 | TACO1, CCDC44 | translational activator of mitochondrially encoded cytochrome c oxidase I |
C00000958
|
| 4070 | TACSTD2, EGP-1, EGP1, GA733-1, GA7331, GP50, M1S1, TROP2 | tumor-associated calcium signal transducer 2 |
C00000958
|
| 6871 | TADA2A, ADA2, ADA2A, TADA2L, hADA2 | transcriptional adaptor 2A |
C00000958
|
| 9014 | TAF1B, MGC:9349, RAF1B, RAFI63, SL1, TAFI63 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
C00000958
|
| 79101 | TAF1D, JOSD3, RAFI41, TAF(I)41, TAFI41 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
C00000958
|
| 85461 | TANC1, ROLSB, TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
C00000958
|
| 26115 | TANC2, ROLSA, rols | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
C00000958
|
| 79613 | TANGO6, TMCO7 | transport and golgi organization 6 homolog (Drosophila) |
C00000958
|
| 10010 | TANK, I-TRAF, ITRAF, TRAF2 | TRAF family member-associated NFKB activator |
C00000958
|
| 51347 | TAOK3, DPK, JIK, MAP3K18 | TAO kinase 3 (EC:2.7.11.1) |
C00000958
|
| 80222 | TARS2, TARSL1, thrRS | threonyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.3) |
C00000958
|
| 50832 | TAS2R4, T2R4 | taste receptor, type 2, member 4 |
C00000958
|
| 55617 | TASP1, C20orf13, dJ585I14.2 | taspase, threonine aspartase, 1 (EC:3.4.25.-) |
C00000958
|
| 83874 | TBC1D10A, EPI64, TBC1D10, dJ130H16.1, dJ130H16.2 | TBC1 domain family, member 10A |
C00000958
|
| 23232 | TBC1D12 | TBC1 domain family, member 12 |
C00000958
|
| 55296 | TBC1D19 | TBC1 domain family, member 19 |
C00000958
|
| 55357 | TBC1D2, PARIS-1, PARIS1, TBC1D2A | TBC1 domain family, member 2 |
C00000958
|
| 221322 | TBC1D32, BROMI, C6orf170, C6orf171, bA301B7.2, bA57L9.1, dJ310J6.1 | TBC1 domain family, member 32 |
C00000958
|
| 79718 | TBL1XR1, C21, DC42, IRA1, TBLR1 | transducin (beta)-like 1 X-linked receptor 1 |
C00000958
|
| 10716 | TBR1, TBR-1, TES-56 | T-box, brain, 1 |
C00000958
|
| 9096 | TBX18 | T-box 18 |
C00000958
|
| 6926 | TBX3, TBX3-ISO, UMS, XHL | T-box 3 |
C00000958
|
| 90843 | TCEAL8 | transcription elongation factor A (SII)-like 8 |
C00000958
|
| 127428 | TCEANC2, C1orf83, RP4-758J24.3 | transcription elongation factor A (SII) N-terminal and central domain containing 2 |
C00000958
|
| 408 | ARRB1, ARB1, ARR1 | arrestin, beta 1 |
C00000958
|
| 55346 | TCP11L1, dJ85M6.3 | t-complex 11, testis-specific-like 1 |
C00000958
|
| 81873 | ARPC5L, ARC16-2 | actin related protein 2/3 complex, subunit 5-like |
C00000958
|
| 81550 | TDRD3 | tudor domain containing 3 |
C00000958
|
| 23424 | TDRD7, CATC4, CTRCT36, PCTAIRE2BP, RP11-508D10.1, TRAP | tudor domain containing 7 |
C00000958
|
| 7004 | TEAD4, EFTR-2, RTEF1, TCF13L1, TEF-3, TEF3, TEFR-1, hRTEF-1B | TEA domain family member 4 |
C00000958
|
| 7006 | TEC, PSCTK4 | tec protein tyrosine kinase (EC:2.7.10.2) |
C00000958
|
| 79736 | TEFM, C17orf42 | transcription elongation factor, mitochondrial |
C00000958
|
| 54386 | TERF2IP, DRIP5, RAP1 | telomeric repeat binding factor 2, interacting protein |
C00000958
|
| 406 | ARNTL, BMAL1, BMAL1c, JAP3, MOP3, PASD3, TIC, bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like |
C00000958
|
| 54790 | TET2, KIAA1546, MDS | tet methylcytosine dioxygenase 2 (EC:1.14.11.n2) |
C00000958
|
| 55852 | TEX2, HT008, TMEM96 | testis expressed 2 |
C00000958
|
| 80210 | ARMC9, ARM, KU-MEL-1 | armadillo repeat containing 9 |
C00000958
|
| 55130 | ARMC4, CILD23 | armadillo repeat containing 4 |
C00000958
|
| 51106 | TFB1M, CGI75, mtTFB, mtTFB1 | transcription factor B1, mitochondrial (EC:2.1.1.-) |
C00000958
|
| 7024 | TFCP2, LBP1C, LSF, LSF1D, SEF, TFCP2C | transcription factor CP2 |
C00000958
|
| 7029 | TFDP2, DP2 | transcription factor Dp-2 (E2F dimerization partner 2) |
C00000958
|
| 7045 | TGFBI, BIGH3, CDB1, CDG2, CDGG1, CSD, CSD1, CSD2, CSD3, EBMD, LCD1 | transforming growth factor, beta-induced, 68kDa |
C00000958
|
| 55156 | ARMC1, Arcp | armadillo repeat containing 1 |
C00000958
|
| 7049 | TGFBR3, BGCAN, betaglycan | transforming growth factor, beta receptor III |
C00000958
|
| 54622 | ARL15, ARFRP2 | ADP-ribosylation factor-like 15 |
C00000958
|
| 168451 | THAP5 | THAP domain containing 5 |
C00000958
|
| 100286461 |
C00000958
|
||
| 9984 | THOC1, HPR1, P84, P84N5 | THO complex 1 |
C00000958
|
| 57187 | THOC2, CXorf3, THO2, dJ506G2.1, hTREX120 | THO complex 2 |
C00000958
|
| 374500 |
C00000958
|
||
| 29087 | THYN1, MDS012, MY105, THY28, THY28KD | thymocyte nuclear protein 1 |
C00000958
|
| 7072 | TIA1, TIA-1, WDM | TIA1 cytotoxic granule-associated RNA binding protein |
C00000958
|
| 7074 | TIAM1 | T-cell lymphoma invasion and metastasis 1 |
C00000958
|
| 166815 | TIGD2 | tigger transposable element derived 2 |
C00000958
|
| 50650 | ARHGEF3, GEF3, STA3, XPLN | Rho guanine nucleotide exchange factor (GEF) 3 |
C00000958
|
| 64283 | ARHGEF28, RGNEF, RIP2, p190RHOGEF | Rho guanine nucleotide exchange factor (GEF) 28 |
C00000958
|
| 9414 | TJP2, C9DUPq21.11, DFNA51, DUP9q21.11, X104, ZO2 | tight junction protein 2 |
C00000958
|
| 7084 | TK2, MTDPS2, MTTK | thymidine kinase 2, mitochondrial (EC:2.7.1.21) |
C00000958
|
| 116238 | TLCD1 | TLC domain containing 1 |
C00000958
|
| 7088 | TLE1, ESG, ESG1, GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
C00000958
|
| 7091 | TLE4, BCE-1, BCE1, E(spI), ESG, ESG4, GRG4 | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
C00000958
|
| 7098 | TLR3, CD283, IIAE2 | toll-like receptor 3 |
C00000958
|
| 4071 | TM4SF1, H-L6, L6, M3S1, TAAL6 | transmembrane 4 L six family member 1 |
C00000958
|
| 23023 | TMCC1 | transmembrane and coiled-coil domain family 1 |
C00000958
|
| 8834 | TMEM11, C17orf35, PM1, PMI | transmembrane protein 11 |
C00000958
|
| 84216 | TMEM117 | transmembrane protein 117 |
C00000958
|
| 144404 | TMEM120B | transmembrane protein 120B |
C00000958
|
| 83935 | TMEM133 | transmembrane protein 133 |
C00000958
|
| 65084 | TMEM135, PMP52 | transmembrane protein 135 |
C00000958
|
| 51524 | TMEM138 | transmembrane protein 138 |
C00000958
|
| 28978 | TMEM14A, C6orf73 | transmembrane protein 14A |
C00000958
|
| 201799 | TMEM154 | transmembrane protein 154 |
C00000958
|
| 26084 | ARHGEF26, CSGEF, HMFN1864, SGEF | Rho guanine nucleotide exchange factor (GEF) 26 |
C00000958
|
| 55858 | TMEM165, CDG2K, FT27, GDT1, TMPT27, TPARL | transmembrane protein 165 |
C00000958
|
| 66074 |
C00000958
|
||
| 134285 | TMEM171, PRP2 | transmembrane protein 171 |
C00000958
|
| 80775 | TMEM177 | transmembrane protein 177 |
C00000958
|
| 55266 | TMEM19 | transmembrane protein 19 |
C00000958
|
| 147007 | TMEM199, C17orf32 | transmembrane protein 199 |
C00000958
|
| 23670 | TMEM2 | transmembrane protein 2 |
C00000958
|
| 85019 | TMEM241, C18orf45, hVVT | transmembrane protein 241 |
C00000958
|
| 140738 | TMEM37, PR, PR1 | transmembrane protein 37 |
C00000958
|
| 148534 | TMEM56 | transmembrane protein 56 |
C00000958
|
| 85025 | TMEM60, C7orf35 | transmembrane protein 60 |
C00000958
|
| 80021 | TMEM62 | transmembrane protein 62 |
C00000958
|
| 91147 | TMEM67, JBTS6, MECKELIN, MKS3, NPHP11, TNEM67 | transmembrane protein 67 |
C00000958
|
| 83857 | TMTC1, OLF, TMTC1A | transmembrane and tetratricopeptide repeat containing 1 |
C00000958
|
| 9411 | ARHGAP29, PARG1, RP11-255E17.1 | Rho GTPase activating protein 29 |
C00000958
|
| 79822 | ARHGAP28 | Rho GTPase activating protein 28 |
C00000958
|
| 7127 | TNFAIP2, B94, EXOC3L3 | tumor necrosis factor, alpha-induced protein 2 |
C00000958
|
| 25816 | TNFAIP8, GG2-1, MDC-3.13, NDED, SCC-S2, SCCS2 | tumor necrosis factor, alpha-induced protein 8 |
C00000958
|
| 8793 | TNFRSF10D, CD264, DCR2, TRAIL-R4, TRAILR4, TRUNDD | tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
C00000958
|
| 51330 | TNFRSF12A, CD266, FN14, TWEAKR | tumor necrosis factor receptor superfamily, member 12A |
C00000958
|
| 55504 | TNFRSF19, TAJ, TAJ-alpha, TRADE, TROY | tumor necrosis factor receptor superfamily, member 19 |
C00000958
|
| 7132 | TNFRSF1A, CD120a, FPF, MS5, TBP1, TNF-R, TNF-R-I, TNF-R55, TNFAR, TNFR1, TNFR1-d2, TNFR55, TNFR60, p55, p55-R, p60 | tumor necrosis factor receptor superfamily, member 1A |
C00000958
|
| 3604 | TNFRSF9, 4-1BB, CD137, CDw137, ILA | tumor necrosis factor receptor superfamily, member 9 |
C00000958
|
| 83478 | ARHGAP24, FILGAP, RC-GAP72, RCGAP72, p73, p73RhoGAP | Rho GTPase activating protein 24 |
C00000958
|
| 23043 | TNIK | TRAF2 and NCK interacting kinase (EC:2.7.11.1) |
C00000958
|
| 23112 | TNRC6B | trinucleotide repeat containing 6B |
C00000958
|
| 64759 | TNS3, TEM6, TENS1 | tensin 3 |
C00000958
|
| 10140 | TOB1, APRO6, PIG49, TOB, TROB, TROB1 | transducer of ERBB2, 1 |
C00000958
|
| 9804 | TOMM20, MAS20, MOM19, TOM20 | translocase of outer mitochondrial membrane 20 homolog (yeast) |
C00000958
|
| 57584 | ARHGAP21, ARHGAP10 | Rho GTPase activating protein 21 |
C00000958
|
| 7155 | TOP2B, TOPIIB, top2beta | topoisomerase (DNA) II beta 180kDa (EC:5.99.1.3) |
C00000958
|
| 9760 | TOX, TOX1 | thymocyte selection-associated high mobility group box |
C00000958
|
| 57569 | ARHGAP20, RARHOGAP | Rho GTPase activating protein 20 |
C00000958
|
| 7158 | TP53BP1, 53BP1, p202 | tumor protein p53 binding protein 1 |
C00000958
|
| 93663 | ARHGAP18, MacGAP, SENEX, bA307O14.2 | Rho GTPase activating protein 18 |
C00000958
|
| 1200 | TPP1, CLN2, LPIC, TPP-1 | tripeptidyl peptidase I (EC:3.4.14.9) |
C00000958
|
| 8460 | TPST1, TANGO13A | tyrosylprotein sulfotransferase 1 (EC:2.8.2.20) |
C00000958
|
| 8459 | TPST2, TANGO13B | tyrosylprotein sulfotransferase 2 (EC:2.8.2.20) |
C00000958
|
| 387590 | TPTEP1, psiTPTE22 | transmembrane phosphatase with tensin homology pseudogene 1 |
C00000958
|
| 10758 | TRAF3IP2, ACT1, C6orf2, C6orf4, C6orf5, C6orf6, CIKS, PSORS13 | TRAF3 interacting protein 2 |
C00000958
|
| 7188 | TRAF5, MGC:39780, RNF84 | TNF receptor-associated factor 5 |
C00000958
|
| 9697 | TRAM2 | translocation associated membrane protein 2 |
C00000958
|
| 9881 | TRANK1, LBA1 | tetratricopeptide repeat and ankyrin repeat containing 1 |
C00000958
|
| 60684 | TRAPPC11, C4orf41, FOIGR, GRY, LGMD2S | trafficking protein particle complex 11 |
C00000958
|
| 51112 | TRAPPC12, TTC-15, TTC15 | trafficking protein particle complex 12 |
C00000958
|
| 51693 | TRAPPC2L, HSPC176 | trafficking protein particle complex 2-like |
C00000958
|
| 27095 | TRAPPC3, BET3 | trafficking protein particle complex 3 |
C00000958
|
| 1787 | TRDMT1, DMNT2, DNMT2, MHSAIIP, PUMET, RNMT1 | tRNA aspartic acid methyltransferase 1 (EC:2.1.1.204) |
C00000958
|
| 94134 | ARHGAP12 | Rho GTPase activating protein 12 |
C00000958
|
| 10221 | TRIB1, C8FW, GIG-2, GIG2, SKIP1, TRB-1, TRB1 | tribbles pseudokinase 1 |
C00000958
|
| 6737 | TRIM21, RNF81, RO52, SSA, SSA1 | tripartite motif containing 21 |
C00000958
|
| 10346 | TRIM22, GPSTAF50, RNF94, STAF50 | tripartite motif containing 22 |
C00000958
|
| 8805 | TRIM24, PTC6, RNF82, TF1A, TIF1, TIF1A, TIF1ALPHA, hTIF1 | tripartite motif containing 24 |
C00000958
|
| 4591 | TRIM37, MUL, POB1, TEF3 | tripartite motif containing 37 |
C00000958
|
| 54765 | TRIM44, DIPB, HSA249128, MC7 | tripartite motif containing 44 |
C00000958
|
| 80128 | TRIM46, GENEY, TRIFIC | tripartite motif containing 46 |
C00000958
|
| 85363 | TRIM5, RNF88, TRIM5alpha | tripartite motif containing 5 |
C00000958
|
| 9824 | ARHGAP11A, GAP_(1-12) | Rho GTPase activating protein 11A |
C00000958
|
| 11078 | TRIOBP, DFNB28, TARA, dJ37E16.4 | TRIO and F-actin binding protein |
C00000958
|
| 79658 | ARHGAP10, GRAF2, PS-GAP, PSGAP | Rho GTPase activating protein 10 |
C00000958
|
| 93587 | TRMT10A, RG9MTD2, TRM10 | tRNA methyltransferase 10 homolog A (S. cerevisiae) |
C00000958
|
| 79979 | TRMT2B, CXorf34, dJ341D10.3 | tRNA methyltransferase 2 homolog B (S. cerevisiae) (EC:2.1.1.35) |
C00000958
|
| 26133 | TRPC4AP, C20orf188, TRRP4AP, TRUSS | transient receptor potential cation channel, subfamily C, member 4 associated protein |
C00000958
|
| 7442 | TRPV1, VR1 | transient receptor potential cation channel, subfamily V, member 1 |
C00000958
|
| 9819 | TSC22D2, TILZ4a, TILZ4b, TILZ4c | TSC22 domain family, member 2 |
C00000958
|
| 23555 | TSPAN15, 2700063A19Rik, NET-7, NET7, TM4SF15 | tetraspanin 15 |
C00000958
|
| 90139 | TSPAN18, TSPAN | tetraspanin 18 |
C00000958
|
| 10098 | TSPAN5, NET-4, NET4, TM4SF9, TSPAN-5 | tetraspanin 5 |
C00000958
|
| 7105 | TSPAN6, T245, TM4SF6, TSPAN-6 | tetraspanin 6 |
C00000958
|
| 55720 | TSR1 | TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
C00000958
|
| 90121 | TSR2, DT1P1A10, WGG1 | TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
C00000958
|
| 7260 | TSSC1 | tumor suppressing subtransferable candidate 1 |
C00000958
|
| 55622 | TTC27 | tetratricopeptide repeat domain 27 |
C00000958
|
| 150737 | TTC30B, IFT70, fleer | tetratricopeptide repeat domain 30B |
C00000958
|
| 123016 | TTC8, BBS8, RP51 | tetratricopeptide repeat domain 8 |
C00000958
|
| 10565 | ARFGEF1, ARFGEP1, BIG1, P200 | ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
C00000958
|
| 25809 | TTLL1, C22orf7, HS323M22B | tubulin tyrosine ligase-like family, member 1 |
C00000958
|
| 51174 | TUBD1, TUBD | tubulin, delta 1 |
C00000958
|
| 114791 | TUBGCP5, GCP5 | tubulin, gamma complex associated protein 5 |
C00000958
|
| 7284 | TUFM, COXPD4, EF-TuMT, EFTU, P43 | Tu translation elongation factor, mitochondrial |
C00000958
|
| 56995 | TULP4, TUSP | tubby like protein 4 |
C00000958
|
| 221830 | TWISTNB | TWIST neighbor |
C00000958
|
| 64411 | ARAP3, CENTD3, DRAG1 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
C00000958
|
| 9352 | TXNL1, TRP32, TXL-1, TXNL, Txl | thioredoxin-like 1 |
C00000958
|
| 54957 | TXNL4B, DLP, Dim2 | thioredoxin-like 4B |
C00000958
|
| 7301 | TYRO3, BYK, Dtk, RSE, Sky, Tif | TYRO3 protein tyrosine kinase (EC:2.7.10.1) |
C00000958
|
| 9716 | AQR, IBP160, fSAP164 | aquarius intron-binding spliceosomal factor |
C00000958
|
| 55198 | APPL2, DIP13B | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
C00000958
|
| 9039 | UBA3, NAE2, UBE1C, hUBA3 | ubiquitin-like modifier activating enzyme 3 |
C00000958
|
| 7318 | UBA7, D8, UBA1B, UBE1L, UBE2 | ubiquitin-like modifier activating enzyme 7 |
C00000958
|
| 283991 | UBALD2, FAM100B | UBA-like domain containing 2 |
C00000958
|
| 51619 | UBE2D4, HBUCE1 | ubiquitin-conjugating enzyme E2D 4 (putative) (EC:6.3.2.19) |
C00000958
|
| 92912 | UBE2Q2 | ubiquitin-conjugating enzyme E2Q family member 2 (EC:6.3.2.19) |
C00000958
|
| 65264 | UBE2Z, USE1 | ubiquitin-conjugating enzyme E2Z (EC:6.3.2.19) |
C00000958
|
| 56061 | UBFD1, UBPH | ubiquitin family domain containing 1 |
C00000958
|
| 29978 | UBQLN2, ALS15, CHAP1, DSK2, N4BP4, PLIC2 | ubiquilin 2 |
C00000958
|
| 56893 | UBQLN4, A1U, A1Up, C1orf6, CIP75, UBIN | ubiquilin 4 |
C00000958
|
| 197131 | UBR1, JBS | ubiquitin protein ligase E3 component n-recognin 1 |
C00000958
|
| 23304 | UBR2, C6orf133, RP3-392M17.3, bA49A4.1, dJ242G1.1, dJ392M17.3 | ubiquitin protein ligase E3 component n-recognin 2 |
C00000958
|
| 130507 | UBR3, ZNF650 | ubiquitin protein ligase E3 component n-recognin 3 (putative) (EC:6.3.2.-) |
C00000958
|
| 7343 | UBTF, NOR-90, UBF, UBF-1, UBF1 | upstream binding transcription factor, RNA polymerase I |
C00000958
|
| 23376 | UFL1, KIAA0776, Maxer, NLBP, RCAD, RP3-393D12.1 | UFM1-specific ligase 1 |
C00000958
|
| 23780 | APOL2, APOL-II, APOL3 | apolipoprotein L, 2 |
C00000958
|
| 7358 | UGDH, GDH, UDP-GlcDH, UDPGDH, UGD | UDP-glucose 6-dehydrogenase (EC:1.1.1.22) |
C00000958
|
| 25989 | ULK3 | unc-51 like kinase 3 (EC:2.7.11.1) |
C00000958
|
| 25972 | UNC50, GMH1, UNCL, URP | unc-50 homolog (C. elegans) |
C00000958
|
| 65109 | UPF3B, HUPF3B, MRXS14, RENT3B, UPF3X | UPF3 regulator of nonsense transcripts homolog B (yeast) |
C00000958
|
| 139596 | UPRT, FUR1, UPP | uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) (EC:2.4.2.9) |
C00000958
|
| 7381 | UQCRB, MC3DN3, QCR7, QP-C, QPC, UQBC, UQBP, UQCR6, UQPC | ubiquinol-cytochrome c reductase binding protein (EC:1.10.2.2) |
C00000958
|
| 8975 | USP13, ISOT3, IsoT-3 | ubiquitin specific peptidase 13 (isopeptidase T-3) (EC:3.4.19.12) |
C00000958
|
| 23358 | USP24 | ubiquitin specific peptidase 24 (EC:3.4.19.12) |
C00000958
|
| 29761 | USP25, USP21 | ubiquitin specific peptidase 25 (EC:3.4.19.12) |
C00000958
|
| 84669 | USP32, NY-REN-60, USP10 | ubiquitin specific peptidase 32 (EC:3.4.19.12) |
C00000958
|
| 9736 | USP34 | ubiquitin specific peptidase 34 (EC:3.4.19.12) |
C00000958
|
| 57602 | USP36, DUB1 | ubiquitin specific peptidase 36 (EC:3.4.19.12) |
C00000958
|
| 8078 | USP5, ISOT | ubiquitin specific peptidase 5 (isopeptidase T) (EC:3.4.19.12) |
C00000958
|
| 9712 | USP6NL, RNTRE, TRE2NL, USP6NL-IT1 | USP6 N-terminal like |
C00000958
|
| 8239 | USP9X, DFFRX, FAF, FAM | ubiquitin specific peptidase 9, X-linked (EC:3.4.19.12) |
C00000958
|
| 10813 | UTP14A, NYCO16, SDCCAG16, dJ537K23.3 | UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
C00000958
|
| 57050 | UTP3, CRL1, CRLZ1, SAS10 | UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
C00000958
|
| 55813 | UTP6, C17orf40, HCA66 | UTP6, small subunit (SSU) processome component, homolog (yeast) |
C00000958
|
| 83464 | APH1B, APH-1B, PRO1328, PSFL, TAAV688 | APH1B gamma secretase subunit |
C00000958
|
| 7405 | UVRAG, DHTX, VPS38, p63 | UV radiation resistance associated |
C00000958
|
| 8673 | VAMP8, EDB, VAMP-8 | vesicle-associated membrane protein 8 |
C00000958
|
| 81839 | VANGL1, KITENIN, LPP2, STB2, STBM2 | VANGL planar cell polarity protein 1 |
C00000958
|
| 7410 | VAV2, VAV-2 | vav 2 guanine nucleotide exchange factor |
C00000958
|
| 323 | APBB2, FE65L, FE65L1 | amyloid beta (A4) precursor protein-binding, family B, member 2 |
C00000958
|
| 7417 | VDAC2, POR | voltage-dependent anion channel 2 |
C00000958
|
| 55317 | AP5S1, C20orf29 | adaptor-related protein complex 5, sigma 1 subunit |
C00000958
|
| 154807 | VKORC1L1 | vitamin K epoxide reductase complex, subunit 1-like 1 |
C00000958
|
| 23230 | VPS13A, CHAC, CHOREIN | vacuolar protein sorting 13 homolog A (S. cerevisiae) |
C00000958
|
| 157680 | VPS13B, CHS1, COH1 | vacuolar protein sorting 13 homolog B (yeast) |
C00000958
|
| 54832 | VPS13C | vacuolar protein sorting 13 homolog C (S. cerevisiae) |
C00000958
|
| 55187 | VPS13D | vacuolar protein sorting 13 homolog D (S. cerevisiae) |
C00000958
|
| 84313 | VPS25, DERP9, EAP20, FAP20 | vacuolar protein sorting 25 homolog (S. cerevisiae) |
C00000958
|
| 26276 | VPS33B | vacuolar protein sorting 33 homolog B (yeast) |
C00000958
|
| 51542 | VPS54, HCC8, SLP-8p, VPS54L, WR, hVps54L | vacuolar protein sorting 54 homolog (S. cerevisiae) |
C00000958
|
| 23355 | VPS8, KIAA0804 | vacuolar protein sorting 8 homolog (S. cerevisiae) |
C00000958
|
| 161 | AP2A2, ADTAB, CLAPA2, HIP-9, HIP9, HYPJ | adaptor-related protein complex 2, alpha 2 subunit |
C00000958
|
| 130340 | AP1S3 | adaptor-related protein complex 1, sigma 3 subunit |
C00000958
|
| 10352 | WARS2, TrpRS | tryptophanyl tRNA synthetase 2, mitochondrial (EC:6.1.1.2) |
C00000958
|
| 10163 | WASF2, IMD2, SCAR2, WASF4, WAVE2, dJ393P12.2 | WAS protein family, member 2 |
C00000958
|
| 11193 | WBP4, FBP21 | WW domain binding protein 4 |
C00000958
|
| 115825 | WDFY2, PROF, RP11-147H23.1, WDF2, ZFYVE22 | WD repeat and FYVE domain containing 2 |
C00000958
|
| 23001 | WDFY3, ALFY, ZFYVE25 | WD repeat and FYVE domain containing 3 |
C00000958
|
| 121601 | ANO4, TMEM16D | anoctamin 4 |
C00000958
|
| 55759 | WDR12, YTM1 | WD repeat domain 12 |
C00000958
|
| 57728 | WDR19, ATD5, CED4, DYF-2, IFT144, NPHP13, ORF26, Oseg6, PWDMP | WD repeat domain 19 |
C00000958
|
| 79446 | WDR25, C14orf67 | WD repeat domain 25 |
C00000958
|
| 10785 | WDR4, TRM82, TRMT82 | WD repeat domain 4 |
C00000958
|
| 54521 | WDR44, RAB11BP, RPH11 | WD repeat domain 44 |
C00000958
|
| 55112 | WDR60, FAP163, SRPS6 | WD repeat domain 60 |
C00000958
|
| 80349 | WDR61, REC14, SKI8 | WD repeat domain 61 |
C00000958
|
| 23335 | WDR7, TRAG | WD repeat domain 7 |
C00000958
|
| 55100 | WDR70 | WD repeat domain 70 |
C00000958
|
| 7474 | WNT5A, hWNT5A | wingless-type MMTV integration site family, member 5A |
C00000958
|
| 11059 | WWP1, AIP5, Tiul1, hSDRP1 | WW domain containing E3 ubiquitin protein ligase 1 |
C00000958
|
| 91526 | ANKRD44, PP6-ARS-B | ankyrin repeat domain 44 |
C00000958
|
| 7514 | XPO1, CRM1, emb, exp1 | exportin 1 (CRM1 homolog, yeast) |
C00000958
|
| 23214 | XPO6, EXP6, RANBP20 | exportin 6 |
C00000958
|
| 9213 | XPR1, SYG1, X3 | xenotropic and polytropic retrovirus receptor 1 |
C00000958
|
| 7518 | XRCC4 | X-ray repair complementing defective repair in Chinese hamster cells 4 |
C00000958
|
| 9942 | XYLB | xylulokinase homolog (H. influenzae) (EC:2.7.1.17) |
C00000958
|
| 64132 | XYLT2, PXYLT2, XT-II, XT2, xylT-II | xylosyltransferase II (EC:2.4.2.26) |
C00000958
|
| 57002 | YAE1D1, C7orf36 | Yae1 domain containing 1 |
C00000958
|
| 8565 | YARS, CMTDIC, TYRRS, YRS, YTS | tyrosyl-tRNA synthetase (EC:6.1.1.1) |
C00000958
|
| 646531 |
C00000958
|
||
| 8089 | YEATS4, 4930573H17Rik, B230215M10Rik, GAS41, NUBI-1, YAF9 | YEATS domain containing 4 |
C00000958
|
| 84272 | YIPF4, FinGER4 | Yip1 domain family, member 4 |
C00000958
|
| 79693 | YRDC, DRIP3, IRIP, RP11-109P14.4, SUA5 | yrdC N(6)-threonylcarbamoyltransferase domain containing |
C00000958
|
| 91746 | YTHDC1, YT521, YT521-B | YTH domain containing 1 |
C00000958
|
| 64848 | YTHDC2, CAHL | YTH domain containing 2 (EC:3.6.4.13) |
C00000958
|
| 54915 | YTHDF1, C20orf21 | YTH domain family, member 1 |
C00000958
|
| 7532 | YWHAG, 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
C00000958
|
| 113763 | ZBED6CL, C7orf29 | ZBED6 C-terminal like |
C00000958
|
| 253461 | ZBTB38, CIBZ, ZNF921 | zinc finger and BTB domain containing 38 |
C00000958
|
| 375248 | ANKRD36, UNQ2430 | ankyrin repeat domain 36 |
C00000958
|
| 85364 | ZCCHC3, C20orf99 | zinc finger, CCHC domain containing 3 |
C00000958
|
| 203430 | ZCCHC5, Mar3, Mart3, ZHC5 | zinc finger, CCHC domain containing 5 |
C00000958
|
| 84240 | ZCCHC9, PPP1R41 | zinc finger, CCHC domain containing 9 |
C00000958
|
| 54503 | ZDHHC13, HIP14L, HIP3RP | zinc finger, DHHC-type containing 13 (EC:2.3.1.225) |
C00000958
|
| 84287 | ZDHHC16, APH2 | zinc finger, DHHC-type containing 16 (EC:2.3.1.225) |
C00000958
|
| 90637 | ZFAND2A, AIRAP | zinc finger, AN1-type domain 2A |
C00000958
|
| 54469 | ZFAND6, AWP1, ZA20D3, ZFAND5B | zinc finger, AN1-type domain 6 |
C00000958
|
| 441951 | ZFAS1, C20orf199, HSUP1, HSUP2, NCRNA00275, ZNFX1-AS1 | ZNFX1 antisense RNA 1 |
C00000958
|
| 57623 | ZFAT, AITD3, ZFAT1, ZNF406 | zinc finger and AT hook domain containing |
C00000958
|
| 678 | ZFP36L2, BRF2, ERF-2, ERF2, RNF162C, TIS11D | ZFP36 ring finger protein-like 2 |
C00000958
|
| 9765 | ZFYVE16, PPP1R69 | zinc finger, FYVE domain containing 16 |
C00000958
|
| 84936 | ZFYVE19, MPFYVE | zinc finger, FYVE domain containing 19 |
C00000958
|
| 23051 | ZHX3, TIX1 | zinc fingers and homeoboxes 3 |
C00000958
|
| 22852 | ANKRD26, THC2, bA145E8.1 | ankyrin repeat domain 26 |
C00000958
|
| 57178 | ZMIZ1, MIZ, RAI17, TRAFIP10, ZIMP10, hZIMP10 | zinc finger, MIZ-type containing 1 |
C00000958
|
| 55608 | ANKRD10 | ankyrin repeat domain 10 |
C00000958
|
| 9202 | ZMYM4, CDIR, MYM, ZNF198L3, ZNF262 | zinc finger, MYM-type 4 |
C00000958
|
| 23613 | ZMYND8, PRKCBP1, PRO2893, RACK7 | zinc finger, MYND-type containing 8 |
C00000958
|
| 7559 | ZNF12, GIOT-3, HZF11, KOX3, ZNF325 | zinc finger protein 12 |
C00000958
|
| 7561 | ZNF14, GIOT-4, KOX6 | zinc finger protein 14 |
C00000958
|
| 90338 | ZNF160, F11, HKr18, HZF5, KR18 | zinc finger protein 160 |
C00000958
|
| 7764 | ZNF217, ZABC1 | zinc finger protein 217 |
C00000958
|
| 286101 | ZNF252P, ZNF252 | zinc finger protein 252, pseudogene |
C00000958
|
| 55609 | ZNF280C, SUHW3, ZNF633 | zinc finger protein 280C |
C00000958
|
| 54816 | ZNF280D, SUHW4, ZNF634 | zinc finger protein 280D |
C00000958
|
| 57336 | ZNF287, ZKSCAN13, ZSCAN45 | zinc finger protein 287 |
C00000958
|
| 162979 | ZNF296, ZFP296, ZNF342 | zinc finger protein 296 |
C00000958
|
| 55900 | ZNF302, HSD16, MST154, MSTP154, ZNF135L, ZNF140L, ZNF327 | zinc finger protein 302 |
C00000958
|
| 7580 | ZNF32, KOX30 | zinc finger protein 32 |
C00000958
|
| 6940 | ZNF354A, EZNF, HKL1, KID-1, KID1, TCF17 | zinc finger protein 354A |
C00000958
|
| 57037 | ANKMY2, ZMYND20 | ankyrin repeat and MYND domain containing 2 |
C00000958
|
| 7587 | ZNF37A, KOX21, ZNF37 | zinc finger protein 37A |
C00000958
|
| 84911 | ZNF382, KS1 | zinc finger protein 382 |
C00000958
|
| 79797 | ZNF408 | zinc finger protein 408 |
C00000958
|
| 353088 | ZNF429 | zinc finger protein 429 |
C00000958
|
| 220929 | ZNF438, bA330O11.1 | zinc finger protein 438 |
C00000958
|
| 79973 | ZNF442 | zinc finger protein 442 |
C00000958
|
| 25888 | ZNF473, HZFP100, ZN473 | zinc finger protein 473 |
C00000958
|
| 220992 | ZNF485 | zinc finger protein 485 |
C00000958
|
| 57473 | ZNF512B, GM632 | zinc finger protein 512B |
C00000958
|
| 9849 | ZNF518A, ZNF518 | zinc finger protein 518A |
C00000958
|
| 90233 | ZNF551 | zinc finger protein 551 |
C00000958
|
| 126295 | ZNF57, ZNF424 | zinc finger protein 57 |
C00000958
|
| 64763 | ZNF574, FP972 | zinc finger protein 574 |
C00000958
|
| 147949 | ZNF583 | zinc finger protein 583 |
C00000958
|
| 23060 | ZNF609 | zinc finger protein 609 |
C00000958
|
| 90441 | ZNF622, ZPR9 | zinc finger protein 622 |
C00000958
|
| 9831 | ZNF623 | zinc finger protein 623 |
C00000958
|
| 57547 | ZNF624 | zinc finger protein 624 |
C00000958
|
| 121274 | ZNF641 | zinc finger protein 641 |
C00000958
|
| 339500 | ZNF678 | zinc finger protein 678 |
C00000958
|
| 148213 | ZNF681 | zinc finger protein 681 |
C00000958
|
| 51058 | ZNF691, Zfp691 | zinc finger protein 691 |
C00000958
|
| 58492 | ZNF77, pT1 | zinc finger protein 77 |
C00000958
|
| 163049 | ZNF791 | zinc finger protein 791 |
C00000958
|
| 91752 | ZNF804A, C2orf10 | zinc finger protein 804A |
C00000958
|
| 152485 | ZNF827 | zinc finger protein 827 |
C00000958
|
| 91603 | ZNF830, CCDC16, OMCG1 | zinc finger protein 830 |
C00000958
|
| 7639 | ZNF85, HPF4, HTF1 | zinc finger protein 85 |
C00000958
|
| 84083 | ZRANB3, 4933425L19Rik, AH2 | zinc finger, RAN-binding domain containing 3 |
C00000958
|
| 7310 |
C00000958
|
||
| 7746 | ZSCAN9, PRD51, ZNF193 | zinc finger and SCAN domain containing 9 |
C00000958
|
| 79699 | ZYG11B, ZYG11 | zyg-11 family member B, cell cycle regulator |
C00000958
|
| 19 | ABCA1, ABC-1, ABC1, CERP, HDLDT1, TGD | ATP-binding cassette, sub-family A (ABC1), member 1 |
C00002903
|
| 54467 | ANKIB1 | ankyrin repeat and IBR domain containing 1 |
C00000958
|
| 1244 | ABCC2, ABC30, CMOAT, DJS, MRP2, cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
C00002903
|
| 8714 | ABCC3, ABC31, EST90757, MLP2, MOAT-D, MRP3, cMOAT2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
C00002903
|
| 10257 | ABCC4, EST170205, MOAT-B, MOATB, MRP4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
C00002903
|
| 83451 | ABHD11, WBSCR21 | abhydrolase domain containing 11 |
C00002903
|
| 116236 | ABHD15 | abhydrolase domain containing 15 (EC:3.1.1.-) |
C00002903
|
| 51434 | ANAPC7, APC7 | anaphase promoting complex subunit 7 |
C00000958
|
| 3983 | ABLIM1, ABLIM, LIMAB1, LIMATIN, abLIM-1 | actin binding LIM protein 1 |
C00002903
|
| 31 | ACACA, ACAC, ACACAD, ACC, ACC1, ACCA | acetyl-CoA carboxylase alpha (EC:6.4.1.2 6.3.4.14) |
C00002903
|
| 8348315 |
C00002903
|
||
| 122970 | ACOT4, PTE-Ib, PTE1B, PTE2B | acyl-CoA thioesterase 4 (EC:3.1.2.2) |
C00002903
|
| 11332 | ACOT7, ACH1, ACT, BACH, CTE-II, LACH, LACH1, RP1-120G22.10, hBACH | acyl-CoA thioesterase 7 (EC:3.1.2.2) |
C00002903
|
| 54 | ACP5, SPENCDI, TRAP | acid phosphatase 5, tartrate resistant (EC:3.1.3.2) |
C00002903
|
| 97 | ACYP1, ACYPE | acylphosphatase 1, erythrocyte (common) type (EC:3.6.1.7) |
C00002903
|
| 8754 | ADAM9, CORD9, MCMP, MDC9, Mltng | ADAM metallopeptidase domain 9 |
C00002903
|
| 120 | ADD3, ADDL | adducin 3 (gamma) |
C00002903
|
| 125 | ADH1B, ADH2 | alcohol dehydrogenase 1B (class I), beta polypeptide (EC:1.1.1.1) |
C00002903
|
| 9370 | ADIPOQ, ACDC, ACRP30, ADIPQTL1, ADPN, APM-1, APM1, GBP28 | adiponectin, C1Q and collagen domain containing |
C00002903
|
| 135 | ADORA2A, A2aR, ADORA2, RDC8 | adenosine A2a receptor |
C00002903
|
| 177 | AGER, RAGE | advanced glycosylation end product-specific receptor |
C00002903
|
| 137964 | AGPAT6, 1-AGPAT_6, LPAAT-zeta, LPAATZ, TSARG7 | 1-acylglycerol-3-phosphate O-acyltransferase 6 (EC:2.3.1.15) |
C00002903
|
| 10551 | AGR2, AG2, GOB-4, HAG-2, PDIA17, XAG-2 | anterior gradient 2 |
C00002903
|
| 155465 | AGR3, AG3, BCMP11, HAG3, PDIA18, hAG-3 | anterior gradient 3 |
C00002903
|
| 183 | AGT, ANHU, SERPINA8 | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
C00002903
|
| 213 | ALB, PRO0883, PRO0903, PRO1341 | albumin |
C00002903
|
| 216 | ALDH1A1, ALDC, ALDH-E1, ALDH1, ALDH11, PUMB1, RALDH1 | aldehyde dehydrogenase 1 family, member A1 (EC:1.2.1.36) |
C00002903
|
| 220 | ALDH1A3, ALDH1A6, ALDH6, MCOP8, RALDH3 | aldehyde dehydrogenase 1 family, member A3 (EC:1.2.1.5) |
C00002903
|
| 217 | ALDH2, ALDH-E2, ALDHI, ALDM | aldehyde dehydrogenase 2 family (mitochondrial) (EC:1.2.1.3) |
C00002903
|
| 222 | ALDH3B2, ALDH8 | aldehyde dehydrogenase 3 family, member B2 (EC:1.2.1.5) |
C00002903
|
| 8659 | ALDH4A1, ALDH4, P5CD, P5CDh | aldehyde dehydrogenase 4 family, member A1 (EC:1.2.1.88) |
C00002903
|
| 230 | ALDOC, ALDC | aldolase C, fructose-bisphosphate (EC:4.1.2.13) |
C00002903
|
| 79796 | ALG9, CDG1L, DIBD1, LOH11CR1J | ALG9, alpha-1,2-mannosyltransferase (EC:2.4.1.259 2.4.1.261) |
C00002903
|
| 239 | ALOX12, 12-LOX, 12S-LOX, LOG12 | arachidonate 12-lipoxygenase (EC:1.13.11.31) |
C00002903
|
| 246 | ALOX15, 12-LOX, 15-LOX-1, 15LOX-1 | arachidonate 15-lipoxygenase (EC:1.13.11.31 1.13.11.33) |
C00002903
|
| 240 | ALOX5, 5-LO, 5-LOX, 5LPG, LOG5 | arachidonate 5-lipoxygenase (EC:1.13.11.34) |
C00002903
|
| 702262 |
C00002903
|
||
| 262 | AMD1, ADOMETDC, AMD, SAMDC | adenosylmethionine decarboxylase 1 (EC:4.1.1.50) |
C00002903
|
| 267 | AMFR, GP78, RNF45 | autocrine motility factor receptor, E3 ubiquitin protein ligase (EC:6.3.2.19) |
C00002903
|
| 253650 | ANKRD18A | ankyrin repeat domain 18A |
C00002903
|
| 84250 | ANKRD32, BRCTD1, BRCTx | ankyrin repeat domain 32 |
C00002903
|
| 54443 | ANLN, Scraps, scra | anillin, actin binding protein |
C00002903
|
| 55107 | ANO1, DOG1, ORAOV2, TAOS2, TMEM16A | anoctamin 1, calcium activated chloride channel |
C00002903
|
| 81611 | ANP32E, LANP-L, LANPL | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
C00002903
|
| 311 | ANXA11, ANX11, CAP50 | annexin A11 |
C00002903
|
| 29945 | ANAPC4, APC4 | anaphase promoting complex subunit 4 |
C00000958
|
| 306 | ANXA3, ANX3 | annexin A3 (EC:3.1.4.43) |
C00002903
|
| 308 | ANXA5, ANX5, ENX2, PP4, RPRGL3 | annexin A5 |
C00002903
|
| 8943 | AP3D1, ADTD, hBLVR | adaptor-related protein complex 3, delta 1 subunit |
C00002903
|
| 328 | APEX1, APE, APE1, APEN, APEX, APX, HAP1, REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 (EC:4.2.99.18) |
C00002903
|
| 8539 | API5, AAC-11, AAC11 | apoptosis inhibitor 5 |
C00002903
|
| 378708 | APITD1, CENP-S, CENPS, FAAP16, MHF1 | apoptosis-inducing, TAF9-like domain 1 |
C00002903
|
| 335 | APOA1 | apolipoprotein A-I |
C00002903
|
| 10393 | ANAPC10, APC10, DOC1 | anaphase promoting complex subunit 10 |
C00000958
|
| 9582 | APOBEC3B, A3B, APOBEC1L, ARCD3, ARP4, DJ742C19.2, PHRBNL, bK150C2.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
C00002903
|
| 348 | APOE, AD2, LDLCQ5, LPG | apolipoprotein E |
C00002903
|
| 351 | APP, AAA, ABETA, ABPP, AD1, APPI, CTFgamma, CVAP, PN-II, PN2 | amyloid beta (A4) precursor protein |
C00002903
|
| 369 | ARAF, A-RAF, ARAF1, PKS2, RAFA1 | v-raf murine sarcoma 3611 viral oncogene homolog (EC:2.7.11.1) |
C00002903
|
| 394 | ARHGAP5, GFI2, RhoGAP5, p190-B, p190BRhoGAP | Rho GTPase activating protein 5 |
C00002903
|
| 5926 | ARID4A, RBBP-1, RBBP1, RBP-1, RBP1 | AT rich interactive domain 4A (RBP1-like) |
C00002903
|
| 403 | ARL3, ARFL3 | ADP-ribosylation factor-like 3 |
C00002903
|
| 409 | ARRB2, ARB2, ARR2, BARR2 | arrestin, beta 2 |
C00002903
|
| 427 | ASAH1, AC, ACDase, ASAH, PHP, PHP32, SMAPME | N-acylsphingosine amidohydrolase (acid ceramidase) 1 (EC:3.5.1.23) |
C00002903
|
| 429 | ASCL1, ASH1, HASH1, MASH1, bHLHa46 | achaete-scute family bHLH transcription factor 1 |
C00002903
|
| 55723 | ASF1B, CIA-II | anti-silencing function 1B histone chaperone |
C00002903
|
| 440 | ASNS, TS11 | asparagine synthetase (glutamine-hydrolyzing) (EC:6.3.5.4) |
C00002903
|
| 445 | ASS1, ASS, CTLN1 | argininosuccinate synthase 1 (EC:6.3.4.5) |
C00002903
|
| 29028 | ATAD2, ANCCA, PRO2000 | ATPase family, AAA domain containing 2 (EC:3.6.1.3) |
C00002903
|
| 466 | ATF1, EWS-ATF1, FUS/ATF-1, TREB36 | activating transcription factor 1 |
C00002903
|
| 270 | AMPD1, MAD, MADA, MMDD | adenosine monophosphate deaminase 1 (EC:3.5.4.6) |
C00000958
|
| 268 | AMH, MIF, MIS | anti-Mullerian hormone |
C00000958
|
| 489 | ATP2A3, SERCA3 | ATPase, Ca++ transporting, ubiquitous (EC:3.6.3.8) |
C00002903
|
| 516 | ATP5G1, ATP5A, ATP5G | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
C00002903
|
| 57198 | ATP8B2, ATPID | ATPase, aminophospholipid transporter, class I, type 8B, member 2 (EC:3.6.3.1) |
C00002903
|
| 9212 | AURKB, AIK2, AIM-1, AIM1, ARK2, AurB, IPL1, PPP1R48, STK12, STK5, aurkb-sv1, aurkb-sv2 | aurora kinase B (EC:2.7.11.1) |
C00002903
|
| 26053 | AUTS2, FBRSL2 | autism susceptibility candidate 2 |
C00002903
|
| 8313 | AXIN2, AXIL, ODCRCS | axin 2 |
C00002903
|
| 115701 | ALPK2, HAK | alpha-kinase 2 |
C00000958
|
| 84002 | B3GNT5, B3GN-T5, beta3Gn-T5 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 (EC:2.4.1.206) |
C00002903
|
| 572 | BAD, BBC2, BCL2L8 | BCL2-associated agonist of cell death |
C00002903
|
| 397543 |
C00002903
|
||
| 8314 | BAP1, HUCEP-13, UCHL2 | BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (EC:3.4.19.12) |
C00002903
|
| 580 | BARD1 | BRCA1 associated RING domain 1 |
C00002903
|
| 27113 | BBC3, JFY-1, JFY1, PUMA | BCL2 binding component 3 |
C00002903
|
| 582 | BBS1, BBS2L2 | Bardet-Biedl syndrome 1 |
C00002903
|
| 10134 | BCAP31, 6C6-AG, BAP31, CDM, DDCH, DXS1357E | B-cell receptor-associated protein 31 |
C00002903
|
| 8412 | BCAR3, NSP2, SH2D3B | breast cancer anti-estrogen resistance 3 |
C00002903
|
| 54828 | BCAS3, GAOB1, MAAB | breast carcinoma amplified sequence 3 |
C00002903
|
| 590 | BCHE, CHE1, CHE2, E1 | butyrylcholinesterase (EC:3.1.1.8) |
C00002903
|
| 597 | BCL2A1, ACC-1, ACC-2, BCL2L5, BFL1, GRS, HBPA1 | BCL2-related protein A1 |
C00002903
|
| 10018 | BCL2L11, BAM, BIM, BOD | BCL2-like 11 (apoptosis facilitator) |
C00002903
|
| 83596 | BCL2L12 | BCL2-like 12 (proline rich) |
C00002903
|
| 79053 | ALG8, CDG1H | ALG8, alpha-1,3-glucosyltransferase (EC:2.4.1.265) |
C00000958
|
| 624 | BDKRB2, B2R, BK-2, BK2, BKR2, BRB2 | bradykinin receptor B2 |
C00002903
|
| 8678 | BECN1, ATG6, VPS30, beclin1 | beclin 1, autophagy related |
C00002903
|
| 10282 | BET1, HBET1 | Bet1 golgi vesicular membrane trafficking protein |
C00002903
|
| 632 | BGLAP, BGP, OC, OCN | bone gamma-carboxyglutamate (gla) protein |
C00002903
|
| 100172339 |
C00002903
|
||
| 29929 | ALG6, CDG1C | ALG6, alpha-1,3-glucosyltransferase (EC:2.4.1.267) |
C00000958
|
| 638 | BIK, BIP1, BP4, NBK | BCL2-interacting killer (apoptosis-inducing) |
C00002903
|
| 329 | BIRC2, API1, HIAP2, Hiap-2, MIHB, RNF48, c-IAP1, cIAP1 | baculoviral IAP repeat containing 2 |
C00002903
|
| 332 | BIRC5, API4, EPR-1 | baculoviral IAP repeat containing 5 |
C00002903
|
| 641 | BLM, BS, RECQ2, RECQL2, RECQL3 | Bloom syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00002903
|
| 29760 | BLNK, AGM4, BASH, BLNK-S, LY57, SLP-65, SLP65, bca | B-cell linker |
C00002903
|
| 90427 | BMF | Bcl2 modifying factor |
C00002903
|
| 650 | BMP2, BDA2, BMP2A | bone morphogenetic protein 2 |
C00002903
|
| 653 | BMP5 | bone morphogenetic protein 5 |
C00002903
|
| 654 | BMP6, VGR, VGR1 | bone morphogenetic protein 6 |
C00002903
|
| 655 | BMP7, OP-1 | bone morphogenetic protein 7 |
C00002903
|
| 659 | BMPR2, BMPR-II, BMPR3, BMR2, BRK-3, PPH1, T-ALK | bone morphogenetic protein receptor, type II (serine/threonine kinase) (EC:2.7.11.30) |
C00002903
|
| 662 | BNIP1, NIP1, SEC20, TRG-8 | BCL2/adenovirus E1B 19kDa interacting protein 1 |
C00002903
|
| 663 | BNIP2, BNIP-2, NIP2 | BCL2/adenovirus E1B 19kDa interacting protein 2 |
C00002903
|
| 666 | BOK, BCL2L9, BOKL | BCL2-related ovarian killer |
C00002903
|
| 2972 | BRF1, BRF, BRF-1, GTF3B, TAF3B2, TAF3C, TAFIII90, TF3B90, TFIIIB90, hBRF | BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
C00002903
|
| 682 | BSG, 5F7, CD147, EMMPRIN, M6, OK, TCSF | basigin |
C00002903
|
| 7832 | BTG2, PC3, TIS21 | BTG family, member 2 |
C00002903
|
| 701 | BUB1B, BUB1beta, BUBR1, Bub1A, MAD3L, MVA1, SSK1, hBUBR1 | BUB1 mitotic checkpoint serine/threonine kinase B (EC:2.7.11.1) |
C00002903
|
| 9184 | BUB3, BUB3L, hBUB3 | BUB3 mitotic checkpoint protein |
C00002903
|
| 26098 | C10orf137, EDRF1 | chromosome 10 open reading frame 137 |
C00002903
|
| 220042 | C11orf82, noxin | chromosome 11 open reading frame 82 |
C00002903
|
| 89927 | C16orf45 | chromosome 16 open reading frame 45 |
C00002903
|
| 79762 | C1orf115, RP11-322F10.4 | chromosome 1 open reading frame 115 |
C00002903
|
| 199920 | C1orf168 | chromosome 1 open reading frame 168 |
C00002903
|
| 112770 | C1orf85, NCU-G1 | chromosome 1 open reading frame 85 |
C00002903
|
| 79680 | C22orf29, BOP | chromosome 22 open reading frame 29 |
C00002903
|
| 205327 | C2orf69 | chromosome 2 open reading frame 69 |
C00002903
|
| 201725 | C4orf46 | chromosome 4 open reading frame 46 |
C00002903
|
| 727 | C5, C5a, C5b, CPAMD4 | complement component 5 |
C00002903
|
| 254778 | C8orf46 | chromosome 8 open reading frame 46 |
C00002903
|
| 55071 | C9orf40 | chromosome 9 open reading frame 40 |
C00002903
|
| 760 | CA2, CA-II, CAC, CAII, Car2 | carbonic anhydrase II (EC:4.2.1.1) |
C00002903
|
| 27101 | CACYBP, GIG5, RP1-102G20.6, S100A6BP, SIP | calcyclin binding protein |
C00002903
|
| 799 | CALCR, CRT, CT-R, CTR, CTR1 | calcitonin receptor |
C00002903
|
| 811 | CALR, CRT, RO, SSA, cC1qR | calreticulin |
C00002903
|
| 55450 | CAMK2N1, PRO1489, RP11-401M16.1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 |
C00002903
|
| 10486 | CAP2 | CAP, adenylate cyclase-associated protein, 2 (yeast) |
C00002903
|
| 10753 | CAPN9, GC36, nCL-4 | calpain 9 (EC:3.4.22.-) |
C00002903
|
| 57082 | CASC5, AF15Q14, CT29, D40, KNL1, PPP1R55, Spc7, hKNL-1, hSpc105 | cancer susceptibility candidate 5 |
C00002903
|
| 843 | CASP10, ALPS2, FLICE2, MCH4 | caspase 10, apoptosis-related cysteine peptidase (EC:3.4.22.63) |
C00002903
|
| 835 | CASP2, CASP-2, ICH1, NEDD-2, NEDD2, PPP1R57 | caspase 2, apoptosis-related cysteine peptidase (EC:3.4.22.55) |
C00002903
|
| 837 | CASP4, ICE(rel)II, ICEREL-II, ICH-2, Mih1/TX, TX | caspase 4, apoptosis-related cysteine peptidase (EC:3.4.22.57) |
C00002903
|
| 199857 | ALG14 | ALG14, UDP-N-acetylglucosaminyltransferase subunit |
C00000958
|
| 9994 | CASP8AP2, CED-4, FLASH, RIP25 | caspase 8 associated protein 2 |
C00002903
|
| 857 | CAV1, BSCL3, CGL3, MSTP085, PPH3, VIP21, CAV | caveolin 1, caveolae protein, 22kDa |
C00002903
|
| 865 | CBFB, PEBP2B | core-binding factor, beta subunit |
C00002903
|
| 23468 | CBX5, HP1, HP1A | chromobox homolog 5 |
C00002903
|
| 150275 | CCDC117, dJ366L4.1 | coiled-coil domain containing 117 |
C00002903
|
| 64770 | CCDC14 | coiled-coil domain containing 14 |
C00002903
|
| 6346 | CCL1, I-309, P500, SCYA1, SISe, TCA3 | chemokine (C-C motif) ligand 1 |
C00002903
|
| 6356 | CCL11, SCYA11 | chemokine (C-C motif) ligand 11 |
C00002903
|
| 6348 | CCL3, G0S19-1, LD78ALPHA, MIP-1-alpha, MIP1A, SCYA3 | chemokine (C-C motif) ligand 3 |
C00002903
|
| 8900 | CCNA1 | cyclin A1 |
C00002903
|
| 890 | CCNA2, CCN1, CCNA | cyclin A2 |
C00002903
|
| 9133 | CCNB2, HsT17299 | cyclin B2 |
C00002903
|
| 894 | CCND2, KIAK0002 | cyclin D2 |
C00002903
|
| 896 | CCND3 | cyclin D3 |
C00002903
|
| 224 | ALDH3A2, ALDH10, FALDH, SLS | aldehyde dehydrogenase 3 family, member A2 (EC:1.2.1.3) |
C00000958
|
| 9134 | CCNE2, CYCE2 | cyclin E2 |
C00002903
|
| 729230 | CCR2, CC-CKR-2, CCR-2, CCR2A, CCR2B, CD192, CKR2, CKR2A, CKR2B, CMKBR2, MCP-1-R | chemokine (C-C motif) receptor 2 |
C00002903
|
| 30835 | CD209, CDSIGN, CLEC4L, DC-SIGN, DC-SIGN1 | CD209 molecule |
C00002903
|
| 100133941 | CD24, CD24A | CD24 molecule |
C00002903
|
| 948 | CD36, BDPLT10, CHDS7, FAT, GP3B, GP4, GPIV, PASIV, SCARB3 | CD36 molecule (thrombospondin receptor) |
C00002903
|
| 952 | CD38, T10 | CD38 molecule (EC:3.2.2.5) |
C00002903
|
| 960 | CD44, CDW44, CSPG8, ECMR-III, HCELL, HUTCH-I, IN, LHR, MC56, MDU2, MDU3, MIC4, Pgp1 | CD44 molecule (Indian blood group) |
C00002903
|
| 969 | CD69, AIM, BL-AC/P26, CLEC2C, EA1, GP32/28, MLR-3 | CD69 molecule |
C00002903
|
| 942 | CD86, B7-2, B7.2, B70, CD28LG2, LAB72 | CD86 molecule |
C00002903
|
| 976 | CD97, TM7LN1 | CD97 molecule |
C00002903
|
| 8872 | CDC123, C10orf7, D123 | cell division cycle 123 |
C00002903
|
| 34411 |
C00002903
|
||
| 994 | CDC25B | cell division cycle 25B (EC:3.1.3.48) |
C00002903
|
| 64400 | AKTIP, FT1, FTS | AKT interacting protein |
C00000958
|
| 31052 |
C00002903
|
||
| 990 | CDC6, CDC18L, HsCDC18, HsCDC6 | cell division cycle 6 |
C00002903
|
| 8317 | CDC7, CDC7L1, HsCDC7, Hsk1, huCDC7 | cell division cycle 7 (EC:2.7.11.1) |
C00002903
|
| 157313 | CDCA2, Repo-Man | cell division cycle associated 2 |
C00002903
|
| 55038 | CDCA4, HEPP, SEI-3/HEPP | cell division cycle associated 4 |
C00002903
|
| 113130 | CDCA5, SORORIN | cell division cycle associated 5 |
C00002903
|
| 55536 | CDCA7L, JPO2, R1, RAM2 | cell division cycle associated 7-like |
C00002903
|
| 55143 | CDCA8, BOR, BOREALIN, DasraB, MESRGP | cell division cycle associated 8 |
C00002903
|
| 999 | CDH1, Arc-1, CD324, CDHE, ECAD, LCAM, UVO | cadherin 1, type 1, E-cadherin (epithelial) |
C00002903
|
| 1010 | CDH12, CDHB | cadherin 12, type 2 (N-cadherin 2) |
C00002903
|
| 1000 | CDH2, CD325, CDHN, CDw325, NCAD | cadherin 2, type 1, N-cadherin (neuronal) |
C00002903
|
| 1003 | CDH5, 7B4, CD144 | cadherin 5, type 2 (vascular endothelium) |
C00002903
|
| 983 | CDK1, CDC2, CDC28A, P34CDC2 | cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22) |
C00002903
|
| 1017 | CDK2, p33(CDK2) | cyclin-dependent kinase 2 (EC:2.7.11.22) |
C00002903
|
| 1024 | CDK8, K35 | cyclin-dependent kinase 8 (EC:2.7.11.23 2.7.11.22) |
C00002903
|
| 1028 | CDKN1C, BWCR, BWS, KIP2, WBS, p57, p57Kip2 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
C00002903
|
| 1030 | CDKN2B, CDK4I, INK4B, MTS2, P15, TP15, p15INK4b | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
C00002903
|
| 1033 | CDKN3, CDI1, CIP2, KAP, KAP1 | cyclin-dependent kinase inhibitor 3 (EC:3.1.3.16 3.1.3.48) |
C00002903
|
| 81620 | CDT1, DUP, RIS2 | chromatin licensing and DNA replication factor 1 |
C00002903
|
| 55573 | CDV3, H41 | CDV3 homolog (mouse) |
C00002903
|
| 1044 | CDX1 | caudal type homeobox 1 |
C00002903
|
| 634 | CEACAM1, BGP, BGP1, BGPI | carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
C00002903
|
| 1056 | CEL, BAL, BSDL, BSSL, CELL, CEase, FAP, FAPP, LIPA, MODY8 | carboxyl ester lipase (EC:3.1.1.13 3.1.1.3) |
C00002903
|
| 1952 | CELSR2, CDHF10, EGFL2, Flamingo1, MEGF3 | cadherin, EGF LAG seven-pass G-type receptor 2 |
C00002903
|
| 1063 | CENPF, CENF, PRO1779, hcp-1 | centromere protein F, 350/400kDa |
C00002903
|
| 64946 | CENPH | centromere protein H |
C00002903
|
| 91687 | CENPL, C1orf155, CENP-L, RP3-383J4.1, dJ383J4.3 | centromere protein L |
C00002903
|
| 79019 | CENPM, C22orf18, CENP-M, PANE1, bK250D10.2 | centromere protein M |
C00002903
|
| 55839 | CENPN, BM039, C16orf60, CENP-N, ICEN32 | centromere protein N |
C00002903
|
| 79172 | CENPO, CENP-O, ICEN-36, MCM21R | centromere protein O |
C00002903
|
| 55166 | CENPQ, C6orf139, CENP-Q | centromere protein Q |
C00002903
|
| 387103 | CENPW, C6orf173, CENP-W, CUG2 | centromere protein W |
C00002903
|
| 22995 | CEP152, MCPH4, MCPH9, SCKL5 | centrosomal protein 152kDa |
C00002903
|
| 55165 | CEP55, C10orf3, CT111, URCC6 | centrosomal protein 55kDa |
C00002903
|
| 9702 | CEP57, MVA2, PIG8, TSP57 | centrosomal protein 57kDa |
C00002903
|
| 84131 | CEP78, C9orf81, IP63 | centrosomal protein 78kDa |
C00002903
|
| 10715 | CERS1, LAG1, LASS1, UOG1 | ceramide synthase 1 |
C00002903
|
| 29956 | CERS2, L3, LASS2, SP260, TMSG1 | ceramide synthase 2 |
C00002903
|
| 79603 | CERS4, LASS4, Trh1 | ceramide synthase 4 |
C00002903
|
| 253782 | CERS6, CERS5, LASS6 | ceramide synthase 6 |
C00002903
|
| 1082 | CGB, CGB3, CGB5, CGB7, CGB8, hCGB | chorionic gonadotropin, beta polypeptide |
C00002903
|
| 10036 | CHAF1A, CAF-1, CAF1, CAF1B, CAF1P150, P150 | chromatin assembly factor 1, subunit A (p150) |
C00002903
|
| 1111 | CHEK1, CHK1 | checkpoint kinase 1 (EC:2.7.11.1) |
C00002903
|
| 11200 | CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53, hCds1 | checkpoint kinase 2 (EC:2.7.11.1) |
C00002903
|
| 1113 | CHGA, CGA | chromogranin A (parathyroid secretory protein 1) |
C00002903
|
| 1116 | CHI3L1, ASRT7, CGP-39, GP-39, GP39, HC-gp39, HCGP-3P, YKL-40, YKL40, YYL-40, hCGP-39 | chitinase 3-like 1 (cartilage glycoprotein-39) |
C00002903
|
| 1137 | CHRNA4, BFNC, EBN, EBN1, NACHR, NACHRA4, NACRA4 | cholinergic receptor, nicotinic, alpha 4 (neuronal) |
C00002903
|
| 1140 | CHRNB1, ACHRB, CHRNB, CMS1D, CMS2A, SCCMS | cholinergic receptor, nicotinic, beta 1 (muscle) |
C00002903
|
| 10000 | AKT3, MPPH, PKB-GAMMA, PKBG, PRKBG, RAC-PK-gamma, RAC-gamma, STK-2 | v-akt murine thymoma viral oncogene homolog 3 (EC:2.7.11.1) |
C00000958
|
| 11113 | CIT, CRIK, STK21 | citron (rho-interacting, serine/threonine kinase 21) (EC:2.7.11.1) |
C00002903
|
| 26586 | CKAP2, LB1, TMAP, se20-10 | cytoskeleton associated protein 2 |
C00002903
|
| 150468 | CKAP2L | cytoskeleton associated protein 2-like |
C00002903
|
| 1163 | CKS1B, CKS1, PNAS-16, PNAS-18, ckshs1 | CDC28 protein kinase regulatory subunit 1B |
C00002903
|
| 9076 | CLDN1, CLD1, ILVASC, SEMP1 | claudin 1 |
C00002903
|
| 8644 | AKR1C3, DD3, DDX, HA1753, HAKRB, HAKRe, HSD17B5, PGFS, hluPGFS | aldo-keto reductase family 1, member C3 (EC:1.3.1.20 1.1.1.188 1.1.1.239 1.1.1.64 1.1.1.112 1.1.1.357) |
C00000958
|
| 56942 | CMC2, 2310061C15Rik, C16orf61 | C-x(9)-C motif containing 2 |
C00002903
|
| 84319 | CMSS1, C3orf26 | cms1 ribosomal small subunit homolog (yeast) |
C00002903
|
| 7555 | CNBP, CNBP1, DM2, PROMM, RNF163, ZCCHC22, ZNF9 | CCHC-type zinc finger, nucleic acid binding protein |
C00002903
|
| 81578 | COL21A1, COLA1L, dJ682J15.1, dJ708F5.1 | collagen, type XXI, alpha 1 |
C00002903
|
| 1280 | COL2A1, ANFH, AOM, COL11A3, SEDC, STL1 | collagen, type II, alpha 1 |
C00002903
|
| 1289 | COL5A1 | collagen, type V, alpha 1 |
C00002903
|
| 1312 | COMT | catechol-O-methyltransferase (EC:2.1.1.6) |
C00002903
|
| 8533 | COPS3, CSN3, SGN3 | COP9 signalosome subunit 3 |
C00002903
|
| 57175 | CORO1B, CORONIN-2 | coronin, actin binding protein, 1B |
C00002903
|
| 23603 | CORO1C, HCRNN4 | coronin, actin binding protein, 1C |
C00002903
|
| 7464 | CORO2A, CLIPINB, IR10, WDR2 | coronin, actin binding protein, 2A |
C00002903
|
| 33136 |
C00002903
|
||
| 1382 | CRABP2, CRABP-II, RBP6 | cellular retinoic acid binding protein 2 |
C00002903
|
| 148327 | CREB3L4, AIBZIP, ATCE1, CREB3, CREB4, JAL, hJAL | cAMP responsive element binding protein 3-like 4 |
C00002903
|
| 1387 | CREBBP, CBP, KAT3A, RSTS | CREB binding protein (EC:2.3.1.48) |
C00002903
|
| 1396 | CRIP1, CRHP, CRIP, CRP-1, CRP1 | cysteine-rich protein 1 (intestinal) |
C00002903
|
| 1397 | CRIP2, CRIP, CRP2, ESP1 | cysteine-rich protein 2 |
C00002903
|
| 1401 | CRP, PTX1 | C-reactive protein, pentraxin-related |
C00002903
|
| 1434 | CSE1L, CAS, CSE1, XPO2 | CSE1 chromosome segregation 1-like (yeast) |
C00002903
|
| 1435 | CSF1, CSF-1, MCSF | colony stimulating factor 1 (macrophage) (EC:2.7.10.1) |
C00002903
|
| 1436 | CSF1R, C-FMS, CD115, CSF-1R, CSFR, FIM2, FMS, HDLS, M-CSF-R | colony stimulating factor 1 receptor (EC:2.7.10.1) |
C00002903
|
| 1437 | CSF2, GMCSF | colony stimulating factor 2 (granulocyte-macrophage) |
C00002903
|
| 53944 | CSNK1G1, CK1gamma1 | casein kinase 1, gamma 1 (EC:2.7.11.1) |
C00002903
|
| 81566 | CSRNP2, C12orf2, C12orf22, FAM130A1, PPP1R72, TAIP-12 | cysteine-serine-rich nuclear protein 2 |
C00002903
|
| 1472 | CST4 | cystatin S |
C00002903
|
| 1475 | CSTA, AREI, STF1, STFA | cystatin A (stefin A) |
C00002903
|
| 51496 | CTDSPL2, HSPC129 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 (EC:3.1.3.-) |
C00002903
|
| 8727 | CTNNAL1, CLLP, alpha-CATU | catenin (cadherin-associated protein), alpha-like 1 |
C00002903
|
| 1500 | CTNND1, CAS, CTNND, P120CAS, P120CTN, p120, p120(CAS), p120(CTN) | catenin (cadherin-associated protein), delta 1 |
C00002903
|
| 1501 | CTNND2, GT24, NPRAP | catenin (cadherin-associated protein), delta 2 |
C00002903
|
| 9646 | CTR9, SH2BP1, TSBP, p150, p150TSP | CTR9, Paf1/RNA polymerase II complex component |
C00002903
|
| 1509 | CTSD, CLN10, CPSD | cathepsin D (EC:3.4.23.5) |
C00002903
|
| 1512 | CTSH, ACC-4, ACC-5, CPSB, minichain | cathepsin H (EC:3.4.22.16) |
C00002903
|
| 1513 | CTSK, CTS02, CTSO, CTSO1, CTSO2, PKND, PYCD | cathepsin K (EC:3.4.22.38) |
C00002903
|
| 1515 | CTSV, CATL2, CTSL2, CTSU | cathepsin V (EC:3.4.22.43) |
C00002903
|
| 55917 | CTTNBP2NL | CTTNBP2 N-terminal like |
C00002903
|
| 8065 | CUL5, VACM-1, VACM1 | cullin 5 |
C00002903
|
| 1523 | CUX1, CASP, CDP, CDP/Cut, CDP1, COY1, CUTL1, CUX, Clox, Cux/CDP, GOLIM6, Nbla10317, p100, p110, p200, p75 | cut-like homeobox 1 |
C00002903
|
| 6376 | CX3CL1, ABCD-3, C3Xkine, CXC3, CXC3C, NTN, NTT, SCYD1, fractalkine, neurotactin | chemokine (C-X3-C motif) ligand 1 |
C00002903
|
| 6387 | CXCL12, IRH, PBSF, SCYB12, SDF1, TLSF, TPAR1, SDF1A, SDF1B | chemokine (C-X-C motif) ligand 12 |
C00002903
|
| 55122 | AKIRIN2, C6orf166, FBI1, dJ486L4.2 | akirin 2 |
C00000958
|
| 51706 | CYB5R1, B5R.1, B5R1, B5R2, NQO3A2, humb5R2 | cytochrome b5 reductase 1 (EC:1.6.2.2) |
C00002903
|
| 1583 | CYP11A1, CYP11A, CYPXIA1, P450SCC | cytochrome P450, family 11, subfamily A, polypeptide 1 (EC:1.14.15.6) |
C00002903
|
| 1588 | CYP19A1, ARO, ARO1, CPV1, CYAR, CYP19, CYPXIX, P-450AROM | cytochrome P450, family 19, subfamily A, polypeptide 1 (EC:1.14.14.1) |
C00002903
|
| 56672 | AKIP1, BCA3, C11orf17 | A kinase (PRKA) interacting protein 1 |
C00000958
|
| 1593 | CYP27A1, CP27, CTX, CYP27 | cytochrome P450, family 27, subfamily A, polypeptide 1 (EC:1.14.13.15) |
C00002903
|
| 1557 | CYP2C19, CPCJ, CYP2C, P450C2C, P450IIC19 | cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00002903
|
| 1559 | CYP2C9, CPC9, CYP2C, CYP2C10, CYPIIC9, P450IIC9 | cytochrome P450, family 2, subfamily C, polypeptide 9 (EC:1.14.13.48 1.14.13.49 1.14.13.80) |
C00002903
|
| 1565 | CYP2D6, CPD6, CYP2D, CYP2D7AP, CYP2D7BP, CYP2D7P2, CYP2D8P2, CYP2DL1, CYPIID6, P450-DB1, P450C2D, P450DB1 | cytochrome P450, family 2, subfamily D, polypeptide 6 (EC:1.14.14.1) |
C00002903
|
| 11214 | AKAP13, AKAP-13, AKAP-Lbc, ARHGEF13, BRX, HA-3, Ht31, LBC, PRKA13, PROTO-LB, PROTO-LBC, c-lbc, p47 | A kinase (PRKA) anchor protein 13 |
C00000958
|
| 8529 | CYP4F2, CPF2 | cytochrome P450, family 4, subfamily F, polypeptide 2 (EC:1.14.13.30) |
C00002903
|
| 1605 | DAG1, 156DAG, A3a, AGRNR, DAG, MDDGC7, MDDGC9 | dystroglycan 1 (dystrophin-associated glycoprotein 1) |
C00002903
|
| 1616 | DAXX, BING2, DAP6, EAP1 | death-domain associated protein |
C00002903
|
| 10926 | DBF4, ASK, CHIF, DBF4A, ZDBF1 | DBF4 homolog (S. cerevisiae) |
C00002903
|
| 1627 | DBN1, D0S117E | drebrin 1 |
C00002903
|
| 26094 | DCAF4, WDR21, WDR21A | DDB1 and CUL4 associated factor 4 |
C00002903
|
| 100528464 |
C00002903
|
||
| 1633 | DCK | deoxycytidine kinase (EC:2.7.1.74) |
C00002903
|
| 9201 | DCLK1, CL1, CLICK1, DCAMKL1, DCDC3A, DCLK | doublecortin-like kinase 1 (EC:2.7.11.1) |
C00002903
|
| 64858 | DCLRE1B, APOLLO, SNM1B, SNMIB | DNA cross-link repair 1B |
C00002903
|
| 1638 | DCT, TRP-2, TYRP2 | dopachrome tautomerase (EC:5.3.3.12) |
C00002903
|
| 1639 | DCTN1, DAP-150, DP-150, P135 | dynactin 1 |
C00002903
|
| 1641 | DCX, DBCN, DC, LISX, SCLH, XLIS | doublecortin |
C00002903
|
| 23564 | DDAH2, DDAH, DDAHII, G6a, NG30 | dimethylarginine dimethylaminohydrolase 2 (EC:3.5.3.18) |
C00002903
|
| 1643 | DDB2, DDBB, UV-DDB2 | damage-specific DNA binding protein 2, 48kDa |
C00002903
|
| 1644 | DDC, AADC | dopa decarboxylase (aromatic L-amino acid decarboxylase) (EC:4.1.1.28) |
C00002903
|
| 326709 |
C00002903
|
||
| 54541 | DDIT4, Dig2, REDD-1, REDD1 | DNA-damage-inducible transcript 4 |
C00002903
|
| 84962 | AJUBA, JUB | ajuba LIM protein |
C00000958
|
| 55635 | DEPDC1, DEP.8, DEPDC1-V2, DEPDC1A, SDP35 | DEP domain containing 1 |
C00002903
|
| 420357 |
C00002903
|
||
| 64798 | DEPTOR, DEP.6, DEPDC6 | DEP domain containing MTOR-interacting protein |
C00002903
|
| 51071 | DERA, DEOC | deoxyribose-phosphate aldolase (putative) (EC:4.1.2.4) |
C00002903
|
| 1676 | DFFA, DFF-45, DFF1, ICAD | DNA fragmentation factor, 45kDa, alpha polypeptide |
C00002903
|
| 1717 | DHCR7, SLOS | 7-dehydrocholesterol reductase (EC:1.3.1.21) |
C00002903
|
| 10202 | DHRS2, HEP27, SDR25C1 | dehydrogenase/reductase (SDR family) member 2 |
C00002903
|
| 9249 | DHRS3, DD83.1, RDH17, Rsdr1, SDR1, SDR16C1, retSDR1 | dehydrogenase/reductase (SDR family) member 3 (EC:1.1.1.300) |
C00002903
|
| 9785 | DHX38, DDX38, PRP16, PRPF16 | DEAH (Asp-Glu-Ala-His) box polypeptide 38 (EC:3.6.4.13) |
C00002903
|
| 1736 | DKC1, CBF5, DKC, DKCX, NAP57, NOLA4, XAP101 | dyskeratosis congenita 1, dyskerin |
C00002903
|
| 79469 | DLEU2L, BCMSUNL | deleted in lymphocytic leukemia 2-like |
C00002903
|
| 23234 | DNAJC9, HDJC9, JDD1, SB73 | DnaJ (Hsp40) homolog, subfamily C, member 9 |
C00002903
|
| 1788 | DNMT3A, DNMT3A2, M.HsaIIIA | DNA (cytosine-5-)-methyltransferase 3 alpha (EC:2.1.1.37) |
C00002903
|
| 55715 | DOK4 | docking protein 4 |
C00002903
|
| 29980 | DONSON, B17, C21orf60 | downstream neighbor of SON |
C00002903
|
| 1828 | DSG1, CDHF4, DG1, DSG, EPKHIA, PPKS1, SPPK1 | desmoglein 1 |
C00002903
|
| 79980 | DSN1, C20orf172, KNL3, MIS13, dJ469A13.2, hKNL-3 | DSN1, MIS12 kinetochore complex component |
C00002903
|
| 11034 | DSTN, ACTDP, ADF, bA462D18.2 | destrin (actin depolymerizing factor) |
C00002903
|
| 1837 | DTNA, D18S892E, DRP3, DTN, DTN-A, LVNC1 | dystrobrevin, alpha |
C00002903
|
| 1843 | DUSP1, CL100, HVH1, MKP-1, MKP1, PTPN10 | dual specificity phosphatase 1 (EC:3.1.3.16 3.1.3.48) |
C00002903
|
| 11221 | DUSP10, MKP-5, MKP5 | dual specificity phosphatase 10 (EC:3.1.3.16 3.1.3.48) |
C00002903
|
| 1845 | DUSP3, VHR | dual specificity phosphatase 3 (EC:3.1.3.16 3.1.3.48) |
C00002903
|
| 1854 | DUT, dUTPase | deoxyuridine triphosphatase (EC:3.6.1.23) |
C00002903
|
| 8655 | DYNLL1, DLC1, DLC8, DNCL1, DNCLC1, LC8, LC8a, PIN, hdlc1 | dynein, light chain, LC8-type 1 |
C00002903
|
| 6990 | DYNLT3, RP3, TCTE1L, TCTEX1L | dynein, light chain, Tctex-type 3 |
C00002903
|
| 1869 | E2F1, E2F-1, RBAP1, RBBP3, RBP3 | E2F transcription factor 1 |
C00002903
|
| 1870 | E2F2, E2F-2 | E2F transcription factor 2 |
C00002903
|
| 144455 | E2F7 | E2F transcription factor 7 |
C00002903
|
| 79733 | E2F8, E2F-8 | E2F transcription factor 8 |
C00002903
|
| 1877 | E4F1, E4F | E4F transcription factor 1 |
C00002903
|
| 10682 | EBP, CDPX2, CHO2, CPX, CPXD | emopamil binding protein (sterol isomerase) (EC:5.3.3.5) |
C00002903
|
| 8726 | EED, HEED, WAIT1 | embryonic ectoderm development |
C00002903
|
| 1917 | EEF1A2, EEF1AL, EF-1-alpha-2, EF1A, HS1, STN, STNL | eukaryotic translation elongation factor 1 alpha 2 |
C00002903
|
| 80303 | EFHD1, MST133, MSTP133, SWS2 | EF-hand domain family, member D1 |
C00002903
|
| 54583 | EGLN1, C1orf12, ECYT3, HIF-PH2, HIFPH2, HPH-2, HPH2, PHD2, SM20, ZMYND6 | egl-9 family hypoxia-inducible factor 1 (EC:1.14.11.29) |
C00002903
|
| 1960 | EGR3, EGR-3, PILOT | early growth response 3 |
C00002903
|
| 9538 | EI24, EPG4, PIG8, TP53I8 | etoposide induced 2.4 |
C00002903
|
| 8890 | EIF2B4, EIF-2B, EIF2B, EIF2Bdelta | eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
C00002903
|
| 1965 | EIF2S1, EIF-2, EIF-2A, EIF-2alpha, EIF2, EIF2A | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
C00002903
|
| 1977 | EIF4E, AUTS19, CBP, EIF4E1, EIF4EL1, EIF4F | eukaryotic translation initiation factor 4E |
C00002903
|
| 1978 | EIF4EBP1, 4E-BP1, 4EBP1, BP-1, PHAS-I | eukaryotic translation initiation factor 4E binding protein 1 |
C00002903
|
| 1982 | EIF4G2, AAG1, DAP5, NAT1, P97 | eukaryotic translation initiation factor 4 gamma, 2 |
C00002903
|
| 1983 | EIF5, EIF-5, EIF-5A | eukaryotic translation initiation factor 5 |
C00002903
|
| 1984 | EIF5A, EIF-5A, EIF5A1, eIF5AI | eukaryotic translation initiation factor 5A |
C00002903
|
| 60481 | ELOVL5, HELO1, dJ483K16.1 | ELOVL fatty acid elongase 5 (EC:2.3.1.199) |
C00002903
|
| 79071 | ELOVL6, FACE, FAE, LCE | ELOVL fatty acid elongase 6 (EC:2.3.1.199) |
C00002903
|
| 79993 | ELOVL7 | ELOVL fatty acid elongase 7 (EC:2.3.1.199) |
C00002903
|
| 23587 | ELP5, C17orf81, DERP6, MST071, MSTP071 | elongator acetyltransferase complex subunit 5 |
C00002903
|
| 51016 | EMC9, C14orf122, FAM158A | ER membrane protein complex subunit 9 |
C00002903
|
| 2019 | EN1 | engrailed homeobox 1 |
C00002903
|
| 8507 | ENC1, CCL28, ENC-1, KLHL35, KLHL37, NRPB, PIG10, TP53I10 | ectodermal-neural cortex 1 (with BTB domain) |
C00002903
|
| 23052 | ENDOD1 | endonuclease domain containing 1 |
C00002903
|
| 2021 | ENDOG | endonuclease G |
C00002903
|
| 2023 | ENO1, ENO1L1, MPB1, NNE, PPH | enolase 1, (alpha) (EC:4.2.1.11) |
C00002903
|
| 2026 | ENO2, NSE | enolase 2 (gamma, neuronal) (EC:4.2.1.11) |
C00002903
|
| 2027 | ENO3, GSD13, MSE | enolase 3 (beta, muscle) (EC:4.2.1.11) |
C00002903
|
| 2033 | EP300, KAT3B, RSTS2, p300 | E1A binding protein p300 (EC:2.3.1.48) |
C00002903
|
| 57669 | EPB41L5, BE37, YMO1 | erythrocyte membrane protein band 4.1 like 5 |
C00002903
|
| 2041 | EPHA1, EPH, EPHT, EPHT1 | EPH receptor A1 (EC:2.7.10.1) |
C00002903
|
| 2052 | EPHX1, EPHX, EPOX, HYL1, MEH | epoxide hydrolase 1, microsomal (xenobiotic) (EC:3.3.2.9) |
C00002903
|
| 2056 | EPO, EP, MVCD2 | erythropoietin |
C00002903
|
| 2060 | EPS15, AF-1P, AF1P, MLLT5 | epidermal growth factor receptor pathway substrate 15 |
C00002903
|
| 2064 | ERBB2, CD340, HER-2, HER-2/neu, HER2, MLN_19, NEU, NGL, TKR1 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (EC:2.7.10.1) |
C00002903
|
| 2065 | ERBB3, ErbB-3, HER3, LCCS2, MDA-BF-1, c-erbB-3, c-erbB3, erbB3-S, p180-ErbB3, p45-sErbB3, p85-sErbB3 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 (EC:2.7.10.1) |
C00002903
|
| 2066 | ERBB4, ALS19, HER4, p180erbB4 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 (EC:2.7.10.1) |
C00002903
|
| 2071 | ERCC3, BTF2, GTF2H, RAD25, TFIIH, XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (EC:3.6.4.12) |
C00002903
|
| 54821 | ERCC6L, PICH, RAD26L | excision repair cross-complementing rodent repair deficiency, complementation group 6-like (EC:3.6.4.12) |
C00002903
|
| 9700 | ESPL1, ESP1, SEPA | extra spindle pole bodies homolog 1 (S. cerevisiae) (EC:3.4.22.49) |
C00002903
|
| 83715 | ESPN, DFNB36 | espin |
C00002903
|
| 2099 | ESR1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | estrogen receptor 1 |
C00002903
|
| 55224 | ETNK2, EKI2, HMFT1716 | ethanolamine kinase 2 (EC:2.7.1.82) |
C00002903
|
| 2118 | ETV4, E1A-F, E1AF, PEA3, PEAS3 | ets variant 4 |
C00002903
|
| 9156 | EXO1, HEX1, hExoI | exonuclease 1 |
C00002903
|
| 23404 | EXOSC2, RRP4, Rrp4p, hRrp4p, p7 | exosome component 2 |
C00002903
|
| 2145 | EZH1, KMT6B | enhancer of zeste homolog 1 (Drosophila) (EC:2.1.1.43) |
C00002903
|
| 2146 | EZH2, ENX-1, ENX1, EZH1, EZH2b, KMT6, KMT6A, WVS, WVS2 | enhancer of zeste homolog 2 (Drosophila) (EC:2.1.1.43) |
C00002903
|
| 2167 | FABP4, A-FABP, AFABP, ALBP, aP2 | fatty acid binding protein 4, adipocyte |
C00002903
|
| 2173 | FABP7, B-FABP, BLBP, FABPB, MRG | fatty acid binding protein 7, brain |
C00002903
|
| 3992 | FADS1, D5D, FADS6, FADSD5, LLCDL1, TU12 | fatty acid desaturase 1 (EC:1.14.19.-) |
C00002903
|
| 11170 | FAM107A, DRR1, TU3A | family with sequence similarity 107, member A |
C00002903
|
| 83641 | FAM107B, C10orf45 | family with sequence similarity 107, member B |
C00002903
|
| 642273 | FAM110C | family with sequence similarity 110, member C |
C00002903
|
| 63901 | FAM111A, GCLEB, KCS2 | family with sequence similarity 111, member A |
C00002903
|
| 374393 | FAM111B, CANP | family with sequence similarity 111, member B |
C00002903
|
| 317662 | FAM149B1, KIAA0974 | family with sequence similarity 149, member B1 |
C00002903
|
| 145483 | FAM161B, C14orf44, c14_5547 | family with sequence similarity 161, member B |
C00002903
|
| 165215 | FAM171B, KIAA1946 | family with sequence similarity 171, member B |
C00002903
|
| 56204 | FAM214A, KIAA1370 | family with sequence similarity 214, member A |
C00002903
|
| 55731 | FAM222B, C17orf63 | family with sequence similarity 222, member B |
C00002903
|
| 131583 | FAM43A | family with sequence similarity 43, member A |
C00002903
|
| 54855 | FAM46C | family with sequence similarity 46, member C |
C00002903
|
| 79850 | FAM57A, CT120 | family with sequence similarity 57, member A |
C00002903
|
| 54478 | FAM64A, CATS, RCS1 | family with sequence similarity 64, member A |
C00002903
|
| 9750 | FAM65B, C6orf32, DIFF40, DIFF48, MYONAP, PL48 | family with sequence similarity 65, member B |
C00002903
|
| 2175 | FANCA, FA, FA-H, FA1, FAA, FACA, FAH, FANCH | Fanconi anemia, complementation group A |
C00002903
|
| 2177 | FANCD2, FA-D2, FA4, FACD, FAD, FAD2, FANCD | Fanconi anemia, complementation group D2 |
C00002903
|
| 2189 | FANCG, FAG, XRCC9 | Fanconi anemia, complementation group G |
C00002903
|
| 55215 | FANCI, KIAA1794 | Fanconi anemia, complementation group I |
C00002903
|
| 55120 | FANCL, FAAP43, PHF9, POG | Fanconi anemia, complementation group L |
C00002903
|
| 9255 | AIMP1, EMAP2, EMAPII, HLD3, SCYE1, p43 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
C00000958
|
| 64853 | AIDA, C1orf80, RP11-378J18.7 | axin interactor, dorsalization associated |
C00000958
|
| 114907 | FBXO32, Fbx32, MAFbx | F-box protein 32 |
C00002903
|
| 26271 | FBXO5, EMI1, FBX5, Fbxo31 | F-box protein 5 |
C00002903
|
| 2222 | FDFT1, DGPT, ERG9, SQS, SS | farnesyl-diphosphate farnesyltransferase 1 (EC:2.5.1.21) |
C00002903
|
| 2224 | FDPS, FPPS, FPS | farnesyl diphosphate synthase (EC:2.5.1.1 2.5.1.10) |
C00002903
|
| 2237 | FEN1, FEN-1, MF1, RAD2 | flap structure-specific endonuclease 1 |
C00002903
|
| 121512 | FGD4, CMT4H, FRABP, ZFYVE6 | FYVE, RhoGEF and PH domain containing 4 |
C00002903
|
| 2249 | FGF4, HBGF-4, HST, HST-1, HSTF1, K-FGF, KFGF | fibroblast growth factor 4 |
C00002903
|
| 2260 | FGFR1, BFGFR, CD331, CEK, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 | fibroblast growth factor receptor 1 (EC:2.7.10.1) |
C00002903
|
| 2274 | FHL2, AAG11, DRAL, FHL-2, SLIM-3, SLIM3 | four and a half LIM domains 2 |
C00002903
|
| 63979 | FIGNL1 | fidgetin-like 1 |
C00002903
|
| 51024 | FIS1, TTC11 | fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
C00002903
|
| 51303 | FKBP11, FKBP19 | FK506 binding protein 11, 19 kDa (EC:5.2.1.8) |
C00002903
|
| 2288 | FKBP4, FKBP51, FKBP52, FKBP59, HBI, Hsp56, PPIase, p52 | FK506 binding protein 4, 59kDa (EC:5.2.1.8) |
C00002903
|
| 2289 | FKBP5, AIG6, FKBP51, FKBP54, P54, PPIase, Ptg-10 | FK506 binding protein 5 (EC:5.2.1.8) |
C00002903
|
| 10211 | FLOT1 | flotillin 1 |
C00002903
|
| 2321 | FLT1, FLT, FLT-1, VEGFR-1, VEGFR1 | fms-related tyrosine kinase 1 (EC:2.7.10.1) |
C00002903
|
| 2335 | FN1, CIG, ED-B, FINC, FN, FNZ, GFND, GFND2, LETS, MSF | fibronectin 1 |
C00002903
|
| 3169 | FOXA1, HNF3A, TCF3A | forkhead box A1 |
C00002903
|
| 3170 | FOXA2, HNF3B, TCF3B | forkhead box A2 |
C00002903
|
| 3171 | FOXA3, FKHH3, HNF3G, TCF3G | forkhead box A3 |
C00002903
|
| 2297 | FOXD1, FKHL8, FREAC4 | forkhead box D1 |
C00002903
|
| 2305 | FOXM1, FKHL16, FOXM1B, HFH-11, HFH11, HNF-3, INS-1, MPHOSPH2, MPP-2, MPP2, PIG29, TGT3, TRIDENT | forkhead box M1 |
C00002903
|
| 1112 | FOXN3, C14orf116, CHES1, PRO1635 | forkhead box N3 |
C00002903
|
| 2308 | FOXO1, FKH1, FKHR, FOXO1A | forkhead box O1 |
C00002903
|
| 2309 | FOXO3, AF6q21, FKHRL1, FKHRL1P2, FOXO2, FOXO3A | forkhead box O3 |
C00002903
|
| 494532 |
C00002903
|
||
| 4303 | FOXO4, AFX, AFX1, MLLT7 | forkhead box O4 |
C00002903
|
| 80020 | FOXRED2, ERFAD | FAD-dependent oxidoreductase domain containing 2 |
C00002903
|
| 10468 | FST, FS | follistatin |
C00002903
|
| 2517 | FUCA1, FUCA | fucosidase, alpha-L- 1, tissue (EC:3.2.1.51) |
C00002903
|
| 50486 | G0S2, RP1-28O10.2 | G0/G1switch 2 |
C00002903
|
| 55568 | GALNT10, GALNACT10, PPGALNACT10, PPGANTASE10 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) (EC:2.4.1.41) |
C00002903
|
| 8693 | GALNT4, GALNAC-T4, GALNACT4 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) (EC:2.4.1.41) |
C00002903
|
| 51809 | GALNT7, GALNAC-T7, GalNAcT7 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (EC:2.4.1.41) |
C00002903
|
| 2596 | GAP43, B-50, PP46 | growth associated protein 43 |
C00002903
|
| 283431 | GAS2L3, G2L3 | growth arrest-specific 2 like 3 |
C00002903
|
| 2643 | GCH1, DYT14, DYT5, DYT5a, GCH, GTP-CH-1, GTPCH1, HPABH4B | GTP cyclohydrolase 1 (EC:3.5.4.16) |
C00002903
|
| 2644 | GCHFR, GFRP, HsT16933, P35 | GTP cyclohydrolase I feedback regulator |
C00002903
|
| 2645 | GCK, FGQTL3, GK, GLK, HHF3, HK4, HKIV, HXKP, LGLK, MODY2 | glucokinase (hexokinase 4) (EC:2.7.1.2) |
C00002903
|
| 2729 | GCLC, GCL, GCS, GLCL, GLCLC | glutamate-cysteine ligase, catalytic subunit (EC:6.3.2.2) |
C00002903
|
| 2730 | GCLM, GLCLR | glutamate-cysteine ligase, modifier subunit (EC:6.3.2.2) |
C00002903
|
| 2657 | GDF1, DORV, DTGA3, RAI | growth differentiation factor 1 |
C00002903
|
| 185 | AGTR1, AG2S, AGTR1A, AGTR1B, AT1, AT1AR, AT1B, AT1BR, AT1R, AT2R1, AT2R1A, AT2R1B, HAT1R | angiotensin II receptor, type 1 |
C00000958
|
| 79153 | GDPD3 | glycerophosphodiester phosphodiesterase domain containing 3 |
C00002903
|
| 81544 | GDPD5, GDE2, PP1665 | glycerophosphodiester phosphodiesterase domain containing 5 |
C00002903
|
| 2674 | GFRA1, GDNFR, GDNFRA, GFR-ALPHA-1, RET1L, RETL1, TRNR1 | GDNF family receptor alpha 1 |
C00002903
|
| 2675 | GFRA2, GDNFRB, NRTNR-ALPHA, NTNRA, RETL2, TRNR2 | GDNF family receptor alpha 2 |
C00002903
|
| 8836 | GGH, GH | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (EC:3.4.19.9) |
C00002903
|
| 51659 | GINS2, HSPC037, PSF2, Pfs2 | GINS complex subunit 2 (Psf2 homolog) |
C00002903
|
| 64785 | GINS3, PSF3 | GINS complex subunit 3 (Psf3 homolog) |
C00002903
|
| 2697 | GJA1, AVSD3, CMDR, CX43, DFNB38, GJAL, HLHS1, HSS, ODDD | gap junction protein, alpha 1, 43kDa |
C00002903
|
| 2717 | GLA, GALA | galactosidase, alpha (EC:3.2.1.22) |
C00002903
|
| 2735 | GLI1, GLI | GLI family zinc finger 1 |
C00002903
|
| 51053 | GMNN, Gem | geminin, DNA replication inhibitor |
C00002903
|
| 2782 | GNB1 | guanine nucleotide binding protein (G protein), beta polypeptide 1 |
C00002903
|
| 8733 | GPAA1, GAA1, hGAA1 | glycosylphosphatidylinositol anchor attachment 1 |
C00002903
|
| 2852 | GPER1, CEPR, CMKRL2, DRY12, FEG-1, GPCR-Br, GPER, GPR30, LERGU, LERGU2, LyGPR | G protein-coupled estrogen receptor 1 |
C00002903
|
| 2822 | GPLD1, GPIPLD, GPIPLDM, PIGPLD, PIGPLD1 | glycosylphosphatidylinositol specific phospholipase D1 (EC:3.1.4.50) |
C00002903
|
| 350383 | GPR142, PGR2 | G protein-coupled receptor 142 |
C00002903
|
| 9052 | GPRC5A, GPCR5A, RAI3, RAIG1 | G protein-coupled receptor, family C, group 5, member A |
C00002903
|
| 2877 | GPX2, GI-GPx, GPRP, GPRP-2, GPx-2, GPx-GI, GSHPX-GI, GSHPx-2 | glutathione peroxidase 2 (gastrointestinal) (EC:1.11.1.9) |
C00002903
|
| 493869 | GPX8, EPLA847, GPx-8, GSHPx-8, UNQ847 | glutathione peroxidase 8 (putative) (EC:1.11.1.9) |
C00002903
|
| 57655 | GRAMD1A, KIAA1533 | GRAM domain containing 1A |
C00002903
|
| 2887 | GRB10, GRB-IR, Grb-10, IRBP, MEG1, RSS | growth factor receptor-bound protein 10 |
C00002903
|
| 8540 | AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY | alkylglycerone phosphate synthase (EC:2.5.1.26) |
C00000958
|
| 9687 | GREB1 | growth regulation by estrogen in breast cancer 1 |
C00002903
|
| 57822 | GRHL3, SOM, TFCP2L4 | grainyhead-like 3 (Drosophila) |
C00002903
|
| 2907 | GRINA, HNRGW, LFG1, NMDARA1, TMBIM3 | glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
C00002903
|
| 83903 | GSG2, HASPIN | germ cell associated 2 (haspin) (EC:2.7.11.1) |
C00002903
|
| 2934 | GSN, ADF, AGEL | gelsolin |
C00002903
|
| 2935 | GSPT1, 551G9.2, ETF3A, GST1, eRF3a | G1 to S phase transition 1 |
C00002903
|
| 2936 | GSR | glutathione reductase (EC:1.8.1.7) |
C00002903
|
| 2959 | GTF2B, TF2B, TFIIB | general transcription factor IIB |
C00002903
|
| 2961 | GTF2E2, FE, TF2E2, TFIIE-B | general transcription factor IIE, polypeptide 2, beta 34kDa |
C00002903
|
| 2968 | GTF2H4, P52, TFB2, TFIIH | general transcription factor IIH, polypeptide 4, 52kDa |
C00002903
|
| 2971 | GTF3A, AP2, TFIIIA | general transcription factor IIIA |
C00002903
|
| 51512 | GTSE1, B99 | G-2 and S-phase expressed 1 |
C00002903
|
| 178 | AGL, GDE | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase (EC:2.4.1.25 3.2.1.33) |
C00000958
|
| 3015 | H2AFZ, H2A.Z-1, H2A.z, H2A/z, H2AZ | H2A histone family, member Z |
C00002903
|
| 3033 | HADH, HAD, HADH1, HADHSC, HCDH, HHF4, MSCHAD, SCHAD | hydroxyacyl-CoA dehydrogenase (EC:1.1.1.35) |
C00002903
|
| 93323 | HAUS8, DGT4, HICE1, NY-SAR-48 | HAUS augmin-like complex, subunit 8 |
C00002903
|
| 3055 | HCK, JTK9, p59Hck, p61Hck | hemopoietic cell kinase (EC:2.7.10.2) |
C00002903
|
| 3065 | HDAC1, GON-10, HD1, RPD3, RPD3L1 | histone deacetylase 1 (EC:3.5.1.98) |
C00002903
|
| 8841 | HDAC3, HD3, RPD3, RPD3-2 | histone deacetylase 3 (EC:3.5.1.98) |
C00002903
|
| 3068 | HDGF, HMG1L2 | hepatoma-derived growth factor |
C00002903
|
| 3070 | HELLS, LSH, PASG, SMARCA6 | helicase, lymphoid-specific |
C00002903
|
| 64399 | HHIP, HIP | hedgehog interacting protein |
C00002903
|
| 8479 | HIRIP3 | HIRA interacting protein 3 |
C00002903
|
| 3017 | HIST1H2BD, H2B.1B, H2B/b, H2BFB, HIRIP2, dJ221C16.6 | histone cluster 1, H2bd |
C00002903
|
| 8344 | HIST1H2BE, H2B.h, H2B/h, H2BFH, dJ221C16.8 | histone cluster 1, H2be |
C00002903
|
| 8339 | HIST1H2BG, H2B.1A, H2B/a, H2BFA, dJ221C16.8 | histone cluster 1, H2bg |
C00002903
|
| 8351 | HIST1H3D, H3/b, H3FB | histone cluster 1, H3d |
C00002903
|
| 8357 | HIST1H3H, H3/k, H3F1K, H3FK | histone cluster 1, H3h |
C00002903
|
| 8361 | HIST1H4F, H4, H4/c, H4FC | histone cluster 1, H4f |
C00002903
|
| 8349 | HIST2H2BE, GL105, H2B, H2B.1, H2BFQ, H2BGL105, H2BQ | histone cluster 2, H2be |
C00002903
|
| 55355 | HJURP, FAKTS, URLC9, hFLEG1 | Holliday junction recognition protein |
C00002903
|
| 3134 | HLA-F, CDA12, HLA-5.4, HLA-CDA12, HLAF | major histocompatibility complex, class I, F |
C00002903
|
| 3148 | HMGB2, HMG2 | high mobility group box 2 |
C00002903
|
| 3156 | HMGCR, LDLCQ3 | 3-hydroxy-3-methylglutaryl-CoA reductase (EC:1.1.1.34) |
C00002903
|
| 3151 | HMGN2, HMG17 | high mobility group nucleosomal binding domain 2 |
C00002903
|
| 6927 | HNF1A, HNF-1A, HNF1, IDDM20, LFB1, MODY3, TCF-1, TCF1 | HNF1 homeobox A |
C00002903
|
| 6928 | HNF1B, FJHN, HNF-1B, HNF1beta, HNF2, HPC11, LF-B3, LFB3, MODY5, TCF-2, TCF2, VHNF1 | HNF1 homeobox B |
C00002903
|
| 9455 | HOMER2, ACPD, CPD, HOMER-2, VESL-2 | homer homolog 2 (Drosophila) |
C00002903
|
| 3198 | HOXA1, BSAS, HOX1, HOX1F | homeobox A1 |
C00002903
|
| 3223 | HOXC6, CP25, HHO.C8, HOX3, HOX3C | homeobox C6 |
C00002903
|
| 3251 | HPRT1, HGPRT, HPRT | hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8) |
C00002903
|
| 3265 | HRAS, C-BAS/HAS, C-H-RAS, C-HA-RAS1, CTLO, H-RASIDX, HAMSV, HRAS1, K-RAS, N-RAS, RASH1 | Harvey rat sarcoma viral oncogene homolog |
C00002903
|
| 3297 | HSF1, HSTF1 | heat shock transcription factor 1 |
C00002903
|
| 39078 |
C00002903
|
||
| 7184 | HSP90B1, ECGP, GP96, GRP94, TRA1 | heat shock protein 90kDa beta (Grp94), member 1 |
C00002903
|
| 3303 | HSPA1A, HSP70-1, HSP70-1A, HSP70I, HSP72, HSPA1 | heat shock 70kDa protein 1A |
C00002903
|
| 3309 | HSPA5, BIP, GRP78, MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
C00002903
|
| 3315 | HSPB1, CMT2F, HMN2B, HS.76067, HSP27, HSP28, Hsp25, SRP27 | heat shock 27kDa protein 1 |
C00002903
|
| 116987 | AGAP1, AGAP-1, CENTG2, GGAP1, cnt-g2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
C00000958
|
| 4299 | AFF1, AF4, MLLT2, PBM1 | AF4/FMR2 family, member 1 |
C00000958
|
| 3397 | ID1, ID, bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
C00002903
|
| 3399 | ID3, HEIR-1, bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
C00002903
|
| 3620 | IDO1, IDO, IDO-1, INDO | indoleamine 2,3-dioxygenase 1 (EC:1.13.11.52) |
C00002903
|
| 3423 | IDS, MPS2, SIDS | iduronate 2-sulfatase (EC:3.1.6.13) |
C00002903
|
| 159 | ADSS, ADEH, ADSS_2 | adenylosuccinate synthase (EC:6.3.4.4) |
C00000958
|
| 3475 | IFRD1, PC4, TIS7 | interferon-related developmental regulator 1 |
C00002903
|
| 7866 | IFRD2, IFNRP, SKMc15, SM15 | interferon-related developmental regulator 2 |
C00002903
|
| 55764 | IFT122, CED, CED1, SPG, WDR10, WDR10p, WDR140 | intraflagellar transport 122 homolog (Chlamydomonas) |
C00002903
|
| 112752 | IFT43, C14orf179, CED3 | intraflagellar transport 43 homolog (Chlamydomonas) |
C00002903
|
| 3482 | IGF2R, CD222, CIMPR, M6P-R, MPR1, MPRI | insulin-like growth factor 2 receptor |
C00002903
|
| 3484 | IGFBP1, AFBP, IBP1, IGF-BP25, PP12, hIGFBP-1 | insulin-like growth factor binding protein 1 |
C00002903
|
| 3485 | IGFBP2, IBP2, IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa |
C00002903
|
| 3486 | IGFBP3, BP-53, IBP3 | insulin-like growth factor binding protein 3 |
C00002903
|
| 3487 | IGFBP4, BP-4, HT29-IGFBP, IBP4, IGFBP-4 | insulin-like growth factor binding protein 4 |
C00002903
|
| 3488 | IGFBP5, IBP5 | insulin-like growth factor binding protein 5 |
C00002903
|
| 157 | ADRBK2, BARK2, GRK3 | adrenergic, beta, receptor kinase 2 (EC:2.7.11.15) |
C00000958
|
| 3592 | IL12A, CLMF, IL-12A, NFSK, NKSF1, P35 | interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
C00002903
|
| 3593 | IL12B, CLMF, CLMF2, IL-12B, NKSF, NKSF2 | interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
C00002903
|
| 3596 | IL13, IL-13, P600 | interleukin 13 |
C00002903
|
| 55540 | IL17RB, CRL4, EVI27, IL17BR, IL17RH1 | interleukin 17 receptor B |
C00002903
|
| 154 | ADRB2, ADRB2R, ADRBR, B2AR, BAR, BETA2AR | adrenoceptor beta 2, surface |
C00000958
|
| 50604 | IL20, IL-20, IL10D, ZCYTO10 | interleukin 20 |
C00002903
|
| 3567 | IL5, EDF, IL-5, TRF | interleukin 5 (colony-stimulating factor, eosinophil) |
C00002903
|
| 3572 | IL6ST, CD130, CDW130, GP130, IL-6RB | interleukin 6 signal transducer (gp130, oncostatin M receptor) |
C00002903
|
| 10994 | ILVBL, 209L8, AHAS, ILV2H | ilvB (bacterial acetolactate synthase)-like (EC:2.2.1.-) |
C00002903
|
| 3615 | IMPDH2, IMPD2, IMPDH-II | IMP (inosine 5'-monophosphate) dehydrogenase 2 (EC:1.1.1.205) |
C00002903
|
| 3619 | INCENP | inner centromere protein antigens 135/155kDa |
C00002903
|
| 8821 | INPP4B | inositol polyphosphate-4-phosphatase, type II, 105kDa (EC:3.1.3.66) |
C00002903
|
| 3630 | INS, IDDM2, ILPR, IRDN, MODY10 | insulin |
C00002903
|
| 3642 | INSM1, IA-1, IA1 | insulinoma-associated 1 |
C00002903
|
| 25896 | INTS7, C1orf73, INT7 | integrator complex subunit 7 |
C00002903
|
| 9670 | IPO13, IMP13, KAP13, LGL2, RANBP13 | importin 13 |
C00002903
|
| 55705 | IPO9, Imp9 | importin 9 |
C00002903
|
| 124152 | IQCK | IQ motif containing K |
C00002903
|
| 10788 | IQGAP2 | IQ motif containing GTPase activating protein 2 |
C00002903
|
| 359948 | IRF2BP2 | interferon regulatory factor 2 binding protein 2 |
C00002903
|
| 3664 | IRF6, LPS, OFC6, PIT, PPS, PPS1, VWS, VWS1 | interferon regulatory factor 6 |
C00002903
|
| 10379 | IRF9, IRF-9, ISGF3, ISGF3G, p48 | interferon regulatory factor 9 |
C00002903
|
| 3667 | IRS1, HIRS-1 | insulin receptor substrate 1 |
C00002903
|
| 3672 | ITGA1, CD49a, VLA1 | integrin, alpha 1 |
C00002903
|
| 3673 | ITGA2, BR, CD49B, GPIa, HPA-5, VLA-2, VLAA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
C00002903
|
| 3678 | ITGA5, CD49e, FNRA, VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
C00002903
|
| 147 | ADRA1B, ADRA1, ALPHA1BAR | adrenoceptor alpha 1B |
C00000958
|
| 3684 | ITGAM, CD11B, CR3A, MAC-1, MAC1A, MO1A, SLEB6 | integrin, alpha M (complement component 3 receptor 3 subunit) |
C00002903
|
| 3688 | ITGB1, CD29, FNRB, GPIIA, MDF2, MSK12, VLA-BETA, VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
C00002903
|
| 100380740 |
C00000958
|
||
| 3690 | ITGB3, BDPLT16, BDPLT2, CD61, GP3A, GPIIIa, GT | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
C00002903
|
| 3708 | ITPR1, INSP3R1, IP3R, IP3R1, SCA15, SCA16, SCA29 | inositol 1,4,5-trisphosphate receptor, type 1 |
C00002903
|
| 3716 | JAK1, JAK1A, JAK1B, JTK3 | Janus kinase 1 (EC:2.7.10.2) |
C00002903
|
| 3726 | JUNB, AP-1 | jun B proto-oncogene |
C00002903
|
| 3727 | JUND, AP-1 | jun D proto-oncogene |
C00002903
|
| 3728 | JUP, ARVD12, CTNNG, DP3, DPIII, PDGB, PKGB | junction plakoglobin |
C00002903
|
| 222658 | KCTD20, C6orf69, dJ108K11.3 | potassium channel tetramerization domain containing 20 |
C00002903
|
| 5927 | KDM5A, RBBP-2, RBBP2, RBP2, JARID1A | lysine (K)-specific demethylase 5A |
C00002903
|
| 10765 | KDM5B, CT31, JARID1B, PLU-1, PLU1, PUT1, RBBP2H1A | lysine (K)-specific demethylase 5B |
C00002903
|
| 9817 | KEAP1, INrf2, KLHL19 | kelch-like ECH-associated protein 1 |
C00002903
|
| 10657 | KHDRBS1, Sam68, p62, p68 | KH domain containing, RNA binding, signal transduction associated 1 |
C00002903
|
| 9768 | NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | KIAA0101 |
C00002903
|
| 57221 | A7322, ARFGEF3, BIG3, C6orf92, PPP1R33, dJ171N11.1 | KIAA1244 |
C00002903
|
| 57535 | EIG121 | KIAA1324 |
C00002903
|
| 57650 | CIP2A, p90 | KIAA1524 |
C00002903
|
| 3832 | KIF11, EG5, HKSP, KNSL1, MCLMR, TRIP5 | kinesin family member 11 |
C00002903
|
| 10112 | KIF20A, MKLP2, RAB6KIFL | kinesin family member 20A |
C00002903
|
| 9493 | KIF23, CHO1, KNSL5, MKLP-1, MKLP1 | kinesin family member 23 |
C00002903
|
| 11004 | KIF2C, KNSL6, MCAK | kinesin family member 2C |
C00002903
|
| 8462 | KLF11, FKLF, FKLF1, MODY7, TIEG2, Tieg3 | Kruppel-like factor 11 |
C00002903
|
| 10365 | KLF2, LKLF | Kruppel-like factor 2 |
C00002903
|
| 687 | KLF9, BTEB, BTEB1 | Kruppel-like factor 9 |
C00002903
|
| 3817 | KLK2, KLK2A2, hGK-1, hK2 | kallikrein-related peptidase 2 (EC:3.4.21.35) |
C00002903
|
| 354 | KLK3, APS, KLK2A1, PSA, hK3 | kallikrein-related peptidase 3 (EC:3.4.21.77) |
C00002903
|
| 346689 | KLRG2, CLEC15B | killer cell lectin-like receptor subfamily G, member 2 |
C00002903
|
| 22914 | KLRK1, CD314, D12S2489E, KLR, NKG2-D, NKG2D | killer cell lectin-like receptor subfamily K, member 1 |
C00002903
|
| 779317 |
C00002903
|
||
| 3838 | KPNA2, IPOA1, QIP2, RCH1, SRP1alpha | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
C00002903
|
| 3845 | KRAS, C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RASK2 | Kirsten rat sarcoma viral oncogene homolog |
C00002903
|
| 54474 | KRT20, CD20, CK-20, CK20, K20, KRT21 | keratin 20 |
C00002903
|
| 3852 | KRT5, CK5, DDD, DDD1, EBS2, K5, KRT5A | keratin 5 |
C00002903
|
| 3897 | L1CAM, CAML1, CD171, HSAS, HSAS1, MASA, MIC5, N-CAM-L1, N-CAML1, NCAM-L1, S10, SPG1 | L1 cell adhesion molecule |
C00002903
|
| 79944 | L2HGDH, C14orf160 | L-2-hydroxyglutarate dehydrogenase (EC:1.1.99.2) |
C00002903
|
| 3930 | LBR, DHCR14B, LMN2R, PHA, TDRD18 | lamin B receptor |
C00002903
|
| 51176 | LEF1, LEF-1, TCF10, TCF1ALPHA, TCF7L3 | lymphoid enhancer-binding factor 1 |
C00002903
|
| 7044 | LEFTY2, EBAF, LEFTA, LEFTYA, TGFB4 | left-right determination factor 2 |
C00002903
|
| 3956 | LGALS1, GAL1, GBP | lectin, galactoside-binding, soluble, 1 |
C00002903
|
| 9355 | LHX2, LH2, hLhx2 | LIM homeobox 2 |
C00002903
|
| 3976 | LIF, CDF, DIA, HILDA, MLPLI | leukemia inhibitory factor |
C00002903
|
| 3978 | LIG1 | ligase I, DNA, ATP-dependent (EC:6.5.1.1) |
C00002903
|
| 8825 | LIN7A, LIN-7A, LIN7, MALS-1, TIP-33, VELI1 | lin-7 homolog A (C. elegans) |
C00002903
|
| 145978 | LINC00052, NCRNA00052, TMEM83 | long intergenic non-protein coding RNA 52 |
C00002903
|
| 9516 | LITAF, PIG7, SIMPLE, TP53I7 | lipopolysaccharide-induced TNF factor |
C00002903
|
| 29995 | LMCD1 | LIM and cysteine-rich domains 1 |
C00002903
|
| 4000 | LMNA, CDCD1, CDDC, CMD1A, CMT2B1, EMD2, FPL, FPLD, FPLD2, HGPS, IDC, LDP1, LFP, LGMD1B, LMN1, LMNC, LMNL1, PRO1 | lamin A/C |
C00002903
|
| 84823 | LMNB2, LAMB2, LMN2 | lamin B2 |
C00002903
|
| 4023 | LPL, HDLCQ11, LIPD | lipoprotein lipase (EC:3.1.1.34) |
C00002903
|
| 64748 | LPPR2, PRG4 | lipid phosphate phosphatase-related protein type 2 (EC:3.1.3.4) |
C00002903
|
| 26018 | LRIG1, LIG-1, LIG1 | leucine-rich repeats and immunoglobulin-like domains 1 |
C00002903
|
| 7804 | LRP8, APOER2, HSZ75190, LRP-8, MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
C00002903
|
| 4085 | MAD2L1, HSMAD2, MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) |
C00002903
|
| 9587 | MAD2L1BP, CMT2, RP1-261G23.6 | MAD2L1 binding protein |
C00002903
|
| 8567 | MADD, DENN, IG20, RAB3GEP | MAP-kinase activating death domain |
C00002903
|
| 4094 | MAF, CCA4, c-MAF | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
C00002903
|
| 55110 | MAGOHB, MGN2, mago, magoh | mago-nashi homolog B (Drosophila) |
C00002903
|
| 54682 | MANSC1, 9130403P13Rik, LOH12CR3 | MANSC domain containing 1 (EC:3.2.1.14) |
C00002903
|
| 4130 | MAP1A, MAP1L, MTAP1A | microtubule-associated protein 1A |
C00002903
|
| 4131 | MAP1B, FUTSCH, MAP5 | microtubule-associated protein 1B |
C00002903
|
| 23536 | ADAT1, HADAT1 | adenosine deaminase, tRNA-specific 1 (EC:3.5.4.34) |
C00000958
|
| 5607 | MAP2K5, HsT17454, MAPKK5, MEK5, PRKMK5 | mitogen-activated protein kinase kinase 5 (EC:2.7.12.2) |
C00002903
|
| 4217 | MAP3K5, ASK1, MAPKKK5, MEKK5 | mitogen-activated protein kinase kinase kinase 5 (EC:2.7.11.25) |
C00002903
|
| 4134 | MAP4 | microtubule-associated protein 4 |
C00002903
|
| 5603 | MAPK13, MAPK_13, MAPK-13, PRKM13, SAPK4, p38delta | mitogen-activated protein kinase 13 (EC:2.7.11.24) |
C00002903
|
| 4137 | MAPT, DDPAC, FTDP-17, MAPTL, MSTD, MTBT1, MTBT2, PPND, TAU | microtubule-associated protein tau |
C00002903
|
| 84930 | MASTL, GREATWALL, GW, GWL, MAST-L, RP11-85G18.2, THC2, hGWL | microtubule associated serine/threonine kinase-like (EC:2.7.11.1) |
C00002903
|
| 4148 | MATN3, DIPOA, EDM5, HOA, OADIP, OS2 | matrilin 3 |
C00002903
|
| 8932 | MBD2, DMTase, NY-CO-41 | methyl-CpG binding domain protein 2 |
C00002903
|
| 4162 | MCAM, CD146, MUC18 | melanoma cell adhesion molecule |
C00002903
|
| 55388 | MCM10, CNA43, DNA43 | minichromosome maintenance complex component 10 |
C00002903
|
| 4171 | MCM2, BM28, CCNL1, CDCL1, D3S3194, MITOTIN, cdc19 | minichromosome maintenance complex component 2 (EC:3.6.4.12) |
C00002903
|
| 4172 | MCM3, HCC5, P1-MCM3, P1.h, RLFB | minichromosome maintenance complex component 3 (EC:3.6.4.12) |
C00002903
|
| 4173 | MCM4, CDC21, CDC54, NKCD, NKGCD, P1-CDC21, hCdc21 | minichromosome maintenance complex component 4 (EC:3.6.4.12) |
C00002903
|
| 4174 | MCM5, CDC46, P1-CDC46 | minichromosome maintenance complex component 5 (EC:3.6.4.12) |
C00002903
|
| 4175 | MCM6, MCG40308, Mis5, P105MCM | minichromosome maintenance complex component 6 (EC:3.6.4.12) |
C00002903
|
| 4176 | MCM7, CDC47, MCM2, P1.1-MCM3, P1CDC47, P85MCM, PNAS146 | minichromosome maintenance complex component 7 (EC:3.6.4.12) |
C00002903
|
| 84515 | MCM8, C20orf154, dJ967N21.5 | minichromosome maintenance complex component 8 (EC:3.6.4.12) |
C00002903
|
| 4193 | MDM2, ACTFS, HDMX, hdm2 | MDM2 oncogene, E3 ubiquitin protein ligase |
C00002903
|
| 64769 | MEAF6, C1orf149, CENP-28, EAF6, NY-SAR-91 | MYST/Esa1-associated factor 6 |
C00002903
|
| 4208 | MEF2C, C5DELq14.3, DEL5q14.3 | myocyte enhancer factor 2C |
C00002903
|
| 25840 | METTL7A, AAM-B | methyltransferase like 7A |
C00002903
|
| 9927 | MFN2, CMT2A, CMT2A2, CPRP1, HSG, MARF | mitofusin 2 |
C00002903
|
| 11343 | MGLL, HU-K5, HUK5, MAGL, MGL | monoglyceride lipase (EC:3.1.1.23) |
C00002903
|
| 4256 | MGP, MGLAP, NTI | matrix Gla protein |
C00002903
|
| 4277 | MICB, PERB11.2 | MHC class I polypeptide-related sequence B |
C00002903
|
| 406892 | MIR100, MIRN100, miR-100 | microRNA 100 |
C00002903
|
| 406895 | MIR103A1, MIR103-1, MIRN103-1 | microRNA 103a-1 |
C00002903
|
| 406896 | MIR103A2, MIR103-2, MIRN103-2 | microRNA 103a-2 |
C00002903
|
| 406937 | MIR145, MIRN145, miR-145, miRNA145 | microRNA 145 |
C00002903
|
| 406938 | MIR146A, MIRN146, MIRN146A, miR-146a, miRNA146A | microRNA 146a |
C00002903
|
| 574447 | MIR146B, MIRN146B, miRNA146B | microRNA 146b |
C00002903
|
| 406950 | MIR16-1, MIRN16-1, miRNA16-1 | microRNA 16-1 |
C00002903
|
| 406952 | MIR17, MIR17-5p, MIR91, MIRN17, MIRN91, hsa-mir-17, miR-17, miR17-3p, miRNA17, miRNA91 | microRNA 17 |
C00002903
|
| 406954 | MIR181A2, MIRN181A, MIRN181A2, hsa-mir-181a-2 | microRNA 181a-2 |
C00002903
|
| 406957 | MIR181C, MIRN181C, mir-181c | microRNA 181c |
C00002903
|
| 406972 | MIR196A1, MIRN196-1, MIRN196A1 | microRNA 196a-1 |
C00002903
|
| 406979 | MIR19A, MIRN19A, hsa-mir-19a, miR-19a, miRNA19A | microRNA 19a |
C00002903
|
| 406985 | MIR200C, MIRN200C | microRNA 200c |
C00002903
|
| 574448 | MIR202, MIRN202, hsa-mir-202 | microRNA 202 |
C00002903
|
| 406986 | MIR203, MIRN203, miR-203, miRNA203 | microRNA 203 |
C00002903
|
| 406988 | MIR205, MIRN205 | microRNA 205 |
C00002903
|
| 406989 | MIR206, MIRN206, miRNA206 | microRNA 206 |
C00002903
|
| 574032 | MIR20B, MIRN20B, hsa-mir-20b | microRNA 20b |
C00002903
|
| 406991 | MIR21, MIRN21, hsa-mir-21, miR-21, miRNA21 | microRNA 21 |
C00002903
|
| 407010 | MIR23A, MIRN23A, hsa-mir-23a, miRNA23A | microRNA 23a |
C00002903
|
| 407011 | MIR23B, MIRN23B, hsa-mir-23b, miRNA23B | microRNA 23b |
C00002903
|
| 407014 | MIR25, MIRN25, hsa-mir-25, miR-25 | microRNA 25 |
C00002903
|
| 407015 | MIR26A1, MIR26A, MIRN26A1 | microRNA 26a-1 |
C00002903
|
| 407021 | MIR29A, MIRN29, MIRN29A, hsa-mir-29, hsa-mir-29a, miRNA29A | microRNA 29a |
C00002903
|
| 407026 | MIR29C, MIRN29C, miRNA29C | microRNA 29c |
C00002903
|
| 442894 | MIR302B, MIRN302B | microRNA 302b |
C00002903
|
| 442896 | MIR302D, MIRN302D | microRNA 302d |
C00002903
|
| 407029 | MIR30A, MIRN30A | microRNA 30a |
C00002903
|
| 407031 | MIR30C1, MIRN30C1 | microRNA 30c-1 |
C00002903
|
| 407033 | MIR30D, MIRN30D | microRNA 30d |
C00002903
|
| 407034 | MIR30E, MIRN30E | microRNA 30e |
C00002903
|
| 442897 | MIR323A, MIR323, MIRN323, hsa-mir-323, hsa-mir-323a | microRNA 323a |
C00002903
|
| 442908 | MIR340, MIRN340, hsa-mir-340 | microRNA 340 |
C00002903
|
| 442909 | MIR342, MIRN342, hsa-mir-342 | microRNA 342 |
C00002903
|
| 442910 | MIR345, MIRN345, hsa-mir-345 | microRNA 345 |
C00002903
|
| 407040 | MIR34A, MIRN34A, miRNA34A, mir-34 | microRNA 34a |
C00002903
|
| 574030 | MIR362, MIRN362, hsa-mir-362 | microRNA 362 |
C00002903
|
| 574031 | MIR363, MIR-363, MIRN363, hsa-mir-363 | microRNA 363 |
C00002903
|
| 494324 | MIR375, MIRN375, hsa-mir-375, miRNA375 | microRNA 375 |
C00002903
|
| 494336 | MIR424, MIR322, MIRN424, hsa-mir-424, miRNA424 | microRNA 424 |
C00002903
|
| 574452 | MIR494, MIRN494, hsa-mir-494 | microRNA 494 |
C00002903
|
| 574456 | MIR497, MIRN497, hsa-mir-497 | microRNA 497 |
C00002903
|
| 574506 | MIR503, MIRN503, hsa-mir-503 | microRNA 503 |
C00002903
|
| 574507 | MIR504, MIRN504, hsa-mir-504 | microRNA 504 |
C00002903
|
| 574512 | MIR507, MIRN507, hsa-mir-507 | microRNA 507 |
C00002903
|
| 574479 | MIR517A, MIRN517A | microRNA 517a |
C00002903
|
| 574474 | MIR518B, MIRN518B | microRNA 518b |
C00002903
|
| 574477 | MIR518C, MIRN518C | microRNA 518c |
C00002903
|
| 574469 | MIR519B, MIRN519B | microRNA 519b |
C00002903
|
| 574470 | MIR525, MIRN525, hsa-mir-525 | microRNA 525 |
C00002903
|
| 693154 | MIR569, MIRN569, hsa-mir-569 | microRNA 569 |
C00002903
|
| 693155 | MIR570, MIRN570, hsa-mir-570 | microRNA 570 |
C00002903
|
| 693156 | MIR571, MIRN571, hsa-mir-571 | microRNA 571 |
C00002903
|
| 693157 | MIR572, MIRN572, hsa-mir-572 | microRNA 572 |
C00002903
|
| 693159 | MIR574, MIRN574, hsa-mir-574 | microRNA 574 |
C00002903
|
| 693169 | MIR584, MIRN584, hsa-mir-584 | microRNA 584 |
C00002903
|
| 693186 | MIR601, MIRN601, hsa-mir-601 | microRNA 601 |
C00002903
|
| 693200 | MIR615, MIRN615, hsa-mir-615 | microRNA 615 |
C00002903
|
| 693207 | MIR622, MIRN622, hsa-mir-622 | microRNA 622 |
C00002903
|
| 693214 | MIR629, MIRN629, hsa-mir-629 | microRNA 629 |
C00002903
|
| 693216 | MIR631, MIRN631, hsa-mir-631 | microRNA 631 |
C00002903
|
| 693217 | MIR632, MIRN632, hsa-mir-632 | microRNA 632 |
C00002903
|
| 693223 | MIR638, MIRN638, hsa-mir-638 | microRNA 638 |
C00002903
|
| 693224 | MIR639, MIRN639, hsa-mir-639 | microRNA 639 |
C00002903
|
| 693230 | MIR645, MIRN645, hsa-mir-645 | microRNA 645 |
C00002903
|
| 724027 | MIR657, MIRN657, hsa-mir-657 | microRNA 657 |
C00002903
|
| 724029 | MIR659, MIRN659, hsa-mir-659 | microRNA 659 |
C00002903
|
| 724033 | MIR663A, MIR663, MIRN663, hsa-mir-663, hsa-mir-663a | microRNA 663a |
C00002903
|
| 407049 | MIR92A2, MIRN92-2, MIRN92A-2, MIRN92A2 | microRNA 92a-2 |
C00002903
|
| 693235 | MIR92B, MIRN92B, hsa-mir-92b | microRNA 92b |
C00002903
|
| 407053 | MIR96, DFNA50, MIRN96, hsa-mir-96, miR-96, miRNA96 | microRNA 96 |
C00002903
|
| 406885 | MIRLET7C, LET7C, MIRNLET7C, hsa-let-7c, let-7c | microRNA let-7c |
C00002903
|
| 55320 | MIS18BP1, C14orf106, HSA242977, KNL2, M18BP1 | MIS18 binding protein 1 |
C00002903
|
| 4288 | MKI67, KIA, MIB-1 | marker of proliferation Ki-67 |
C00002903
|
| 79682 | CENPU, CENP50, CENPU50, KLIP1, MLF1IP, PBIP1 | centromere protein U |
C00002903
|
| 23531 | MMD, MMA, MMD1, PAQR11 | monocyte to macrophage differentiation-associated |
C00002903
|
| 4311 | MME, CALLA, CD10, NEP, SFE | membrane metallo-endopeptidase (EC:3.4.24.11) |
C00002903
|
| 4322 | MMP13, CLG3, MANDP1 | matrix metallopeptidase 13 (collagenase 3) (EC:3.4.24.-) |
C00002903
|
| 4325 | MMP16, C8orf57, MMP-X2, MT-MMP2, MT-MMP3, MT3-MMP | matrix metallopeptidase 16 (membrane-inserted) (EC:3.4.24.-) |
C00002903
|
| 104 | ADARB1, ADAR2, DRABA2, DRADA2, RED1 | adenosine deaminase, RNA-specific, B1 (EC:3.5.4.37) |
C00000958
|
| 4314 | MMP3, CHDS6, MMP-3, SL-1, STMY, STMY1, STR1 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) (EC:3.4.24.17) |
C00002903
|
| 574406 | ADAMTSL4-AS1, C1orf138 | ADAMTSL4 antisense RNA 1 |
C00000958
|
| 744 | MPPED2, 239FB, C11orf8 | metallophosphoesterase domain containing 2 |
C00002903
|
| 23164 | MPRIP, M-RIP, MRIP, RHOIP3, RIP3, p116Rip | myosin phosphatase Rho interacting protein |
C00002903
|
| 64968 | MRPS6, C21orf101, MRP-S6, RPMS6, S6mt | mitochondrial ribosomal protein S6 |
C00002903
|
| 2956 | MSH6, GTBP, GTMBP, HNPCC5, HSAP, p160 | mutS homolog 6 |
C00002903
|
| 6307 | MSMO1, DESP4, ERG25, SC4MOL | methylsterol monooxygenase 1 (EC:1.14.13.72) |
C00002903
|
| 4481 | MSR1, CD204, SCARA1, SR-A, SRA, phSR1, phSR2 | macrophage scavenger receptor 1 |
C00002903
|
| 9112 | MTA1 | metastasis associated 1 |
C00002903
|
| 4520 | MTF1, MTF-1, ZRF | metal-regulatory transcription factor 1 |
C00002903
|
| 113115 | MTFR2, DUFD1, FAM54A | mitochondrial fission regulator 2 |
C00002903
|
| 4522 | MTHFD1, MTHFC, MTHFD | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase (EC:1.5.1.5 6.3.4.3 3.5.4.9) |
C00002903
|
| 4543 | MTNR1A, MEL-1A-R, MT1 | melatonin receptor 1A |
C00002903
|
| 4544 | MTNR1B, FGQTL2, MEL-1B-R, MT2 | melatonin receptor 1B |
C00002903
|
| 2475 | MTOR, FRAP, FRAP1, FRAP2, RAFT1, RAPT1 | mechanistic target of rapamycin (serine/threonine kinase) (EC:2.7.11.1) |
C00002903
|
| 136319 | MTPN, GCDP, V-1 | myotrophin |
C00002903
|
| 4582 | MUC1, CA_15-3, CD227, EMA, H23AG, KL-6, MAM6, MCKD1, MUC-1, MUC-1/SEC, MUC-1/X, MUC1/ZD, PEM, PEMT, PUM | mucin 1, cell surface associated |
C00002903
|
| 4586 | MUC5AC, MUC5, TBM, leB | mucin 5AC, oligomeric mucus/gel-forming |
C00002903
|
| 4594 | MUT, MCM | methylmalonyl CoA mutase (EC:5.4.99.2) |
C00002903
|
| 10514 | MYBBP1A, P160, PAP2 | MYB binding protein (P160) 1a |
C00002903
|
| 4605 | MYBL2, B-MYB, BMYB | v-myb avian myeloblastosis viral oncogene homolog-like 2 |
C00002903
|
| 79784 | MYH14, DFNA4, DFNA4A, MHC16, MYH17, NMHC_II-C, NMHC-II-C, PNMHH, myosin | myosin, heavy chain 14, non-muscle (EC:3.6.4.1) |
C00002903
|
| 399687 | MYO18A, MYSPDZ, SPR210 | myosin XVIIIA |
C00002903
|
| 4644 | MYO5A, GS1, MYH12, MYO5, MYR12 | myosin VA (heavy chain 12, myoxin) |
C00002903
|
| 440145 | MZT1, C13orf37, MOZART1 | mitotic spindle organizing protein 1 |
C00002903
|
| 9683 | N4BP1 | NEDD4 binding protein 1 |
C00002903
|
| 4664 | NAB1 | NGFI-A binding protein 1 (EGR1 binding protein 1) |
C00002903
|
| 64859 | NABP1, OBFC2A, SOSS-B2, SSB2 | nucleic acid binding protein 1 |
C00002903
|
| 4671 | NAIP, BIRC1, NLRB1, psiNAIP | NLR family, apoptosis inhibitory protein |
C00002903
|
| 79923 | NANOG | Nanog homeobox |
C00002903
|
| 340719 | NANOS1, NOS1, SPGF12 | nanos homolog 1 (Drosophila) |
C00002903
|
| 4678 | NASP, FLB7527, PRO1999 | nuclear autoantigenic sperm protein (histone-binding) |
C00002903
|
| 26960 | NBEA, BCL8B, LYST2 | neurobeachin |
C00002903
|
| 64151 | NCAPG, CAPG, CHCG, NY-MEL-3 | non-SMC condensin I complex, subunit G |
C00002903
|
| 54892 | NCAPG2, CAP-G2, CAPG2, LUZP5, MTB, hCAP-G2 | non-SMC condensin II complex, subunit G2 |
C00002903
|
| 11096 | ADAMTS5, ADAM-TS_11, ADAM-TS_5, ADAM-TS5, ADAMTS-11, ADAMTS-5, ADAMTS11, ADMP-2 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
C00000958
|
| 8202 | NCOA3, ACTR, AIB-1, AIB1, CAGH16, CTG26, KAT13B, RAC3, SRC-3, SRC3, TNRC14, TNRC16, TRAM-1, bHLHe42, pCIP | nuclear receptor coactivator 3 (EC:2.3.1.48) |
C00002903
|
| 8031 | NCOA4, ARA70, ELE1, PTC3, RFG | nuclear receptor coactivator 4 |
C00002903
|
| 55706 | NDC1, NET3, TMEM48 | NDC1 transmembrane nucleoporin |
C00002903
|
| 54820 | NDE1, HOM-TES-87, LIS4, NDE, NUDE, NUDE1 | nudE neurodevelopment protein 1 |
C00002903
|
| 10529 | NEBL, LASP2, LNEBL | nebulette |
C00002903
|
| 55707 | NECAP2 | NECAP endocytosis associated 2 |
C00002903
|
| 252969 | NEIL2, NEH2, NEI2 | nei endonuclease VIII-like 2 (E. coli) (EC:4.2.99.18) |
C00002903
|
| 4751 | NEK2, HsPK21, NEK2A, NLK1 | NIMA-related kinase 2 (EC:2.7.11.1) |
C00002903
|
| 4753 | NELL2, NRP2 | NEL-like 2 (chicken) |
C00002903
|
| 10763 | NES | nestin |
C00002903
|
| 4772 | NFATC1, NF-ATC, NFAT2, NFATc | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
C00002903
|
| 4784 | NFIX, MRSHSS, NF1A, SOTOS2 | nuclear factor I/X (CCAAT-binding transcription factor) |
C00002903
|
| 4791 | NFKB2, H2TF1, LYT-10, LYT10, NF-kB2, p105, p52 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
C00002903
|
| 22981 | NINL, NLP, dJ691N24.1 | ninein-like |
C00002903
|
| 79570 | NKAIN1, FAM77C | Na+/K+ transporting ATPase interacting 1 |
C00002903
|
| 4824 | NKX3-1, BAPX2, NKX3, NKX3.1, NKX3A | NK3 homeobox 1 |
C00002903
|
| 10874 | NMU | neuromedin U |
C00002903
|
| 10528 | NOP56, NOL5A, SCA36 | NOP56 ribonucleoprotein |
C00002903
|
| 4842 | NOS1, IHPS1, N-NOS, NC-NOS, NOS, bNOS, nNOS | nitric oxide synthase 1 (neuronal) (EC:1.14.13.39) |
C00002903
|
| 170689 | ADAMTS15 | ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
C00000958
|
| 404036 |
C00002903
|
||
| 81792 | ADAMTS12, PRO4389 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
C00000958
|
| 4853 | NOTCH2, AGS2, HJCYS, hN2 | notch 2 |
C00002903
|
| 50507 | NOX4, KOX, KOX-1, RENOX | NADPH oxidase 4 (EC:1.6.3.-) |
C00002903
|
| 4862 | NPAS2, MOP4, PASD4, bHLHe9 | neuronal PAS domain protein 2 |
C00002903
|
| 64067 | NPAS3, MOP6, PASD6, bHLHe12 | neuronal PAS domain protein 3 |
C00002903
|
| 4864 | NPC1, NPC | Niemann-Pick disease, type C1 |
C00002903
|
| 10577 | NPC2, EDDM1, HE1 | Niemann-Pick disease, type C2 |
C00002903
|
| 4880 | NPPC, CNP, CNP2 | natriuretic peptide C |
C00002903
|
| 4886 | NPY1R, NPY1-R, NPYR | neuropeptide Y receptor Y1 |
C00002903
|
| 1728 | NQO1, DHQU, DIA4, DTD, NMOR1, NMORI, QR1 | NAD(P)H dehydrogenase, quinone 1 (EC:1.6.5.2) |
C00002903
|
| 4835 | NQO2, DHQV, DIA6, NMOR2, QR2 | NAD(P)H dehydrogenase, quinone 2 (EC:1.10.99.2) |
C00002903
|
| 10062 | NR1H3, LXR-a, LXRA, RLD-1 | nuclear receptor subfamily 1, group H, member 3 |
C00002903
|
| 8748 | ADAM20 | ADAM metallopeptidase domain 20 |
C00000958
|
| 8728 | ADAM19, MADDAM, MLTNB | ADAM metallopeptidase domain 19 |
C00000958
|
| 7026 | NR2F2, ARP1, COUPTFB, COUPTFII, NF-E3, NR2F1, SVP40, TFCOUP2 | nuclear receptor subfamily 2, group F, member 2 |
C00002903
|
| 4899 | NRF1, ALPHA-PAL | nuclear respiratory factor 1 |
C00002903
|
| 11270 | NRM, NRM29 | nurim (nuclear envelope membrane protein) |
C00002903
|
| 59277 | NTN4, PRO3091 | netrin 4 |
C00002903
|
| 4521 | NUDT1, MTH1 | nudix (nucleoside diphosphate linked moiety X)-type motif 1 (EC:3.6.1.55 3.6.1.56) |
C00002903
|
| 131870 | NUDT16 | nudix (nucleoside diphosphate linked moiety X)-type motif 16 (EC:3.6.1.62) |
C00002903
|
| 9631 | NUP155, N155 | nucleoporin 155kDa |
C00002903
|
| 23225 | NUP210, GP210, POM210 | nucleoporin 210kDa |
C00002903
|
| 51203 | NUSAP1, ANKT, BM037, LNP, NUSAP, PRO0310p1, Q0310, SAPL | nucleolar and spindle associated protein 1 |
C00002903
|
| 100506658 | OCLN, BLCPMG | occludin (EC:2.1.1.67) |
C00002903
|
| 4968 | OGG1, HMMH, HOGG1, MUTM, OGH1 | 8-oxoguanine DNA glycosylase (EC:4.2.99.18) |
C00002903
|
| 10439 | OLFM1, AMY, NOE1, NOELIN1, OlfA | olfactomedin 1 |
C00002903
|
| 56944 | OLFML3, HNOEL-iso, OLF44 | olfactomedin-like 3 |
C00002903
|
| 4976 | OPA1, MGM1, NPG, NTG, largeG | optic atrophy 1 (autosomal dominant) (EC:3.6.5.5) |
C00002903
|
| 4998 | ORC1, HSORC1, ORC1L, PARC1 | origin recognition complex, subunit 1 |
C00002903
|
| 716175 |
C00002903
|
||
| 5019 | OXCT1, OXCT, SCOT | 3-oxoacid CoA transferase 1 (EC:2.8.3.5) |
C00002903
|
| 5021 | OXTR, OT-R | oxytocin receptor |
C00002903
|
| 10606 | PAICS, ADE2, ADE2H1, AIRC, PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase (EC:6.3.2.6 4.1.1.21) |
C00002903
|
| 5058 | PAK1, PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 (EC:2.7.11.1) |
C00002903
|
| 5069 | PAPPA, ASBABP2, DIPLA1, IGFBP-4ase, PAPA, PAPP-A, PAPPA1 | pregnancy-associated plasma protein A, pappalysin 1 (EC:3.4.24.79) |
C00002903
|
| 54852 | PAQR5, MPRG | progestin and adipoQ receptor family member V |
C00002903
|
| 10038 | PARP2, ADPRT2, ADPRTL2, ADPRTL3, ARTD2, PARP-2, pADPRT-2 | poly (ADP-ribose) polymerase 2 (EC:2.4.2.30) |
C00002903
|
| 55010 | PARPBP, AROM, C12orf48, PARI | PARP1 binding protein |
C00002903
|
| 55872 | PBK, CT84, Nori-3, SPK, TOPK | PDZ binding kinase (EC:2.7.12.2) |
C00002903
|
| 5087 | PBX1 | pre-B-cell leukemia homeobox 1 |
C00002903
|
| 5111 | PCNA | proliferating cell nuclear antigen |
C00002903
|
| 5121 | PCP4, PEP-19 | Purkinje cell protein 4 |
C00002903
|
| 5046 | PCSK6, PACE4, SPC4 | proprotein convertase subtilisin/kexin type 6 |
C00002903
|
| 255738 | PCSK9, FH3, HCHOLA3, LDLCQ1, NARC-1, NARC1, PC9 | proprotein convertase subtilisin/kexin type 9 |
C00002903
|
| 5134 | PDCD2, RP8, ZMYND7 | programmed cell death 2 |
C00002903
|
| 27250 | PDCD4, H731 | programmed cell death 4 (neoplastic transformation inhibitor) |
C00002903
|
| 10081 | PDCD7, ES18, HES18 | programmed cell death 7 |
C00002903
|
| 5138 | PDE2A, CGS-PDE, PDE2A1, PED2A4, cGSPDE | phosphodiesterase 2A, cGMP-stimulated (EC:3.1.4.17) |
C00002903
|
| 55860 | ACTR10, ACTR11, Arp11, HARP11 | actin-related protein 10 homolog (S. cerevisiae) |
C00000958
|
| 5155 | PDGFB, IBGC5, PDGF-2, PDGF2, SIS, SSV, c-sis | platelet-derived growth factor beta polypeptide |
C00002903
|
| 5160 | PDHA1, PDHA, PDHCE1A, PHE1A | pyruvate dehydrogenase (lipoamide) alpha 1 (EC:1.2.4.1) |
C00002903
|
| 9124 | PDLIM1, CLIM1, CLP-36, CLP36, hCLIM1 | PDZ and LIM domain 1 |
C00002903
|
| 5170 | PDPK1, PDK1, PDPK2, PRO0461 | 3-phosphoinositide dependent protein kinase-1 (EC:2.7.11.1) |
C00002903
|
| 23590 | PDSS1, COQ1, COQ10D2, DPS, RP13-16H11.3, SPS, TPRT, TPT, TPT_1, hDPS1 | prenyl (decaprenyl) diphosphate synthase, subunit 1 (EC:2.5.1.91) |
C00002903
|
| 3651 | PDX1, GSF, IDX-1, IPF1, IUF1, MODY4, PDX-1, STF-1 | pancreatic and duodenal homeobox 1 |
C00002903
|
| 5174 | PDZK1, CAP70, CLAMP, NHERF-3, NHERF3, PDZD1 | PDZ domain containing 1 |
C00002903
|
| 10158 | PDZK1IP1, DD96, MAP17, RP1-18D14.5, SPAP | PDZK1 interacting protein 1 |
C00002903
|
| 23527 | ACAP2, CENTB2, CNT-B2 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
C00000958
|
| 27043 | PELP1, MNAR, P160 | proline, glutamate and leucine rich protein 1 |
C00002903
|
| 64065 | PERP, KCP1, KRTCAP1, PIGPC1, RP3-496H19.1, THW, dJ496H19.1 | PERP, TP53 apoptosis effector |
C00002903
|
| 5223 | PGAM1, PGAM-B, PGAMA | phosphoglycerate mutase 1 (brain) (EC:3.1.3.13 5.4.2.4 5.4.2.11) |
C00002903
|
| 5230 | PGK1, MIG10, PGKA | phosphoglycerate kinase 1 (EC:2.7.2.3) |
C00002903
|
| 283209 | PGM2L1, BM32A | phosphoglucomutase 2-like 1 (EC:2.7.1.106) |
C00002903
|
| 90102 | PHLDB2, LL5b, LL5beta | pleckstrin homology-like domain, family B, member 2 |
C00002903
|
| 10745 | PHTF1, PHTF | putative homeodomain transcription factor 1 |
C00002903
|
| 57157 | PHTF2 | putative homeodomain transcription factor 2 |
C00002903
|
| 55300 | PI4K2B, PI4KIIB, PIK42B | phosphatidylinositol 4-kinase type 2 beta (EC:2.7.1.67) |
C00002903
|
| 10401 | PIAS3, ZMIZ5 | protein inhibitor of activated STAT, 3 |
C00002903
|
| 5290 | PIK3CA, CLOVE, CWS5, MCAP, MCM, MCMTC, PI3K, p110-alpha | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (EC:2.7.11.1 2.7.1.153) |
C00002903
|
| 5294 | PIK3CG, PI3CG, PI3K, PI3Kgamma, PIK3, p110gamma, p120-PI3K | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma (EC:2.7.11.1 2.7.1.153) |
C00002903
|
| 5295 | PIK3R1, AGM7, GRB1, p85, p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
C00002903
|
| 8503 | PIK3R3, p55, p55-GAMMA | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
C00002903
|
| 5570 | PKIB, PRKACN2 | protein kinase (cAMP-dependent, catalytic) inhibitor beta |
C00002903
|
| 5315 | PKM, CTHBP, OIP3, PK3, PKM2, TCB, THBP1 | pyruvate kinase, muscle (EC:2.7.1.40) |
C00002903
|
| 11187 | PKP3 | plakophilin 3 |
C00002903
|
| 8398 | PLA2G6, CaI-PLA2, GVI, INAD1, IPLA2-VIA, NBIA2, NBIA2A, NBIA2B, PARK14, PLA2, PNPLA9, iPLA2, iPLA2beta | phospholipase A2, group VI (cytosolic, calcium-independent) (EC:3.1.1.4) |
C00002903
|
| 5329 | PLAUR, CD87, U-PAR, UPAR, URKR | plasminogen activator, urokinase receptor |
C00002903
|
| 5331 | PLCB3 | phospholipase C, beta 3 (phosphatidylinositol-specific) (EC:3.1.4.11) |
C00002903
|
| 5333 | PLCD1, NDNC3, PLC-III | phospholipase C, delta 1 (EC:3.1.4.11) |
C00002903
|
| 5335 | PLCG1, NCKAP3, PLC-II, PLC1, PLC148, PLCgamma1 | phospholipase C, gamma 1 (EC:3.1.4.11) |
C00002903
|
| 5341 | PLEK, P47 | pleckstrin |
C00002903
|
| 79666 | PLEKHF2, EAPF, PHAFIN2, ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
C00002903
|
| 5347 | PLK1, PLK, STPK13 | polo-like kinase 1 (EC:2.7.11.21) |
C00002903
|
| 10733 | PLK4, SAK, STK18 | polo-like kinase 4 (EC:2.7.11.21) |
C00002903
|
| 5352 | PLOD2, LH2, TLH | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (EC:1.14.11.4) |
C00002903
|
| 5355 | PLP2, A4, A4LSB | proteolipid protein 2 (colonic epithelium-enriched) |
C00002903
|
| 84898 | PLXDC2, TEM7R | plexin domain containing 2 |
C00002903
|
| 56937 | PMEPA1, STAG1, TMEPAI | prostate transmembrane protein, androgen induced 1 |
C00002903
|
| 5371 | PML, MYL, PP8675, RNF71, TRIM19 | promyelocytic leukemia |
C00002903
|
| 150379 | PNPLA5, GS2L, dJ388M5, dJ388M5.4 | patatin-like phospholipase domain containing 5 |
C00002903
|
| 10957 | PNRC1, B4-2, PNAS-145, PROL2, PRR2 | proline-rich nuclear receptor coactivator 1 |
C00002903
|
| 5422 | POLA1, POLA, p180 | polymerase (DNA directed), alpha 1, catalytic subunit (EC:2.7.7.7) |
C00002903
|
| 5424 | POLD1, CDC2, CRCS10, MDPL, POLD | polymerase (DNA directed), delta 1, catalytic subunit (EC:2.7.7.7) |
C00002903
|
| 5425 | POLD2 | polymerase (DNA directed), delta 2, accessory subunit (EC:2.7.7.7) |
C00002903
|
| 10714 | POLD3, P66, P68 | polymerase (DNA-directed), delta 3, accessory subunit |
C00002903
|
| 5444 | PON1, ESA, MVCD5, PON | paraoxonase 1 (EC:3.1.1.2 3.1.8.1 3.1.1.81) |
C00002903
|
| 5460 | POU5F1, OCT3, OCT4, OTF-3, OTF3, OTF4, Oct-3, Oct-4 | POU class 5 homeobox 1 |
C00002903
|
| 8612 | PPAP2C, LPP2, PAP-2c, PAP2-g | phosphatidic acid phosphatase type 2C (EC:3.1.3.4) |
C00002903
|
| 5465 | PPARA, NR1C1, PPAR, PPARalpha, hPPAR | peroxisome proliferator-activated receptor alpha |
C00002903
|
| 5467 | PPARD, FAAR, NR1C2, NUC1, NUCI, NUCII, PPARB | peroxisome proliferator-activated receptor delta |
C00002903
|
| 10891 | PPARGC1A, LEM6, PGC-1(alpha), PGC-1v, PGC1, PGC1A, PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
C00002903
|
| 10105 | PPIF, CYP3, CyP-M, Cyp-D, CypD | peptidylprolyl isomerase F (EC:5.2.1.8) |
C00002903
|
| 5493 | PPL | periplakin |
C00002903
|
| 22843 | PPM1E, CaMKP-N, POPX1, PP2CH, caMKN | protein phosphatase, Mg2+/Mn2+ dependent, 1E (EC:3.1.3.16) |
C00002903
|
| 5504 | PPP1R2, IPP-2, IPP2 | protein phosphatase 1, regulatory (inhibitor) subunit 2 (EC:3.1.3.16) |
C00002903
|
| 5525 | PPP2R5A, B56A, PR61A | protein phosphatase 2, regulatory subunit B', alpha |
C00002903
|
| 7799 | PRDM2, HUMHOXY1, KMT8, MTB-ZF, RIZ, RIZ1, RIZ2 | PR domain containing 2, with ZNF domain (EC:2.1.1.43) |
C00002903
|
| 5551 | PRF1, FLH2, HPLH2, P1, PFN1, PFP | perforin 1 (pore forming protein) |
C00002903
|
| 5562 | PRKAA1, AMPK, AMPKa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit (EC:2.7.11.1 2.7.11.26 2.7.11.27 2.7.11.31) |
C00002903
|
| 5563 | PRKAA2, AMPK, AMPK2, AMPKa2, PRKAA | protein kinase, AMP-activated, alpha 2 catalytic subunit (EC:2.7.11.1 2.7.11.27 2.7.11.31) |
C00002903
|
| 5576 | PRKAR2A, PKR2, PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha (EC:2.7.11.1) |
C00002903
|
| 34 | ACADM, ACAD1, MCAD, MCADH | acyl-CoA dehydrogenase, C-4 to C-12 straight chain (EC:1.3.8.7) |
C00000958
|
| 5579 | PRKCB, PKC-beta, PKCB, PRKCB1, PRKCB2 | protein kinase C, beta (EC:2.7.11.13) |
C00002903
|
| 5584 | PRKCI, DXS1179E, PKCI, nPKC-iota | protein kinase C, iota (EC:2.7.11.13) |
C00002903
|
| 1601 | DAB2, DOC-2, DOC2 | Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
C00000958
|
| 150696 | PROM2, PROML2 | prominin 2 |
C00002903
|
| 5627 | PROS1, PROS, PS21, PS22, PS23, PS24, PS25, PSA, THPH5, THPH6 | protein S (alpha) |
C00002903
|
| 9128 | PRPF4, HPRP4, HPRP4P, PRP4, Prp4p, SNRNP60 | pre-mRNA processing factor 4 |
C00002903
|
| 5634 | PRPS2, PRSII | phosphoribosyl pyrophosphate synthetase 2 (EC:2.7.6.1) |
C00002903
|
| 29 | ABR, MDB | active BCR-related |
C00000958
|
| 5652 | PRSS8, CAP1, PROSTASIN | protease, serine, 8 |
C00002903
|
| 5664 | PSEN2, AD3L, AD4, CMD1V, PS2, STM2 | presenilin 2 (Alzheimer disease 4) |
C00002903
|
| 5693 | PSMB5, LMPX, MB1, X | proteasome (prosome, macropain) subunit, beta type, 5 (EC:3.4.25.1) |
C00002903
|
| 5702 | PSMC3, TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
C00002903
|
| 29893 | PSMC3IP, GT198, HOP2, HUMGT198A, ODG3, TBPIP | PSMC3 interacting protein |
C00002903
|
| 84262 | PSMG3, C7orf48, PAC3 | proteasome (prosome, macropain) assembly chaperone 3 |
C00002903
|
| 5727 | PTCH1, BCNS, HPE7, NBCCS, PTC, PTC1, PTCH, PTCH11 | patched 1 |
C00002903
|
| 5728 | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1 | phosphatase and tensin homolog (EC:3.1.3.67 3.1.3.16 3.1.3.48) |
C00002903
|
| 9536 | PTGES, MGST-IV, MGST1-L1, MGST1L1, MPGES, PGES, PIG12, PP102, PP1294, TP53I12, mPGES-1 | prostaglandin E synthase (EC:5.3.99.3) |
C00002903
|
| 80142 | PTGES2, C9orf15, GBF-1, GBF1, PGES2, mPGES-2 | prostaglandin E synthase 2 (EC:5.3.99.3) |
C00002903
|
| 11099 | PTPN21, PTPD1, PTPRL10 | protein tyrosine phosphatase, non-receptor type 21 (EC:3.1.3.48) |
C00002903
|
| 5786 | PTPRA, HEPTP, HLPR, HPTPA, HPTPalpha, LRP, PTPA, PTPRL2, R-PTP-alpha, RPTPA | protein tyrosine phosphatase, receptor type, A (EC:3.1.3.48) |
C00002903
|
| 5792 | PTPRF, LAR | protein tyrosine phosphatase, receptor type, F (EC:3.1.3.48) |
C00002903
|
| 5793 | PTPRG, HPTPG, PTPG, R-PTP-GAMMA, RPTPG | protein tyrosine phosphatase, receptor type, G (EC:3.1.3.48) |
C00002903
|
| 5799 | PTPRN2, IA-2beta, IAR, ICAAR, PTPRP, R-PTP-N2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 (EC:3.1.3.48) |
C00002903
|
| 9232 | PTTG1, EAP1, HPTTG, PTTG, TUTR1 | pituitary tumor-transforming 1 |
C00002903
|
| 5819 | PVRL2, CD112, HVEB, PRR2, PVRR2 | poliovirus receptor-related 2 (herpesvirus entry mediator B) |
C00002903
|
| 5881 | RAC3 | ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
C00002903
|
| 29127 | RACGAP1, CYK4, HsCYK-4, ID-GAP, MgcRacGAP | Rac GTPase activating protein 1 |
C00002903
|
| 10635 | RAD51AP1, PIR51 | RAD51 associated protein 1 |
C00002903
|
| 25788 | RAD54B, RDH54 | RAD54 homolog B (S. cerevisiae) |
C00002903
|
| 8438 | RAD54L, HR54, RAD54A, hHR54, hRAD54 | RAD54-like (S. cerevisiae) |
C00002903
|
| 5894 | RAF1, CRAF, NS5, Raf-1, c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 (EC:2.7.11.1) |
C00002903
|
| 5901 | RAN, ARA24, Gsp1, TC4 | RAN, member RAS oncogene family |
C00002903
|
| 8498 | RANBP3 | RAN binding protein 3 |
C00002903
|
| 5911 | RAP2A, K-REV, KREV, RAP2, RbBP-30 | RAP2A, member of RAS oncogene family |
C00002903
|
| 5922 | RASA2, GAP1M | RAS p21 protein activator 2 |
C00002903
|
| 51655 | RASD1, AGS1, DEXRAS1, MGC:26290 | RAS, dexamethasone-induced 1 |
C00002903
|
| 5923 | RASGRF1, CDC25, CDC25L, GNRP, GRF1, GRF55, H-GRF55, PP13187, ras-GRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 |
C00002903
|
| 10125 | RASGRP1, CALDAG-GEFI, CALDAG-GEFII, RASGRP, V, hRasGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
C00002903
|
| 5932 | RBBP8, COM1, CTIP, JWDS, RIM, SAE2, SCKL2 | retinoblastoma binding protein 8 |
C00002903
|
| 31818 |
C00002903
|
||
| 64783 | RBM15, OTT, OTT1, SPEN | RNA binding motif protein 15 |
C00002903
|
| 221662 | RBM24, RNPC6, dJ259A10.1 | RNA binding motif protein 24 |
C00002903
|
| 91433 | RCCD1 | RCC1 domain containing 1 |
C00002903
|
| 9401 | RECQL4, RECQ4 | RecQ protein-like 4 (EC:3.6.4.12) |
C00002903
|
| 80346 | REEP4, C8orf20 | receptor accessory protein 4 |
C00002903
|
| 5971 | RELB, I-REL, IREL, REL-B | v-rel avian reticuloendotheliosis viral oncogene homolog B |
C00002903
|
| 5649 | RELN, LIS2, PRO1598, RL | reelin |
C00002903
|
| 85021 | REPS1, RALBP1 | RALBP1 associated Eps domain containing 1 |
C00002903
|
| 85004 | RERG | RAS-like, estrogen-regulated, growth inhibitor |
C00002903
|
| 5979 | RET, CDHF12, CDHR16, HSCR1, MEN2A, MEN2B, MTC1, PTC, RET-ELE1, RET51 | ret proto-oncogene (EC:2.7.10.1) |
C00002903
|
| 5982 | RFC2, RFC40 | replication factor C (activator 1) 2, 40kDa |
C00002903
|
| 5983 | RFC3, RFC38 | replication factor C (activator 1) 3, 38kDa |
C00002903
|
| 5984 | RFC4, A1, RFC37 | replication factor C (activator 1) 4, 37kDa |
C00002903
|
| 5985 | RFC5, RFC36 | replication factor C (activator 1) 5, 36.5kDa |
C00002903
|
| 8490 | RGS5, MST092, MST106, MST129, MSTP032, MSTP092, MSTP106, MSTP129 | regulator of G-protein signaling 5 |
C00002903
|
| 64285 | RHBDF1, C16orf8, Dist1, EGFR-RS, gene-89, gene-90, hDist1 | rhomboid 5 homolog 1 (Drosophila) |
C00002903
|
| 83695 | RHNO1, C12orf32, RHINO | RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
C00002903
|
| 23433 | RHOQ, ARHQ, RASL7A, TC10, TC10A | ras homolog family member Q |
C00002903
|
| 26150 | RIBC2, C22orf11 | RIB43A domain with coiled-coils 2 |
C00002903
|
| 80010 | RMI1, BLAP75, C9orf76, FAAP75, RP11-346I8.1 | RecQ mediated genome instability 1 |
C00002903
|
| 116028 | RMI2, BLAP18, C16orf75 | RecQ mediated genome instability 2 |
C00002903
|
| 9781 | RNF144A, RNF144, UBCE7IP4 | ring finger protein 144A |
C00002903
|
| 6047 | RNF4, SLX5, SNURF | ring finger protein 4 |
C00002903
|
| 84900 | RNFT2, TMEM118 | ring finger protein, transmembrane 2 |
C00002903
|
| 26824 | RNU11, RNU11-1, U11 | RNA, U11 small nuclear |
C00002903
|
| 6093 | ROCK1, P160ROCK, ROCK-I | Rho-associated, coiled-coil containing protein kinase 1 (EC:2.7.11.1) |
C00002903
|
| 6117 | RPA1, HSSB, MST075, REPA1, RF-A, RP-A, RPA70 | replication protein A1, 70kDa |
C00002903
|
| 6119 | RPA3, REPA3 | replication protein A3, 14kDa |
C00002903
|
| 56475 | RPRM, REPRIMO | reprimo, TP53 dependent G2 arrest mediator candidate |
C00002903
|
| 6194 | RPS6, S6 | ribosomal protein S6 |
C00002903
|
| 27 | ABL2, ABLL, ARG | c-abl oncogene 2, non-receptor tyrosine kinase (EC:2.7.10.2) |
C00000958
|
| 6198 | RPS6KB1, PS6K, S6K, S6K-beta-1, S6K1, STK14A, p70_S6KA, p70(S6K)-alpha, p70-S6K, p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 (EC:2.7.11.1) |
C00002903
|
| 6240 | RRM1, R1, RIR1, RR1 | ribonucleotide reductase M1 (EC:1.17.4.1) |
C00002903
|
| 6241 | RRM2, R2, RR2, RR2M | ribonucleotide reductase M2 (EC:1.17.4.1) |
C00002903
|
| 50484 | RRM2B, MTDPS8A, MTDPS8B, P53R2 | ribonucleotide reductase M2 B (TP53 inducible) (EC:1.17.4.1) |
C00002903
|
| 91543 | RSAD2, 2510004L01Rik, cig33, cig5, vig1 | radical S-adenosyl methionine domain containing 2 |
C00002903
|
| 51319 | RSRC1, SFRS21, SRrp53 | arginine/serine-rich coiled-coil 1 |
C00002903
|
| 10900 | RUNDC3A, RAP2IP, RPIP-8, RPIP8 | RUN domain containing 3A |
C00002903
|
| 6277 | S100A6, 2A9, 5B10, CABP, CACY, PRA | S100 calcium binding protein A6 |
C00002903
|
| 1903 | S1PR3, EDG-3, EDG3, LPB3, S1P3 | sphingosine-1-phosphate receptor 3 |
C00002903
|
| 29901 | SAC3D1, HSU79266, SHD1 | SAC3 domain containing 1 |
C00002903
|
| 26278 | SACS, ARSACS, DNAJC29 | spastic ataxia of Charlevoix-Saguenay (sacsin) |
C00002903
|
| 57167 | SALL4, DRRS, HSAL4, ZNF797, dJ1112F19.1 | spalt-like transcription factor 4 |
C00002903
|
| 23034 | SAMD4A, SAMD4, SMAUG, SMAUG1, SMG, SMGA | sterile alpha motif domain containing 4A |
C00002903
|
| 57406 | ABHD6 | abhydrolase domain containing 6 (EC:3.1.1.23) |
C00000958
|
| 45384 |
C00002903
|
||
| 54581 | SCAND2P, SCAND2 | SCAN domain containing 2 pseudogene |
C00002903
|
| 949 | SCARB1, CD36L1, CLA-1, CLA1, HDLQTL6, SR-BI, SRB1 | scavenger receptor class B, member 1 |
C00002903
|
| 6383 | SDC2, CD362, HSPG, HSPG1, SYND2 | syndecan 2 |
C00002903
|
| 8991 | SELENBP1, LPSB, SBP56, SP56, hSBP | selenium binding protein 1 |
C00002903
|
| 6402 | SELL, CD62L, LAM1, LECAM1, LEU8, LNHR, LSEL, LYAM1, PLNHR, TQ1 | selectin L |
C00002903
|
| 6403 | SELP, CD62, CD62P, GMP140, GRMP, LECAM3, PADGEM, PSEL | selectin P (granule membrane protein 140kDa, antigen CD62) |
C00002903
|
| 22928 | SEPHS2, SPS2, SPS2b | selenophosphate synthetase 2 (EC:2.7.9.3) |
C00002903
|
| 6414 | SEPP1, SELP, SeP | selenoprotein P, plasma, 1 |
C00002903
|
| 6059 | ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I | ATP-binding cassette, sub-family E (OABP), member 1 |
C00000958
|
| 27244 | SESN1, PA26, SEST1 | sestrin 1 |
C00002903
|
| 83667 | SESN2, HI95, SES2, SEST2 | sestrin 2 |
C00002903
|
| 6419 | SETMAR, HsMar1, METNASE, Mar1 | SET domain and mariner transposase fusion gene (EC:2.1.1.43) |
C00002903
|
| 83443 | SF3B5, SF3b10, Ysf3 | splicing factor 3b, subunit 5, 10kDa |
C00002903
|
| 119392 | SFR1, C10orf78, MEI5, MEIR5, bA373N18.1 | SWI5-dependent recombination repair 1 |
C00002903
|
| 6422 | SFRP1, FRP, FRP-1, FRP1, FrzA, SARP2 | secreted frizzled-related protein 1 |
C00002903
|
| 118980 | SFXN2 | sideroflexin 2 |
C00002903
|
| 6445 | SGCG, A4, DAGA4, DMDA, DMDA1, LGMD2C, MAM, SCARMD2, SCG3, TYPE | sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
C00002903
|
| 23678 | SGK3, CISK, SGK2, SGKL | serum/glucocorticoid regulated kinase family, member 3 (EC:2.7.11.1) |
C00002903
|
| 151648 | SGOL1, NY-BR-85, SGO, Sgo1 | shugoshin-like 1 (S. pombe) |
C00002903
|
| 6451 | SH3BGRL, SH3BGR | SH3 domain binding glutamic acid-rich protein like |
C00002903
|
| 23677 | SH3BP4, BOG25, TTP | SH3-domain binding protein 4 |
C00002903
|
| 6461 | SHB, RP11-3J10.8, bA3J10.2 | Src homology 2 domain containing adaptor protein B |
C00002903
|
| 79801 | SHCBP1, PAL | SHC SH2-domain binding protein 1 |
C00002903
|
| 6472 | SHMT2, GLYA, SHMT | serine hydroxymethyltransferase 2 (mitochondrial) (EC:2.1.2.1) |
C00002903
|
| 6477 | SIAH1, SIAH1A | siah E3 ubiquitin protein ligase 1 (EC:6.3.2.-) |
C00002903
|
| 23411 | SIRT1, SIR2L1 | sirtuin 1 |
C00002903
|
| 22933 | SIRT2, SIR2, SIR2L, SIR2L2 | sirtuin 2 (EC:3.5.1.-) |
C00002903
|
| 23410 | SIRT3, SIR2L3 | sirtuin 3 |
C00002903
|
| 23408 | SIRT5, SIR2L5 | sirtuin 5 |
C00002903
|
| 220134 | SKA1, C18orf24 | spindle and kinetochore associated complex subunit 1 |
C00002903
|
| 348235 | SKA2, FAM33A | spindle and kinetochore associated complex subunit 2 |
C00002903
|
| 221150 | SKA3, C13orf3, RAMA1 | spindle and kinetochore associated complex subunit 3 |
C00002903
|
| 8935 | SKAP2, PRAP, RA70, SAPS, SCAP2, SKAP-HOM, SKAP55R | src kinase associated phosphoprotein 2 |
C00002903
|
| 7884 | SLBP, HBP | stem-loop binding protein |
C00002903
|
| 6560 | SLC12A4, KCC1 | solute carrier family 12 (potassium/chloride transporter), member 4 |
C00002903
|
| 6509 | SLC1A4, ASCT1, SATT | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
C00002903
|
| 6510 | SLC1A5, AAAT, ASCT2, ATBO, M7V1, M7VS1, R16, RDRC | solute carrier family 1 (neutral amino acid transporter), member 5 |
C00002903
|
| 8402 | SLC25A11, OGC, SLC20A4 | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
C00002903
|
| 284129 | SLC26A11 | solute carrier family 26 (anion exchanger), member 11 |
C00002903
|
| 6514 | SLC2A2, GLUT2 | solute carrier family 2 (facilitated glucose transporter), member 2 |
C00002903
|
| 1317 | SLC31A1, COPT1, CTR1 | solute carrier family 31 (copper transporter), member 1 |
C00002903
|
| 64116 | SLC39A8, BIGM103, LZT-Hs6, ZIP8 | solute carrier family 39 (zinc transporter), member 8 |
C00002903
|
| 54946 | SLC41A3, SLC41A1-L2 | solute carrier family 41, member 3 |
C00002903
|
| 51151 | SLC45A2, 1A1, AIM1, MATP, OCA4, SHEP5 | solute carrier family 45, member 2 |
C00002903
|
| 6521 | SLC4A1, AE1, BND3, CD233, DI, EMPB3, EPB3, FR, RTA1A, SW, WD, WD1, WR | solute carrier family 4 (anion exchanger), member 1 |
C00002903
|
| 6528 | SLC5A5, NIS, TDH1 | solute carrier family 5 (sodium/iodide cotransporter), member 5 |
C00002903
|
| 11254 | SLC6A14, BMIQ11 | solute carrier family 6 (amino acid transporter), member 14 |
C00002903
|
| 8140 | SLC7A5, 4F2LC, CD98, D16S469E, E16, LAT1, MPE16, hLAT1 | solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
C00002903
|
| 6548 | SLC9A1, APNH, NHE-1, NHE1 | solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
C00002903
|
| 6550 | SLC9A3, NHE3 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
C00002903
|
| 6578 | SLCO2A1, MATR1, OATP2A1, PGT, PHOAR2, SLC21A2 | solute carrier organic anion transporter family, member 2A1 |
C00002903
|
| 4087 | SMAD2, JV18, JV18-1, MADH2, MADR2, hMAD-2, hSMAD2 | SMAD family member 2 |
C00002903
|
| 4089 | SMAD4, DPC4, JIP, MADH4, MYHRS | SMAD family member 4 |
C00002903
|
| 4090 | SMAD5, DWFC, JV5-1, MADH5 | SMAD family member 5 |
C00002903
|
| 4092 | SMAD7, CRCS3, MADH7, MADH8 | SMAD family member 7 |
C00002903
|
| 4093 | SMAD9, MADH6, MADH9, PPH2, SMAD8, SMAD8A, SMAD8B | SMAD family member 9 |
C00002903
|
| 8243 | SMC1A, CDLS2, DXS423E, SB1.8, SMC1, SMC1L1, SMC1alpha, SMCB | structural maintenance of chromosomes 1A |
C00002903
|
| 10051 | SMC4, CAP-C, CAPC, SMC-4, SMC4L1, hCAP-C | structural maintenance of chromosomes 4 |
C00002903
|
| 6608 | SMO, FZD11, Gx, SMOH | smoothened, frizzled family receptor |
C00002903
|
| 6591 | SNAI2, SLUG, SLUGH1, SNAIL2, WS2D | snail family zinc finger 2 |
C00002903
|
| 5825 | ABCD3, ABC43, PMP70, PXMP1, ZWS2 | ATP-binding cassette, sub-family D (ALD), member 3 |
C00000958
|
| 84973 | SNHG7, NCRNA00061 | small nucleolar RNA host gene 7 (non-protein coding) |
C00002903
|
| 79622 | SNRNP25, C16orf33 | small nuclear ribonucleoprotein 25kDa (U11/U12) |
C00002903
|
| 6627 | SNRPA1, Lea1 | small nuclear ribonucleoprotein polypeptide A' |
C00002903
|
| 29887 | SNX10, OPTB8 | sorting nexin 10 |
C00002903
|
| 8835 | SOCS2, CIS2, Cish2, SOCS-2, SSI-2, SSI2, STATI2 | suppressor of cytokine signaling 2 |
C00002903
|
| 9021 | SOCS3, ATOD4, CIS3, Cish3, SOCS-3, SSI-3, SSI3 | suppressor of cytokine signaling 3 |
C00002903
|
| 89845 | ABCC10, EST182763, MRP7, SIMRP7 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
C00000958
|
| 6654 | SOS1, GF1, GGF1, GINGF, HGF, NS4 | son of sevenless homolog 1 (Drosophila) |
C00002903
|
| 64321 | SOX17, VUR3 | SRY (sex determining region Y)-box 17 |
C00002903
|
| 6657 | SOX2, ANOP3, MCOPS3 | SRY (sex determining region Y)-box 2 |
C00002903
|
| 6658 | SOX3, GHDX, MRGH, PHP, PHPX, SOXB | SRY (sex determining region Y)-box 3 |
C00002903
|
| 6659 | SOX4, EVI16 | SRY (sex determining region Y)-box 4 |
C00002903
|
| 6662 | SOX9, CMD1, CMPD1, SRA1 | SRY (sex determining region Y)-box 9 |
C00002903
|
| 8647 | ABCB11, ABC16, BRIC2, BSEP, PFIC-2, PFIC2, PGY4, SPGP | ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
C00000958
|
| 121340 | SP7, OI11, OI12, OSX, osterix | Sp7 transcription factor |
C00002903
|
| 10615 | SPAG5, DEEPEST, MAP126, hMAP126 | sperm associated antigen 5 |
C00002903
|
| 6678 | SPARC, ON | secreted protein, acidic, cysteine-rich (osteonectin) |
C00002903
|
| 64847 | SPATA20, SSP411, Tisp78 | spermatogenesis associated 20 |
C00002903
|
| 55812 | SPATA7, HSD-3.1, HSD3, LCA3 | spermatogenesis associated 7 |
C00002903
|
| 57405 | SPC25, SPBC25, hSpc25 | SPC25, NDC80 kinetochore complex component |
C00002903
|
| 25803 | SPDEF, PDEF, RP11-375E1__A.3, bA375E1.3 | SAM pointed domain containing ETS transcription factor |
C00002903
|
| 54908 | SPDL1, CCDC99 | spindle apparatus coiled-coil protein 1 |
C00002903
|
| 8877 | SPHK1, SPHK | sphingosine kinase 1 (EC:2.7.1.91) |
C00002903
|
| 139886 | SPIN4 | spindlin family, member 4 |
C00002903
|
| 6696 | SPP1, BNSP, BSPI, ETA-1, OPN | secreted phosphoprotein 1 |
C00002903
|
| 10252 | SPRY1, hSPRY1 | sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
C00002903
|
| 90864 | SPSB3, C16orf31, SSB3 | splA/ryanodine receptor domain and SOCS box containing 3 |
C00002903
|
| 6709 | SPTAN1, EIEE5, NEAS, SPTA2 | spectrin, alpha, non-erythrocytic 1 |
C00002903
|
| 6714 | SRC, ASV, SRC1, c-SRC, p60-Src | v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (EC:2.7.10.2) |
C00002903
|
| 6721 | SREBF2, SREBP-2, SREBP2, bHLHd2 | sterol regulatory element binding transcription factor 2 |
C00002903
|
| 23456 | ABCB10, EST20237, M-ABC2, MTABC2 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
C00000958
|
| 63826 | SRR, ILV1, ISO1 | serine racemase (EC:4.3.1.17 5.1.1.18 4.3.1.18) |
C00002903
|
| 10772 | SRSF10, FUSIP1, FUSIP2, NSSR, SFRS13, SFRS13A, SRp38, SRrp40, TASR, TASR1, TASR2 | serine/arginine-rich splicing factor 10 |
C00002903
|
| 117178 | SSX2IP, ADIP | synovial sarcoma, X breakpoint 2 interacting protein |
C00002903
|
| 6767 | ST13, AAG2, FAM10A1, FAM10A4, HIP, HOP, HSPABP, HSPABP1, P48, PRO0786, SNC6 | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
C00002903
|
| 6482 | ST3GAL1, Gal-NAc6S, SIAT4A, SIATFL, ST3GalA, ST3GalA.1, ST3GalIA, ST3GalIA,1, ST3O | ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (EC:2.4.99.4) |
C00002903
|
| 7903 | ST8SIA4, PST, PST1, SIAT8D, ST8SIA-IV | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (EC:2.4.99.-) |
C00002903
|
| 132949 | AASDH, ACSF4, LYS2, NRPS1098, NRPS998 | aminoadipate-semialdehyde dehydrogenase (EC:6.2.1.-) |
C00000958
|
| 6491 | STIL, MCPH7, SIL | SCL/TAL1 interrupting locus |
C00002903
|
| 6794 | STK11, LKB1, PJS, hLKB1 | serine/threonine kinase 11 (EC:2.7.11.1) |
C00002903
|
| 9262 | STK17B, DRAK2 | serine/threonine kinase 17b (EC:2.7.11.1) |
C00002903
|
| 27347 | STK39, DCHT, PASK, SPAK | serine threonine kinase 39 (EC:2.7.11.1) |
C00002903
|
| 3925 | STMN1, C1orf215, LAP18, Lag, OP18, PP17, PP19, PR22, SMN | stathmin 1 |
C00002903
|
| 29966 | STRN3, SG2NA | striatin, calmodulin binding protein 3 |
C00002903
|
| 23213 | SULF1, HSULF-1, SULF-1 | sulfatase 1 |
C00002903
|
| 6799 | SULT1A2, HAST4, P-PST, ST1A2, STP2, TSPST2 | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (EC:2.8.2.1) |
C00002903
|
| 6818 | SULT1A3, HAST, HAST3, M-PST, ST1A3/ST1A4, ST1A5, STM, TL-PST | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (EC:2.8.2.1) |
C00002903
|
| 6819 | SULT1C2, ST1C1, ST1C2, SULT1C1, humSULTC2 | sulfotransferase family, cytosolic, 1C, member 2 (EC:2.8.2.-) |
C00002903
|
| 6783 | SULT1E1, EST, EST-1, ST1E1, STE | sulfotransferase family 1E, estrogen-preferring, member 1 (EC:2.8.2.4) |
C00002903
|
| 25830 | SULT4A1, BR-STL-1, BRSTL1, DJ388M5.3, NST, SULTX3, hBR-STL-1 | sulfotransferase family 4A, member 1 (EC:2.8.2.-) |
C00002903
|
| 7341 | SUMO1, DAP1, GMP1, OFC10, PIC1, SENP2, SMT3, SMT3C, SMT3H3, UBL1 | small ubiquitin-like modifier 1 |
C00002903
|
| 6839 | SUV39H1, H3-K9-HMTase_1, KMT1A, MG44, SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) (EC:2.1.1.43) |
C00002903
|
| 79723 | SUV39H2, KMT1B | suppressor of variegation 3-9 homolog 2 (Drosophila) (EC:2.1.1.43) |
C00002903
|
| 55638 | SYBU, GOLSYN, OCSYN, SNPHL | syntabulin (syntaxin-interacting) |
C00002903
|
| 9143 | SYNGR3 | synaptogyrin 3 |
C00002903
|
| 6855 | SYP, MRXSYP | synaptophysin |
C00002903
|
| 54843 | SYTL2, CHR11SYT, EXO4, SGA72M, SLP2, SLP2A | synaptotagmin-like 2 |
C00002903
|
| 94122 | SYTL5, slp5 | synaptotagmin-like 5 |
C00002903
|
| 10460 | TACC3, ERIC-1, ERIC1 | transforming, acidic coiled-coil containing protein 3 |
C00002903
|
| 6877 | TAF5, TAF2D, TAFII100 | TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
C00002903
|
| 6888 | TALDO1, TAL, TAL-H, TALDOR, TALH | transaldolase 1 (EC:2.2.1.2) |
C00002903
|
| 51256 | TBC1D7, PIG51, TBC7, dJ257A7.3 | TBC1 domain family, member 7 |
C00002903
|
| 6938 | TCF12, CRS3, HEB, HTF4, HsT17266, bHLHb20 | transcription factor 12 |
C00002903
|
| 6941 | TCF19, SC1, TCF-19 | transcription factor 19 |
C00002903
|
| 25980 | AAR2, C20orf4 | AAR2 splicing factor homolog (S. cerevisiae) |
C00000958
|
| 83439 | TCF7L1, TCF-3, TCF3 | transcription factor 7-like 1 (T-cell specific, HMG-box) |
C00002903
|
| 7013 | TERF1, PIN2, TRBF1, TRF, TRF1, hTRF1-AS, t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 |
C00002903
|
| 7018 | TF, PRO1557, PRO2086, TFQTL1 | transferrin |
C00002903
|
| 7020 | TFAP2A, AP-2, AP-2alpha, AP2TF, BOFS, TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
C00002903
|
| 7022 | TFAP2C, AP2-GAMMA, ERF1, TFAP2G, hAP-2g | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
C00002903
|
| 7027 | TFDP1, DP1, DRTF1, Dp-1 | transcription factor Dp-1 |
C00002903
|
| 7031 | TFF1, BCEI, D21S21, HP1.A, HPS2, pNR-2, pS2 | trefoil factor 1 |
C00002903
|
| 7033 | TFF3, ITF, P1B, TFI | trefoil factor 3 (intestinal) |
C00002903
|
| 7980 | TFPI2, PP5, REF1, TFPI-2 | tissue factor pathway inhibitor 2 |
C00002903
|
| 7039 | TGFA, TFGA | transforming growth factor, alpha |
C00002903
|
| 28971 | AAMDC, C11orf67, CK067 | adipogenesis associated, Mth938 domain containing |
C00000958
|
| 7042 | TGFB2, LDS4, TGF-beta2 | transforming growth factor, beta 2 |
C00002903
|
| 7048 | TGFBR2, AAT3, FAA3, LDS1B, LDS2B, MFS2, RIIC, TAAD2, TGFR-2, TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) (EC:2.7.11.30) |
C00002903
|
| 60436 | TGIF2 | TGFB-induced factor homeobox 2 |
C00002903
|
| 7051 | TGM1, ARCI1, ICR2, KTG, LI, LI1, TGASE, TGK | transglutaminase 1 (EC:2.3.2.13) |
C00002903
|
| 7053 | TGM3, TGE | transglutaminase 3 (EC:2.3.2.13) |
C00002903
|
| 9333 | TGM5, TGASE5, TGASEX, TGM6, TGMX, TGX | transglutaminase 5 (EC:2.3.2.13) |
C00002903
|
| 7056 | THBD, AHUS6, BDCA3, CD141, THPH12, THRM, TM | thrombomodulin |
C00002903
|
| 79875 | THSD4, ADAMTSL-6, ADAMTSL6, FVSY9334, PRO34005 | thrombospondin, type I, domain containing 4 |
C00002903
|
| 8914 | TIMELESS, TIM, TIM1, hTIM | timeless circadian clock |
C00002903
|
| 100287932 | TIMM23, TIM23 | translocase of inner mitochondrial membrane 23 homolog (yeast) |
C00002903
|
| 54962 | TIPIN | TIMELESS interacting protein |
C00002903
|
| 195827 | AAED1, C9orf21 | AhpC/TSA antioxidant enzyme domain containing 1 |
C00000958
|
| 7099 | TLR4, ARMD10, CD284, TLR-4, TOLL | toll-like receptor 4 |
C00002903
|
| 53346 | TM6SF1 | transmembrane 6 superfamily member 1 |
C00002903
|
| 147798 | TMC4 | transmembrane channel-like 4 |
C00002903
|
| 79022 | TMEM106C | transmembrane protein 106C |
C00002903
|
| 84314 | TMEM107, GRVS638, PRO1268 | transmembrane protein 107 |
C00002903
|
| 23306 | TMEM194A, TMEM194 | transmembrane protein 194A |
C00002903
|
| 55151 | TMEM38B, C9orf87, D4Ertd89e, OI14, TRIC-B, TRICB, bA219P18.1 | transmembrane protein 38B |
C00002903
|
| 169200 | TMEM64 | transmembrane protein 64 |
C00002903
|
| 27346 | TMEM97, MAC30 | transmembrane protein 97 |
C00002903
|
| 7112 | TMPO, CMD1T, LAP2, LEMD4, PRO0868, TP | thymopoietin |
C00002903
|
| 7113 | TMPRSS2, PP9284, PRSS10 | transmembrane protease, serine 2 (EC:3.4.21.-) |
C00002903
|
| 64699 | TMPRSS3, DFNB10, DFNB8, ECHOS1, TADG12 | transmembrane protease, serine 3 (EC:3.4.21.-) |
C00002903
|
| 735524 |
C00002903
|
||
| 160335 | TMTC2, IBDBP1 | transmembrane and tetratricopeptide repeat containing 2 |
C00002903
|
| 81542 | TMX1, PDIA11, TMX, TXNDC, TXNDC1 | thioredoxin-related transmembrane protein 1 |
C00002903
|
| 7128 | TNFAIP3, A20, OTUD7C, TNFA1P2 | tumor necrosis factor, alpha-induced protein 3 (EC:3.4.19.12) |
C00002903
|
| 8797 | TNFRSF10A, APO2, CD261, DR4, TRAILR-1, TRAILR1 | tumor necrosis factor receptor superfamily, member 10a |
C00002903
|
| 8795 | TNFRSF10B, CD262, DR5, KILLER, KILLER/DR5, TRAIL-R2, TRAILR2, TRICK2, TRICK2A, TRICK2B, TRICKB, ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b |
C00002903
|
| 8792 | TNFRSF11A, CD265, FEO, LOH18CR1, ODFR, OFE, OPTB7, OSTS, PDB2, RANK, TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
C00002903
|
| 27242 | TNFRSF21, BM-018, CD358, DR6 | tumor necrosis factor receptor superfamily, member 21 |
C00002903
|
| 8600 | TNFSF11, CD254, ODF, OPGL, OPTB2, RANKL, TRANCE, hRANKL2, sOdf | tumor necrosis factor (ligand) superfamily, member 11 |
C00002903
|
| 10318 | TNIP1, ABIN-1, NAF1, VAN, nip40-1 | TNFAIP3 interacting protein 1 |
C00002903
|
| 10188 | TNK2, ACK, ACK-1, ACK1, p21cdc42Hs | tyrosine kinase, non-receptor, 2 (EC:2.7.11.1 2.7.10.2) |
C00002903
|
| 7145 | TNS1, MST091, MST122, MST127, MSTP122, MSTP127, MXRA6, TNS | tensin 1 |
C00002903
|
| 11073 | TOPBP1, TOP2BP1 | topoisomerase (DNA) II binding protein 1 |
C00002903
|
| 7159 | TP53BP2, 53BP2, ASPP2, BBP, P53BP2, PPP1R13A | tumor protein p53 binding protein, 2 |
C00002903
|
| 9540 | TP53I3, PIG3 | tumor protein p53 inducible protein 3 |
C00002903
|
| 94241 | TP53INP1, SIP, TP53DINP1, TP53INP1A, TP53INP1B, Teap, p53DINP1 | tumor protein p53 inducible nuclear protein 1 |
C00002903
|
| 8626 | TP63, AIS, B(p51A), B(p51B), EEC3, KET, LMS, NBP, OFC8, RHS, SHFM4, TP53CP, TP53L, TP73L, p40, p51, p53CP, p63, p73H, p73L | tumor protein p63 |
C00002903
|
| 7161 | TP73, P73 | tumor protein p73 |
C00002903
|
| 127262 | TPRG1L, FAM79A, RP11-46F15.3, h-mover, mover | tumor protein p63 regulated 1-like |
C00002903
|
| 7178 | TPT1, HRF, TCTP, p02, p23 | tumor protein, translationally-controlled 1 |
C00002903
|
| 7179 | TPTE, CT44, PTEN2 | transmembrane phosphatase with tensin homology (EC:3.1.3.48) |
C00002903
|
| 22974 | TPX2, C20orf1, C20orf2, DIL-2, DIL2, FLS353, GD:C20orf1, HCA519, HCTP4, REPP86, p100 | TPX2, microtubule-associated |
C00002903
|
| 7185 | TRAF1, EBI6, MGC:10353 | TNF receptor-associated factor 1 |
C00002903
|
| 7186 | TRAF2, MGC:45012, TRAP, TRAP3 | TNF receptor-associated factor 2 |
C00002903
|
| 51605 | TRMT6, GCD10, Gcd10p | tRNA methyltransferase 6 homolog (S. cerevisiae) |
C00002903
|
| 10024 | TROAP, TASTIN | trophinin associated protein |
C00002903
|
| 2539 | G6PD, G6PD1 | glucose-6-phosphate dehydrogenase (EC:1.1.1.49) |
C00002648
|
| 7249 | TSC2, LAM, TSC4 | tuberous sclerosis 2 |
C00002903
|
| 8848 | TSC22D1, Ptg-2, TGFB1I4, TSC22 | TSC22 domain family, member 1 |
C00002903
|
| 1831 | TSC22D3, DIP, DSIPI, GILZ, TSC-22R, hDIP | TSC22 domain family, member 3 |
C00002903
|
| 7251 | TSG101, TSG10, VPS23 | tumor susceptibility 101 |
C00002903
|
| 7252 | TSHB, TSH-B, TSH-BETA | thyroid stimulating hormone, beta |
C00002903
|
| 7253 | TSHR, CHNG1, LGR3, hTSHR-I | thyroid stimulating hormone receptor |
C00002903
|
| 7247 | TSN, BCLF-1, C3PO, RCHF1, REHF-1, TBRBP, TRSLN | translin |
C00002903
|
| 10103 | TSPAN1, NET1, TM4C, TM4SF | tetraspanin 1 |
C00002903
|
| 23554 | TSPAN12, EVR5, NET-2, NET2, TM4SF12 | tetraspanin 12 |
C00002903
|
| 6302 | TSPAN31, SAS | tetraspanin 31 |
C00002903
|
| 23508 | TTC9, TTC9A | tetratricopeptide repeat domain 9 |
C00002903
|
| 8458 | TTF2, HuF2 | transcription termination factor, RNA polymerase II |
C00002903
|
| 7283 | TUBG1, CDCBM4, GCP-1, TUBG, TUBGCP1 | tubulin, gamma 1 |
C00002903
|
| 27229 | TUBGCP4, 76P, GCP-4, GCP4, Grip76 | tubulin, gamma complex associated protein 4 |
C00002903
|
| 10628 | TXNIP, EST01027, HHCPA78, THIF, VDUP1 | thioredoxin interacting protein |
C00002903
|
| 7296 | TXNRD1, GRIM-12, TR, TR1, TRXR1, TXNR | thioredoxin reductase 1 (EC:1.8.1.9) |
C00002903
|
| 7298 | TYMS, HST422, TMS, TS | thymidylate synthetase (EC:2.1.1.45) |
C00002903
|
| 7299 | TYR, ATN, CMM8, OCA1, OCA1A, OCAIA, SHEP3 | tyrosinase (EC:1.14.18.1) |
C00002903
|
| 6675 | UAP1, AGX, AGX1, AGX2, SPAG2 | UDP-N-acetylglucosamine pyrophosphorylase 1 (EC:2.7.7.23 2.7.7.83) |
C00002903
|
| 7323 | UBE2D3, E2(17)KB3, UBC4/5, UBCH5C | ubiquitin-conjugating enzyme E2D 3 (EC:6.3.2.19) |
C00002903
|
| 7328 | UBE2H, E2-20K, GID3, UBC8, UBCH, UBCH2 | ubiquitin-conjugating enzyme E2H (EC:6.3.2.19) |
C00002903
|
| 7332 | UBE2L3, E2-F1, L-UBC, UBCH7, UbcM4 | ubiquitin-conjugating enzyme E2L 3 (EC:6.3.2.19) |
C00002903
|
| 134111 | UBE2QL1 | ubiquitin-conjugating enzyme E2Q family-like 1 (EC:6.3.2.19) |
C00002903
|
| 29089 | UBE2T, PIG50 | ubiquitin-conjugating enzyme E2T (putative) (EC:6.3.2.19) |
C00002903
|
| 9354 | UBE4A, E4, UBOX2, UFD2 | ubiquitination factor E4A |
C00002903
|
| 7371 | UCK2, TSA903, UK, UMPK | uridine-cytidine kinase 2 (EC:2.7.1.48) |
C00002903
|
| 7360 | UGP2, UDPG, UDPGP, UDPGP2, UGP1, UGPP1, UGPP2, pHC379 | UDP-glucose pyrophosphorylase 2 (EC:2.7.7.9) |
C00002903
|
| 54658 | UGT1A1, BILIQTL1, GNT1, HUG-BR1, UDPGT, UDPGT_1-1, UGT1, UGT1A | UDP glucuronosyltransferase 1 family, polypeptide A1 (EC:2.4.1.17) |
C00002903
|
| 54575 | UGT1A10, UDPGT, UGT-1J, UGT1-10, UGT1.10, UGT1J | UDP glucuronosyltransferase 1 family, polypeptide A10 (EC:2.4.1.17) |
C00002903
|
| 54600 | UGT1A9, HLUGP4, LUGP4, UDPGT, UDPGT_1-9, UGT-1I, UGT1-09, UGT1-9, UGT1.9, UGT1AI, UGT1I | UDP glucuronosyltransferase 1 family, polypeptide A9 (EC:2.4.1.17) |
C00002903
|
| 7366 | UGT2B15, HLUG4, UDPGT_2B8, UDPGT2B15, UDPGTH3, UGT2B8 | UDP glucuronosyltransferase 2 family, polypeptide B15 (EC:2.4.1.17) |
C00002903
|
| 7364 | UGT2B7, UDPGT_2B9, UDPGT2B7, UDPGTH2, UGT2B9 | UDP glucuronosyltransferase 2 family, polypeptide B7 (EC:2.4.1.17) |
C00002903
|
| 29128 | UHRF1, ICBP90, Np95, RNF106, hNP95, hUHRF1 | ubiquitin-like with PHD and ring finger domains 1 (EC:6.3.2.19) |
C00002903
|
| 7374 | UNG, DGU, HIGM4, HIGM5, UDG, UNG1, UNG15, UNG2 | uracil-DNA glycosylase (EC:3.2.2.27) |
C00002903
|
| 7398 | USP1, UBP | ubiquitin specific peptidase 1 (EC:3.4.19.12) |
C00002903
|
| 7375 | USP4, UNP, Unph | ubiquitin specific peptidase 4 (proto-oncogene) (EC:3.4.19.12) |
C00002903
|
| 10090 | UST, 2OST | uronyl-2-sulfotransferase |
C00002903
|
| 9218 | VAPA, VAP-33, VAP-A, VAP33, hVAP-33 | VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
C00002903
|
| 7408 | VASP | vasodilator-stimulated phosphoprotein |
C00002903
|
| 10451 | VAV3 | vav 3 guanine nucleotide exchange factor |
C00002903
|
| 7412 | VCAM1, CD106, INCAM-100 | vascular cell adhesion molecule 1 |
C00002903
|
| 7414 | VCL, CMD1W, CMH15, MV, MVCL | vinculin |
C00002903
|
| 7421 | VDR, NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
C00002903
|
| 7431 | VIM, CTRCT30 | vimentin |
C00002903
|
| 79001 | VKORC1, EDTP308, IMAGE3455200, MST134, MST576, VKCFD2, VKOR | vitamin K epoxide reductase complex, subunit 1 (EC:1.1.4.1) |
C00002903
|
| 10406 | WFDC2, EDDM4, HE4, WAP5, dJ461P17.6 | WAP four-disulfide core domain 2 |
C00002903
|
| 7468 | WHSC1, MMSET, NSD2, REIIBP, TRX5, WHS | Wolf-Hirschhorn syndrome candidate 1 (EC:2.1.1.43) |
C00002903
|
| 8839 | WISP2, CCN5, CT58, CTGF-L | WNT1 inducible signaling pathway protein 2 |
C00002903
|
| 7486 | WRN, RECQ3, RECQL2, RECQL3 | Werner syndrome, RecQ helicase-like (EC:3.6.4.12) |
C00002903
|
| 26118 | WSB1, SWIP1, WSB-1 | WD repeat and SOCS box containing 1 |
C00002903
|
| 11060 | WWP2, AIP2, WWp2-like | WW domain containing E3 ubiquitin protein ligase 2 |
C00002903
|
| 7494 | XBP1, TREB5, XBP-1, XBP2 | X-box binding protein 1 |
C00002903
|
| 7515 | XRCC1, RCC | X-ray repair complementing defective repair in Chinese hamster cells 1 |
C00002903
|
| 7516 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 |
C00002903
|
| 388403 | YPEL2 | yippee-like 2 (Drosophila) |
C00002903
|
| 51646 | YPEL5 | yippee-like 5 (Drosophila) |
C00002903
|
| 7529 | YWHAB, GW128, HS1, KCIP-1, YWHAA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
C00002903
|
| 7531 | YWHAE, 14-3-3E, KCIP-1, MDCR, MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
C00002903
|
| 7534 | YWHAZ, 14-3-3-zeta, KCIP-1, YWHAD | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
C00002903
|
| 7704 | ZBTB16, PLZF, ZNF145 | zinc finger and BTB domain containing 16 |
C00002903
|
| 84186 | ZCCHC7, AIR1, RP11-397D12.1 | zinc finger, CCHC domain containing 7 |
C00002903
|
| 55625 | ZDHHC7, DHHC7, SERZ-B, SERZ1, ZNF370 | zinc finger, DHHC-type containing 7 (EC:2.3.1.225) |
C00002903
|
| 6935 | ZEB1, AREB6, BZP, DELTAEF1, FECD6, NIL2A, PPCD3, TCF8, ZFHEP, ZFHX1A | zinc finger E-box binding homeobox 1 |
C00002903
|
| 7763 | ZFAND5, ZA20D2, ZFAND5A, ZNF216 | zinc finger, AN1-type domain 5 |
C00002903
|
| 342357 | ZKSCAN2, ZNF694, ZSCAN31, ZSCAN34 | zinc finger with KRAB and SCAN domains 2 |
C00002903
|
| 9203 | ZMYM3, DXS6673E, MYM, XFIM, ZNF198L2, ZNF261 | zinc finger, MYM-type 3 |
C00002903
|
| 7694 | ZNF135, ZNF61, ZNF78L1, pHZ-17, pT3 | zinc finger protein 135 |
C00002903
|
| 7707 | ZNF148, BERF-1, BFCOL1, HT-BETA, ZBP-89, ZFP148, pHZ-52 | zinc finger protein 148 |
C00002903
|
| 23036 | ZNF292, Nbla00365, ZFP292, ZN-16, Zn-15, bA393I2.3 | zinc finger protein 292 |
C00002903
|
| 7582 | ZNF33B, KOX2, KOX31, ZNF11B | zinc finger protein 33B |
C00002903
|
| 151126 | ZNF385B, ZNF533 | zinc finger protein 385B |
C00002903
|
| 168544 | ZNF467, EZI, Zfp467 | zinc finger protein 467 |
C00002903
|
| 80139 | ZNF703, ZEPPO1, ZNF503L, ZPO1 | zinc finger protein 703 |
C00002903
|
| 7637 | ZNF84, HPF2 | zinc finger protein 84 |
C00002903
|
| 7644 | ZNF91, HPF7, HTF10 | zinc finger protein 91 |
C00002903
|
| 57688 | ZSWIM6 | zinc finger, SWIM-type containing 6 |
C00002903
|
| 23053 | ZSWIM8, KIAA0913 | zinc finger, SWIM-type containing 8 |
C00002903
|
| 55055 | ZWILCH, KNTC1AP, hZwilch | zwilch kinetochore protein |
C00002903
|
| 11130 | ZWINT, HZwint-1, KNTC2AP, ZWINT1 | ZW10 interacting kinetochore protein |
C00002903
|
| 1548 | CYP2A6, CPA6, CYP2A, CYP2A3, CYPIIA6, P450C2A, P450PB | cytochrome P450, family 2, subfamily A, polypeptide 6 (EC:1.14.14.1) |
C00002905
|
| 1555 | CYP2B6, CPB6, CYP2B, CYP2B7, CYP2B7P, CYPIIB6, EFVM, IIB1, P450 | cytochrome P450, family 2, subfamily B, polypeptide 6 (EC:1.14.14.1) |
C00002905
|
| 1571 | CYP2E1, CPE1, CYP2E, P450-J, P450C2E | cytochrome P450, family 2, subfamily E, polypeptide 1 (EC:1.14.13.n7) |
C00002905
|
| 23600 | AMACR, AMACRD, CBAS4, RACE, RM | alpha-methylacyl-CoA racemase (EC:5.1.99.4) |
C00002902
|
| 2921 | CXCL3, CINC-2b, GRO3, GROg, MIP-2b, MIP2B, SCYB3 | chemokine (C-X-C motif) ligand 3 |
C00002902
|
| 2523 | FUT1, H, HH, HSC | fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) (EC:2.4.1.69) |
C00002902
|
| 4670 | HNRNPM, CEAR, HNRNPM4, HNRPM, HNRPM4, HTGR1, NAGR1, hnRNP_M | heterogeneous nuclear ribonucleoprotein M |
C00002902
|
| 3313 | HSPA9, CSA, GRP-75, GRP75, HSPA9B, MOT, MOT2, MTHSP75, PBP74 | heat shock 70kDa protein 9 (mortalin) |
C00002902
|
| 3696 | ITGB8 | integrin, beta 8 |
C00002902
|
| 9189 | ZBED1, ALTE, DREF, TRAMP, hDREF | zinc finger, BED-type containing 1 |
C00002902
|
| 118738 | ZNF488 | zinc finger protein 488 |
C00002902
|
| 393 | ARHGAP4, C1, RGC1, RhoGAP4, SrGAP4, p115 | Rho GTPase activating protein 4 |
C00000947
|
| 2683 | B4GALT1, B4GAL-T1, CDG2D, GGTB2, GT1, GTB, beta4Gal-T1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (EC:2.4.1.22 2.4.1.90 2.4.1.38) |
C00000947
|
| 56751 | BARHL1 | BarH-like homeobox 1 |
C00000947
|
| 388962 | BOLA3, MMDS2 | bolA family member 3 |
C00000947
|
| 79184 | BRCC3, BRCC36, C6.1A, CXorf53 | BRCA1/BRCA2-containing complex, subunit 3 (EC:3.1.2.15) |
C00000947
|
| 930 | CD19, B4, CVID3 | CD19 molecule |
C00000947
|
| 975 | CD81, CVID6, S5.7, TAPA1, TSPAN28 | CD81 molecule |
C00000947
|
| 928 | CD9, BTCC-1, DRAP-27, MIC3, MRP-1, TSPAN-29, TSPAN29 | CD9 molecule |
C00000947
|
| 1054 | CEBPG, GPE1BP, IG/EBP-1 | CCAAT/enhancer binding protein (C/EBP), gamma |
C00000947
|
| 2130 | EWSR1, EWS, bK984G1.4 | EWS RNA-binding protein 1 |
C00000947
|
| 2323 | FLT3LG, FL, FLT3L | fms-related tyrosine kinase 3 ligand |
C00000947
|
| 2495 | FTH1, FHC, FTH, FTHL6, PIG15, PLIF | ferritin, heavy polypeptide 1 (EC:1.16.3.1) |
C00000947
|
| 3054 | HCFC1, CFF, HCF-1, HCF1, HFC1, MRX3, VCAF | host cell factor C1 (VP16-accessory protein) |
C00000947
|
| 3232 | HOXD3, HOX1D, HOX4, HOX4A, Hox-4.1 | homeobox D3 |
C00000947
|
| 3559 | IL2RA, CD25, IDDM10, IL2R, TCGFR | interleukin 2 receptor, alpha |
C00000947
|
| 9235 | IL32, IL-32alpha, IL-32beta, IL-32delta, IL-32gamma, NK4, TAIF, TAIFa, TAIFb, TAIFc, TAIFd | interleukin 32 |
C00000947
|
| 3662 | IRF4, LSIRF, MUM1, NF-EM5 | interferon regulatory factor 4 |
C00000947
|
| 4067 | LYN, JTK8, p53Lyn, p56Lyn | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog (EC:2.7.10.2) |
C00000947
|
| 5605 | MAP2K2, CFC4, MAPKK2, MEK2, MKK2, PRKMK2 | mitogen-activated protein kinase kinase 2 (EC:2.7.12.2) |
C00000947
|
| 4282 | MIF, GIF, GLIF, MMIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) (EC:5.3.3.12 5.3.2.1) |
C00000947
|
| 64975 | MRPL41, BMRP, MRP-L27, MRPL27, RPML27 | mitochondrial ribosomal protein L41 |
C00000947
|
| 4493 | MT1E, MT1, MTD | metallothionein 1E |
C00000947
|
| 4869 | NPM1, B23, NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
C00000947
|
| 5187 | PER1, PER, RIGUI, hPER | period circadian clock 1 |
C00000947
|
| 6128 | RPL6, L6, SHUJUN-2, TAXREB107, TXREB1 | ribosomal protein L6 |
C00000947
|
| 6285 | S100B, NEF, S100, S100-B, S100beta | S100 calcium binding protein B |
C00000947
|
| 6286 | S100P, MIG9 | S100 calcium binding protein P |
C00000947
|
| 6574 | SLC20A1, GLVR1, Glvr-1, PIT1, PiT-1 | solute carrier family 20 (phosphate transporter), member 1 |
C00000947
|
| 6699 | SPRR1B, CORNIFIN, GADD33, SPRR1 | small proline-rich protein 1B |
C00000947
|
| 6777 | STAT5B, STAT5 | signal transducer and activator of transcription 5B |
C00000947
|
| 943 | TNFRSF8, CD30, D1S166E, Ki-1 | tumor necrosis factor receptor superfamily, member 8 |
C00000947
|
| 7150 | TOP1, TOPI | topoisomerase (DNA) I (EC:5.99.1.2) |
C00000947
|
| 2829 | XCR1, CCXCR1, GPR5 | chemokine (C motif) receptor 1 |
C00000947
|
| 51042 | ZNF593, ZT86 | zinc finger protein 593 |
C00000947
|
| 2551 | GABPA, E4TF1-60, E4TF1A, NFT2, NRF2, NRF2A, RCH04A07 | GA binding protein transcription factor, alpha subunit 60kDa |
C00002647
|
| 5591 | PRKDC, DNA-PKcs, DNAPK, DNPK1, HYRC, HYRC1, XRCC7, p350 | protein kinase, DNA-activated, catalytic polypeptide (EC:2.7.11.1) |
C00002647
|
| 19412 |
C00002647
|
||
| 387 | RHOA, ARH12, ARHA, RHO12, RHOH12 | ras homolog family member A |
C00002647
|
| 388 | RHOB, ARH6, ARHB, MST081, MSTP081, RHOH6 | ras homolog family member B |
C00002647
|
| 1081 | CGA, CG-ALPHA, FSHA, GPHA1, GPHa, HCG, LHA, TSHA | glycoprotein hormones, alpha polypeptide |
C00001180
|
| 8288 | EPX, EPO, EPP, EPX-PEN | eosinophil peroxidase (EC:1.11.1.7) |
C00001180
|
| 2623 | GATA1, ERYF1, GATA-1, GF-1, GF1, NF-E1, NFE1, XLANP, XLTDA, XLTT | GATA binding protein 1 (globin transcription factor 1) |
C00001180
|
| 2624 | GATA2, DCML, MONOMAC, NFE1B | GATA binding protein 2 |
C00001180
|
| 2993 | GYPA, CD235a, GPA, GPErik, GPSAT, HGpMiV, HGpMiXI, HGpSta(C), MN, MNS, PAS-2 | glycophorin A (MNS blood group) |
C00001180
|
| 10013 | HDAC6, CPBHM, HD6 | histone deacetylase 6 (EC:3.5.1.98) |
C00001180
|
| 8290 | HIST3H3, H3.4, H3/g, H3FT, H3t | histone cluster 3, H3 |
C00001180
|
| 121504 | HIST4H4, H4/p | histone cluster 4, H4 |
C00001180
|
| 3145 | HMBS, PBG-D, PBGD, PORC, UPS | hydroxymethylbilane synthase (EC:2.5.1.61) |
C00001180
|
| 10524 | KAT5, ESA1, HTATIP, HTATIP1, PLIP, TIP, TIP60, ZC2HC5, cPLA2 | K(lysine) acetyltransferase 5 (EC:2.3.1.48) |
C00001180
|
| 4254 | KITLG, FPH2, KL-1, Kitl, MGF, SCF, SF, SHEP7 | KIT ligand |
C00001180
|
| 4778 | NFE2, NF-E2, p45 | nuclear factor, erythroid 2 |
C00001180
|
| 11315 | PARK7, DJ-1, DJ1 | parkinson protein 7 |
C00001180
|
| 5553 | PRG2, BMPG, MBP, MBP1 | proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
C00001180
|
| 6427 | SRSF2, PR264, SC-35, SC35, SFRS2, SFRS2A, SRp30b | serine/arginine-rich splicing factor 2 |
C00001180
|
| 7043 | TGFB3, ARVD, ARVD1, TGF-beta3 | transforming growth factor, beta 3 |
C00001180
|
| 7096 | TLR1, CD281, TIL, TIL._LPRS5, rsc786 | toll-like receptor 1 |
C00001180
|
| 10333 | TLR6, CD286 | toll-like receptor 6 |
C00001180
|
Related Diseases
Diseases related to proteins in ChEMBL interactions
OMIM (169)
| OMIM | preferred title | UniProt |
|---|---|---|
| #300438 | 17-beta-hydroxysteroid dehydrogenase x deficiency |
Q99714
|
| #100100 | Abdominal muscles, absence of, with urinary tract abnormality and cryptorchidism |
P20309
|
| #202300 | Adrenocortical carcinoma, hereditary; adcc |
P04637
|
| #103470 | Albinism, ocular, with sensorineural deafness |
P14679
|
| #203100 | Albinism, oculocutaneous, type ia; oca1a |
P14679
|
| #606952 | Albinism, oculocutaneous, type ib; oca1b |
P14679
|
| #103780 | Alcohol dependence |
P08172
P14416 P31645 |
| #612069 | Amyotrophic lateral sclerosis 10, with or without frontotemporal dementia; als10 |
Q13148
|
| #614373 | Amyotrophic lateral sclerosis 16, juvenile; als16 |
Q99720
|
| #300068 | Androgen insensitivity syndrome; ais |
P10275
|
| #312300 | Androgen insensitivity, partial; pais |
P10275
|
| #609135 | Aplastic anemia |
O14746
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #600807 | Asthma, susceptibility to |
Q13093
|
| #608584 | Asthma-related traits, susceptibility to, 2 |
Q6W5P4
|
| #614740 | Basal cell carcinoma, susceptibility to, 7; bcc7 |
P04637
|
| #601816 | Bilirubin, serum level of, quantitative trait locus 1; biliqtl1 |
P22309
|
| #210900 | Bloom syndrome; blm |
P54132
|
| #602025 | Body mass index quantitative trait locus 9; bmiq9 |
P41968
|
| %606641 | Body mass index; bmi |
P37231
|
| #600430 | Brachydactyly-mental retardation syndrome; bdmr |
P56524
|
| #114480 | Breast cancer |
P31749
P38398 |
| #604370 | Breast-ovarian cancer, familial, susceptibility to, 1; brovca1 |
P38398
|
| #300615 | Brunner syndrome |
P21397
|
| #113970 | Burkitt lymphoma; bl |
P01106
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #609338 | Carotid intimal medial thickness 1 |
P37231
|
| #604121 | Cerebellar ataxia, deafness, and narcolepsy, autosomal dominant; adcadn |
P26358
|
| #118300 | Charcot-marie-tooth disease and deafness |
Q01453
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #118220 | Charcot-marie-tooth disease, demyelinating, type 1a; cmt1a |
Q01453
|
| #605479 | Cholestasis, benign recurrent intrahepatic, 2; bric2 |
O95342
|
| #601847 | Cholestasis, progressive familial intrahepatic, 2; pfic2 |
O95342
|
| #114500 | Colorectal cancer; crc |
P31749
P84022 |
| #300882 | Cornelia de lange syndrome 5; cdls5 |
Q9BY41
|
| #614466 | Coronary heart disease, susceptibility to, 6; chds6 |
P08254
|
| #615109 | Cowden syndrome 6; cws6 |
P31749
|
| #218800 | Crigler-najjar syndrome, type i |
P22309
P22310 |
| #606785 | Crigler-najjar syndrome, type ii |
P22309
P22310 |
| #162800 | Cyclic neutropenia |
P08246
|
| #612522 | Diabetes mellitus, insulin-dependent, 22; iddm22 |
P51681
|
| #119900 | Digital clubbing, isolated congenital |
P15428
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #237500 | Dubin-johnson syndrome; djs |
Q92887
|
| #613989 | Dyskeratosis congenita, autosomal dominant, 2; dkca2 |
O14746
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #133239 | Esophageal cancer |
P04637
|
| #615363 | Estrogen resistance; estrr |
P03372
|
| #612219 | Ewing sarcoma; es |
P11308
|
| #600274 | Frontotemporal dementia; ftd |
P10636
|
| #613163 | Gaba-transaminase deficiency |
P80404
|
| #230200 | Galactokinase deficiency |
P51570
|
| #613659 | Gastric cancer |
P04626
|
| #137215 | Gastric cancer, hereditary diffuse; hdgc |
P04626
|
| #231095 | Ghosal hematodiaphyseal dysplasia; ghdd |
P24557
|
| #143500 | Gilbert syndrome |
P22309
P22310 |
| #137800 | Glioma susceptibility 1; glm1 |
O75874
P04626 P37231 |
| #232300 | Glycogen storage disease ii |
P10253
|
| #139393 | Guillain-barre syndrome, familial; gbs |
Q01453
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #614519 | Hemorrhage, intracerebral, susceptibility to; ich |
P12821
|
| #114550 | Hepatocellular carcinoma |
P08581
|
| #609423 | Human immunodeficiency virus type 1, susceptibility to |
P41597
P51681 |
| #143100 | Huntington disease; hd |
P42858
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #237900 | Hyperbilirubinemia, transient familial neonatal; hblrtfn |
P22309
|
| #143860 | Hyperchlorhidrosis, isolated |
O43570
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #603373 | Hyperthyroidism, familial gestational |
P16473
|
| #609152 | Hyperthyroidism, nonautoimmune |
P16473
|
| #145900 | Hypertrophic neuropathy of dejerine-sottas |
Q01453
|
| #259100 | Hypertrophic osteoarthropathy, primary, autosomal recessive, 1; phoar1 |
P15428
|
| #146300 | Hypophosphatasia, adult |
P05186
|
| #241510 | Hypophosphatasia, childhood |
P05186
|
| #241500 | Hypophosphatasia, infantile |
P05186
|
| #275200 | Hypothyroidism, congenital, nongoitrous, 1; chng1 |
P16473
|
| #147050 | Ige responsiveness, atopic; iger |
Q13093
|
| #612244 | Inflammatory bowel disease 13; ibd13 |
P08183
|
| #614328 | Inflammatory skin and bowel disease, neonatal; nisbd |
P78536
|
| #603932 | Intervertebral disc disease; idd |
P14780
|
| #608232 | Leukemia, chronic myeloid; cml |
P00519
P11274 |
| #151623 | Li-fraumeni syndrome 1; lfs1 |
P04637
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #604367 | Lipodystrophy, familial partial, type 3; fpld3 |
P37231
|
| #613795 | Loeys-dietz syndrome, type 3; lds3 |
P84022
|
| #613688 | Long qt syndrome 2; lqt2 |
Q12809
|
| #211980 | Lung cancer |
P00533
P04626 P04637 |
| #608516 | Major depressive disorder; mdd |
P08172
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #613839 | Megaloblastic anemia due to dihydrofolate reductase deficiency |
P00374
|
| #615134 | Melanoma, cutaneous malignant, susceptibility to, 9; cmm9 |
O14746
|
| #614104 | Mental retardation, autosomal dominant 7; mrd7 |
Q13627
|
| #300705 | Mental retardation, x-linked 17; mrx17 |
Q99714
|
| %300852 | Mental retardation, x-linked 88; mrx88 |
P50052
|
| #300220 | Mental retardation, x-linked, syndromic 10; mrxs10 |
Q99714
|
| #613073 | Metaphyseal anadysplasia 2; mandp2 |
P14780
|
| #612624 | Microvascular complications of diabetes, susceptibility to, 3; mvcd3 |
P12821
|
| #259600 | Multicentric osteolysis, nodulosis, and arthropathy; mona |
P08253
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #126200 | Multiple sclerosis, susceptibility to; ms |
P08575
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #607948 | Mycobacterium tuberculosis, susceptibility to |
P11473
|
| #159900 | Myoclonic dystonia |
P14416
|
| #613076 | Myopathy, mitochondrial progressive, with congenital cataract, hearing loss, and developmental delay |
P55789
|
| #160900 | Myotonic dystrophy 1; dm1 |
Q9NR56
|
| #614116 | Neuropathy, hereditary sensory, type ie; hsn1e |
P26358
|
| #162500 | Neuropathy, hereditary, with liability to pressure palsies; hnpp |
Q01453
|
| #202700 | Neutropenia, severe congenital, 1, autosomal dominant; scn1 |
P08246
|
| #257200 | Niemann-pick disease, type a |
P17405
|
| #607616 | Niemann-pick disease, type b |
P17405
|
| #257220 | Niemann-pick disease, type c1; npc1 |
O15118
|
| #601665 | Obesity |
P32245
P37231 |
| #164230 | Obsessive-compulsive disorder; ocd |
P31645
|
| #604715 | Orthostatic intolerance |
P23975
|
| #259730 | Osteopetrosis, autosomal recessive 3; optb3 |
P00918
|
| #167000 | Ovarian cancer |
P04626
P38398 |
| #614320 | Pancreatic cancer, susceptibility to, 4; pnca4 |
P38398
|
| #260500 | Papilloma of choroid plexus; cpp |
P04637
|
| #260540 | Parkinson-dementia syndrome |
P10636
|
| #613135 | Parkinsonism-dystonia, infantile; pkdys |
Q01959
|
| #172700 | Pick disease of brain |
P10636
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #614278 | Platelet-activating factor acetylhydrolase deficiency; pafad |
Q13093
|
| #614390 | Pregnancy loss, recurrent, susceptibility to, 2; rprgl2 |
P00734
|
| #176920 | Proteus syndrome |
P31749
|
| #613679 | Prothrombin deficiency, congenital |
P00734
|
| #614742 | Pulmonary fibrosis and/or bone marrow failure, telomere-related, 1; pfbmft1 |
O14746
|
| #178500 | Pulmonary fibrosis, idiopathic; ipf |
O14746
|
| #605074 | Renal cell carcinoma, papillary, 1; rccp1 |
P08581
|
| #267430 | Renal tubular dysgenesis; rtd |
P12821
|
| #607276 | Resting heart rate, variation in |
P08588
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #600852 | Retinitis pigmentosa 17; rp17 |
P22748
|
| #268220 | Rhabdomyosarcoma 2; rms2 |
Q12778
|
| #604906 | Schizophrenia 9; sczd9 |
P49798
|
| #181500 | Schizophrenia; sczd |
P49798
|
| #608971 | Severe combined immunodeficiency, autosomal recessive, t cell-negative, b cell-positive, nk cell-positive |
P08575
|
| #609620 | Short qt syndrome 1; sqt1 |
Q12809
|
| #601800 | Skin/hair/eye pigmentation, variation in, 3; shep3 |
P14679
|
| #313200 | Spinal and bulbar muscular atrophy, x-linked 1; smax1 |
P10275
|
| #253300 | Spinal muscular atrophy, type i; sma1 |
Q16637
|
| #253550 | Spinal muscular atrophy, type ii; sma2 |
Q16637
|
| #253400 | Spinal muscular atrophy, type iii; sma3 |
Q16637
|
| #271150 | Spinal muscular atrophy, type iv; sma4 |
Q16637
|
| #605361 | Spinocerebellar ataxia 14; sca14 |
P05129
|
| #183090 | Spinocerebellar ataxia 2; sca2 |
Q99700
|
| #607250 | Spinocerebellar ataxia, autosomal recessive, with axonal neuropathy; scan1 |
Q9NUW8
|
| #275355 | Squamous cell carcinoma, head and neck; hnscc |
P04637
|
| #601367 | Stroke, ischemic |
P00734
P12821 P16109 P24723 |
| #271980 | Succinic semialdehyde dehydrogenase deficiency; ssadhd |
P51649
|
| #601104 | Supranuclear palsy, progressive, 1; psnp1 |
P10636
|
| #610460 | Thiopurine s-methyltransferase deficiency |
P51580
|
| #187950 | Thrombocythemia 1; thcyt1 |
P40225
|
| #188050 | Thrombophilia due to thrombin defect; thph1 |
P00734
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
| #190300 | Tremor, hereditary essential, 1; etm1 |
P35462
|
| #277440 | Vitamin d-dependent rickets, type 2a; vddr2a |
P11473
|
| #610379 | West nile virus, susceptibility to |
P51681
|
| #309585 | Wilson-turner x-linked mental retardation syndrome; wts |
Q9BY41
|
| #278300 | Xanthinuria, type i |
P47989
|
| #278750 | Xeroderma pigmentosum, variant type; xpv |
Q9Y253
|
| #112100 | Yt blood group antigen |
P22303
|
KEGG DISEASE (131)
| KEGG | name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
P04637 (related) |
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00764 | Cri du chat syndrome |
O14746
(related)
|
| H01132 | Aplastic anemia (AA) |
O14746
(related)
|
| H01299 | Idiopathic pulmonary fibrosis |
O14746
(related)
|
| H00022 | Bladder cancer |
O14746
(marker)
P00533 (related) P04626 (related) P04637 (related) |
| H00024 | Prostate cancer |
O14746
(marker)
P10275 (related) P11308 (related) |
| H00136 | Niemann-Pick disease type C (NPC) |
O15118
(related)
|
| H01302 | Hyperchlorhidrosis isolated (HCHLH) |
O43570
(related)
|
| H00021 | Renal cell carcinoma |
O43570
(marker)
P04637 (marker) P08581 (related) Q16790 (marker) |
| H00624 | Familial cholestasis |
O95342
(related)
|
| H01197 | Dihydrofolate reductase (DHFR) deficiency |
P00374
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
P00519
(related)
P00519 (marker) P01106 (related) P01106 (marker) P11274 (related) P11274 (marker) Q01196 (related) Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
P00519
(related)
P00519 (marker) P04637 (related) P11274 (related) P11274 (marker) Q01196 (related) |
| H00016 | Oral cancer |
P00533
(related)
P00533 (marker) P01106 (related) P04637 (related) P04637 (marker) |
| H00017 | Esophageal cancer |
P00533
(related)
P04637 (related) P04637 (marker) P35354 (related) |
| H00018 | Gastric cancer |
P00533
(related)
P04626 (related) P04637 (related) P08581 (related) P10415 (related) |
| H00028 | Choriocarcinoma |
P00533
(related)
P01106 (related) P03956 (related) P04626 (related) P04637 (related) P08253 (related) P10415 (related) |
| H00030 | Cervical cancer |
P00533
(related)
P04626 (related) P10415 (related) |
| H00042 | Glioma |
P00533
(related)
P00533 (marker) P04637 (related) P04637 (marker) |
| H00055 | Laryngeal cancer |
P00533
(related)
P00533 (marker) P01106 (related) P04637 (related) P04637 (marker) |
| H00223 | Inherited thrombophilia |
P00734
(related)
|
| H01254 | Congenital prothrombin deficiency |
P00734
(related)
|
| H00241 | Combined proximal and distal renal tubular acidosis (RTA type 3) |
P00918
(related)
|
| H00436 | Osteopetrosis |
P00918
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
P01106
(related)
Q03164 (related) |
| H00008 | Burkitt lymphoma |
P01106
(related)
P01106 (marker) P04637 (related) |
| H00010 | Multiple myeloma |
P01106
(related)
P04637 (related) |
| H00013 | Small cell lung cancer |
P01106
(related)
P04637 (related) P10415 (related) |
| H00025 | Penile cancer |
P01106
(related)
P04637 (related) P04637 (marker) P08253 (related) P14780 (related) P35354 (related) |
| H00027 | Ovarian cancer |
P01106
(related)
P04626 (related) P04637 (related) P38398 (related) |
| H00031 | Breast cancer |
P01106
(related)
P04626 (related) P04626 (marker) P04637 (related) P38398 (related) |
| H00036 | Osteosarcoma |
P01106
(related)
P04637 (related) P08684 (marker) |
| H00041 | Kaposi's sarcoma |
P01106
(related)
P04637 (related) P10415 (related) |
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
Q01453 (related) |
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
P37231 (related) |
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00026 | Endometrial Cancer |
P03372
(marker)
P04626 (related) P04637 (related) Q92731 (marker) |
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P04150
(related)
P11511 (related) |
| H00019 | Pancreatic cancer |
P04626
(related)
P04637 (related) P04637 (marker) |
| H00046 | Cholangiocarcinoma |
P04626
(related)
P04637 (related) P08581 (related) P35354 (related) |
| H00005 | Chronic lymphocytic leukemia (CLL) |
P04637
(related)
P10415 (related) P28907 (marker) |
| H00006 | Hairy-cell leukemia |
P04637
(related)
|
| H00009 | Adult T-cell leukemia |
P04637
(related)
|
| H00014 | Non-small cell lung cancer |
P04637
(related)
|
| H00015 | Malignant pleural mesothelioma |
P04637
(related)
|
| H00020 | Colorectal cancer |
P04637
(related)
P04637 (marker) |
| H00029 | Vulvar cancer |
P04637
(related)
|
| H00032 | Thyroid cancer |
P04637
(related)
P37231 (related) |
| H00038 | Malignant melanoma |
P04637
(related)
P14679 (marker) |
| H00039 | Basal cell carcinoma |
P04637
(related)
|
| H00040 | Squamous cell carcinoma |
P04637
(related)
|
| H00044 | Cancer of the anal canal |
P04637
(related)
|
| H00047 | Gallbladder cancer |
P04637
(related)
|
| H00048 | Hepatocellular carcinoma |
P04637
(related)
|
| H00881 | Li-Fraumeni syndrome |
P04637
(related)
|
| H01007 | Choroid plexus papilloma |
P04637
(related)
|
| H00063 | Spinocerebellar ataxia (SCA) |
P05129
(related)
Q99700 (related) Q9NUW8 (related) |
| H00213 | Hypophosphatasia |
P05186
(related)
|
| H00093 | Combined immunodeficiencies (CIDs) |
P06239
(related)
|
| H00079 | Asthma |
P07550
(related)
|
| H00100 | Neutropenic disorders |
P08246
(related)
|
| H00472 | Torg-Winchester syndrome |
P08253
(related)
|
| H00091 | T-B+Severe combined immunodeficiencies (SCIDs) |
P08575
(related)
|
| H00069 | Glycogen storage diseases (GSD) |
P10253
(related)
|
| H00062 | Spinal and bulbar muscular atrophy (SBMA) |
P10275
(related)
|
| H00608 | 46,XY disorders of sex development (Disorders in androgen synthesis or action) |
P10275
(related)
|
| H00609 | 46,XY disorders of sex development (Other) |
P10275
(related)
|
| H00054 | Nasopharyngeal cancer |
P10415
(related)
|
| H00058 | Amyotrophic lateral sclerosis (ALS) |
P10636
(related)
Q13148 (related) |
| H00077 | Progressive supranuclear palsy (PSP) |
P10636
(related)
|
| H00078 | Frontotemporal lobar degeneration (FTLD) |
P10636
(related)
|
| H00023 | Testicular cancer |
P10696
(marker)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00035 | Ewing's sarcoma |
P11308
(related)
|
| H00342 | Tuberculosis |
P11473
(related)
|
| H00784 | Localized autosomal recessive hypotrichosis |
P11473
(related)
|
| H01143 | Vitamin D-dependent rickets |
P11473
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00083 | Allograft rejection |
P12821
(related)
|
| H00575 | Renal tubular dysgenesis |
P12821
(related)
|
| H00168 | Oculocutaneous albinism (OCA) |
P14679
(related)
|
| H00479 | Metaphyseal dysplasias |
P14780
(related)
|
| H00457 | Primary hypertrophic osteoarthropathy (PHO) |
P15428
(related)
|
| H01246 | Isolated congenital nail clubbing (ICNC) |
P15428
(related)
|
| H00250 | Congenital nongoitrous hypothyroidism (CHNG) |
P16473
(related)
|
| H01269 | Congenital hyperthyroidism |
P16473
(related)
|
| H00137 | Niemann-Pick disease (NPD) typeA and B |
P17405
(related)
|
| H00424 | Defects in the degradation of sphingomyelin |
P17405
(related)
|
| H00548 | Brunner syndrome |
P21397
(related)
|
| H00208 | Hyperbilirubinemia |
P22309
(related)
Q92887 (related) |
| H00527 | Retinitis pigmentosa (RP) |
P22748
(related)
|
| H01031 | Orthostatic intolerance (OI) |
P23975
(related)
|
| H00490 | Diaphyseal dysplasia with anemia (Ghosal) |
P24557
(related)
|
| H00539 | PTEN hamartoma tumor syndrome (PHTS) |
P31749
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00409 | Type II diabetes mellitus |
P37231
(related)
|
| H00227 | Congenital amegakaryocytic thrombocytopenia (CAMT) |
P40225
(marker)
|
| H00059 | Huntington's disease (HD) |
P42858
(related)
|
| H00192 | Xanthinuria |
P47989
(related)
|
| H00480 | Non-syndromic X-linked mental retardation |
P50052
(related)
Q99714 (related) |
| H00070 | Galactosemia |
P51570
(related)
|
| H00964 | Thiopurine S-methyltransferase deficiency (TPMT deficiency) |
P51580
(related)
|
| H00835 | Succinic semialdehyde dehydrogenase (SSADH) deficiency |
P51649
(related)
|
| H00094 | DNA repair defects |
P54132
(related)
|
| H00296 | Defects in RecQ helicases |
P54132
(related)
|
| H00561 | Brachydacytly-mental retardation syndrome and Smith-Magenis syndrome |
P56524
(related)
|
| H01257 | GABA-transaminase deficiency |
P80404
(related)
|
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H01296 | Hereditary neuropathy with liability to pressure palsies (HNPP) |
Q01453
(related)
|
| H00408 | Type I diabetes mellitus |
Q04759
(related)
|
| H00037 | Alveolar rhabdomyosarcoma |
Q12778
(related)
Q12778 (marker) |
| H00720 | Long QT syndrome |
Q12809
(related)
|
| H00725 | Short QT syndrome |
Q12809
(related)
|
| H00455 | Spinal muscular atrophy (SMA) |
Q16637
(related)
|
| H01065 | Pentosuria |
Q7Z4W1
(related)
|
| H00658 | Syndromic X-linked mental retardation |
Q99714
(related)
|
| H00925 | 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency |
Q99714
(related)
|
| H00403 | Disorders of nucleotide excision repair |
Q9Y253
(related)
|
Diseases related to CTD interactions
211 disease in interactions with metabolites
| MESH or OMIM | name |
KNApSAcK
metabolite |
|---|---|---|
| D056486 | Drug-Induced Liver Injury |
C00000100
C00000958 C00002903 C00002776 C00000947 |
| D001943 | Breast Neoplasms |
C00000958
C00002903 C00002902 C00000947 |
| D007249 | Inflammation |
C00000958
C00002903 C00015693 C00002902 |
| D009362 | Neoplasm Metastasis |
C00002903
C00002902 C00000947 C00002647 |
| D009361 | Neoplasm Invasiveness |
C00002903
C00002902 C00002647 |
| D000236 | Adenoma |
C00000958
C00002903 C00002902 |
| D050197 | Atherosclerosis |
C00000958
C00002903 C00000947 |
| D012878 | Skin Neoplasms |
C00000958
C00002903 C00002902 |
| D004487 | Edema |
C00002903
C00015693 C00002776 |
| D015464 | Leukemia, Myelogenous, Chronic, BCR-ABL Positive |
C00002903
C00002902 C00002647 |
| D009203 | Myocardial Infarction |
C00000958
C00002903 C00002647 |
| D002471 | Cell Transformation, Neoplastic |
C00002903
C00002902 C00000947 |
| D003110 | Colonic Neoplasms |
C00000958
C00002903 C00002902 |
| D015179 | Colorectal Neoplasms |
C00000958
C00002903 C00002902 |
| D006331 | Heart Diseases |
C00000958
C00002903 C00000947 |
| D009134 | Muscular Atrophy, Spinal |
C00000958
C00002903 |
| D003924 | Diabetes Mellitus, Type 2 |
C00000958
C00002903 |
| D008113 | Liver Neoplasms |
C00002903
C00000947 |
| D008325 | Mammary Neoplasms, Experimental |
C00002903
C00000947 |
| D008106 | Liver Cirrhosis, Experimental |
C00000958
C00002903 |
| D008175 | Lung Neoplasms |
C00000958
C00001180 |
| D008545 | Melanoma |
C00000958
C00002903 |
| D018450 | Disease Progression |
C00000958
C00002903 |
| D009069 | Movement Disorders |
C00002903
C00002647 |
| D008107 | Liver Diseases |
C00002903
C00000947 |
| D002294 | Carcinoma, Squamous Cell |
C00000958
C00002903 |
| D015228 | Hypertriglyceridemia |
C00002903
C00002647 |
| D011230 | Precancerous Conditions |
C00000958
C00002903 |
| D006949 | Hyperlipidemias |
C00002903
C00002647 |
| D011658 | Pulmonary Fibrosis |
C00000958
C00002903 |
| D001008 | Anxiety Disorders |
C00000958
C00001180 |
| D020138 | HYPERHOMOCYSTEINEMIA |
C00002903
C00000947 |
| D058739 | Aberrant Crypt Foci |
C00002903
C00002902 |
| D006943 | Hyperglycemia |
C00002903
C00002647 |
| D006937 | Hypercholesterolemia |
C00002903
C00002647 |
| D018149 | Glucose Intolerance |
C00002903
C00002647 |
| D005234 | Fatty Liver |
C00002903
C00002647 |
| D010190 | Pancreatic Neoplasms |
C00002903
C00002902 |
| D050171 | Dyslipidemias |
C00002903
C00002647 |
| D003921 | Diabetes Mellitus, Experimental |
C00002903
C00002647 |
| D012516 | Osteosarcoma |
C00002902
C00002647 |
| D000860 | Anoxia |
C00002903
C00002776 |
| D006528 | Carcinoma, Hepatocellular |
C00002903
C00002902 |
| D001930 | Brain Injuries |
C00002903
C00002776 |
| D013274 | Stomach Neoplasms |
C00002903
|
| D001862 | Bone Resorption |
C00002903
|
| D001919 | Bradycardia |
C00002903
|
| D020520 | Brain Infarction |
C00002903
|
| D001169 | Arthritis, Experimental |
C00002903
|
| D002545 | Brain Ischemia |
C00002903
|
| D002277 | Carcinoma |
C00002903
|
| D044584 | Carcinoma, Ductal |
C00002903
|
| D017544 | Aortic Aneurysm, Abdominal |
C00002903
|
| D018827 | Carcinoma, Lewis Lung |
C00002903
|
| D002292 | Carcinoma, Renal Cell |
C00002903
|
| C535575 | Carcinoma, squamous cell of head and neck |
C00002903
|
| D006332 | Cardiomegaly |
C00002903
|
| D009202 | Cardiomyopathies |
C00002903
|
| D002318 | Cardiovascular Diseases |
C00002903
|
| D020212 | Carotid Artery Injuries |
C00002903
|
| D002357 | Cartilage Diseases |
C00002903
|
| D000740 | Anemia |
C00002903
|
| D002544 | Cerebral Infarction |
C00002903
|
| C537948 | Ceroid lipofuscinosis, neuronal 1, infantile |
C00002903
|
| D002779 | Cholestasis |
C00002903
|
| D003072 | Cognition Disorders |
C00002903
|
| D003092 | Colitis |
C00002903
|
| D003093 | Colitis, Ulcerative |
C00002903
|
| D003327 | Coronary Disease |
C00002903
|
| D003643 | Death |
C00002903
|
| D003920 | Diabetes Mellitus |
C00002903
|
| D000707 | Anaphylaxis |
C00002903
|
| D003922 | Diabetes Mellitus, Type 1 |
C00002903
|
| D003925 | Diabetic Angiopathies |
C00002903
|
| D058065 | Diabetic Cardiomyopathies |
C00002903
|
| D003928 | Diabetic Nephropathies |
C00002903
|
| D003929 | Diabetic Neuropathies |
C00002903
|
| D003930 | Diabetic Retinopathy |
C00002903
|
| D064420 | Drug-Related Side Effects and Adverse Reactions |
C00002903
|
| D000544 | Alzheimer Disease |
C00002903
|
| D000419 | Albuminuria |
C00002903
|
| D004489 | Edema, Cardiac |
C00002903
|
| D016769 | Embolism and Thrombosis |
C00002903
|
| D004681 | Encephalomyelitis, Autoimmune, Experimental |
C00002903
|
| D020345 | Enterocolitis, Necrotizing |
C00002903
|
| D020031 | Epstein-Barr Virus Infections |
C00002903
|
| D004938 | Esophageal Neoplasms |
C00002903
|
| D005199 | Fanconi Anemia |
C00002903
|
| D011125 | Adenomatous Polyposis Coli |
C00002903
|
| D005355 | Fibrosis |
C00002903
|
| D005512 | Food Hypersensitivity |
C00002903
|
| D005621 | Friedreich Ataxia |
C00002903
|
| D005909 | Glioblastoma |
C00002903
|
| D005910 | Glioma |
C00002903
|
| C538231 | Adenocarcinoma of lung |
C00002903
|
| D006470 | Hemorrhage |
C00002903
|
| D006505 | Hepatitis |
C00002903
|
| D006529 | Hepatomegaly |
C00002903
|
| D006561 | Herpes Simplex |
C00002903
|
| D015658 | HIV Infections |
C00002903
|
| D006930 | Hyperalgesia |
C00002903
|
| D055371 | Acute Lung Injury |
C00002903
|
| D058186 | Acute Kidney Injury |
C00002903
|
| D013375 | Substance Withdrawal Syndrome |
C00000958
|
| D006946 | Hyperinsulinism |
C00002903
|
| D011472 | Prostatitis |
C00000958
|
| D006965 | Hyperplasia |
C00002903
|
| D006973 | Hypertension |
C00002903
|
| D006976 | Hypertension, Pulmonary |
C00002903
|
| D020343 | Hypertensive Encephalopathy |
C00002903
|
| C563161 | Hypertensive Nephropathy |
C00002903
|
| D011041 | Poisoning |
C00000958
|
| D006984 | Hypertrophy |
C00002903
|
| D017379 | Hypertrophy, Left Ventricular |
C00002903
|
| D017380 | Hypertrophy, Right Ventricular |
C00002903
|
| D020244 | Infarction, Middle Cerebral Artery |
C00002903
|
| D007251 | Influenza, Human |
C00002903
|
| D007333 | Insulin Resistance |
C00002903
|
| D007511 | Ischemia |
C00002903
|
| D002546 | Ischemic Attack, Transient |
C00002903
|
| D007674 | Kidney Diseases |
C00002903
|
| D007945 | Leukemia, Lymphoid |
C00002903
|
| D007948 | Leukemia, Monocytic, Acute |
C00002903
|
| D056627 | Peritoneal Fibrosis |
C00000958
|
| D015470 | Leukemia, Myeloid, Acute |
C00002903
|
| D015473 | Leukemia, Promyelocytic, Acute |
C00002903
|
| D015458 | Leukemia, T-Cell |
C00002903
|
| D008103 | Liver Cirrhosis |
C00002903
|
| D009336 | Necrosis |
C00000958
|
| D008108 | Liver Diseases, Alcoholic |
C00002903
|
| D006948 | Hyperkinesis |
C00000958
|
| D008114 | Liver Neoplasms, Experimental |
C00002903
|
| D008171 | Lung Diseases |
C00002903
|
| D008206 | Lymphatic Diseases |
C00002903
|
| D016393 | Lymphoma, B-Cell |
C00002903
|
| D008228 | Lymphoma, Non-Hodgkin |
C00002903
|
| D015674 | Mammary Neoplasms, Animal |
C00002903
|
| D019970 | Cocaine-Related Disorders |
C00000958
|
| D008569 | Memory Disorders |
C00002903
|
| C562839 | Mesothelioma, Malignant |
C00002903
|
| D008659 | Metabolic Diseases |
C00002903
|
| D028361 | Mitochondrial Diseases |
C00002903
|
| D002312 | Cardiomyopathy, Hypertrophic |
C00000958
|
| D009101 | Multiple Myeloma |
C00002903
|
| D009133 | Muscular Atrophy |
C00002903
|
| D015428 | Myocardial Reperfusion Injury |
C00002903
|
| D009205 | Myocarditis |
C00002903
|
| C538339 | Nasopharyngeal carcinoma |
C00002903
|
| D001327 | Autoimmune Diseases |
C00000958
|
| D001249 | Asthma |
C00000958
|
| D009369 | Neoplasms |
C00002903
|
| D009374 | Neoplasms, Experimental |
C00002903
|
| D009389 | Neovascularization, Pathologic |
C00002903
|
| D009410 | Nerve Degeneration |
C00002903
|
| D019636 | Neurodegenerative Diseases |
C00002903
|
| D020078 | Neurogenic Inflammation |
C00002903
|
| C541083 | Non-alcoholic Fatty Liver Disease |
C00002903
|
| D009765 | Obesity |
C00002903
|
| D009293 | Opioid-Related Disorders |
C00002903
|
| D010003 | Osteoarthritis |
C00002903
|
| D020370 | Osteoarthritis, Knee |
C00002903
|
| D015663 | OSTEOPOROSIS, POSTMENOPAUSAL |
C00002903
|
| D010051 | Ovarian Neoplasms |
C00002903
|
| D012220 | Rhinitis |
C00000856
|
| D010195 | Pancreatitis |
C00002903
|
| D010302 | Parkinson Disease, Secondary |
C00002903
|
| D020734 | Parkinsonian Disorders |
C00002903
|
| D010538 | Peritonitis |
C00002903
|
| D015452 | Precursor B-Cell Lymphoblastic Leukemia-Lymphoma |
C00002903
|
| D054218 | Precursor T-Cell Lymphoblastic Leukemia-Lymphoma |
C00002903
|
| D011225 | Pre-Eclampsia |
C00002903
|
| D011297 | Prenatal Exposure Delayed Effects |
C00002903
|
| D015175 | Prolactinoma |
C00002903
|
| D011471 | Prostatic Neoplasms |
C00002903
|
| C536282 | Pulmonary arterial hypertension |
C00002903
|
| D011654 | Pulmonary Edema |
C00002903
|
| D051436 | Renal Insufficiency, Chronic |
C00002903
|
| D015427 | Reperfusion Injury |
C00002903
|
| D018357 | Respiratory Syncytial Virus Infections |
C00002903
|
| D012140 | Respiratory Tract Diseases |
C00002903
|
| D018805 | Sepsis |
C00002903
|
| D012751 | Sezary Syndrome |
C00002903
|
| D012771 | Shock, Hemorrhagic |
C00002903
|
| D001172 | Arthritis, Rheumatoid |
C00002903
|
| D013276 | Stomach Ulcer |
C00002903
|
| D020521 | Stroke |
C00002903
|
| D013724 | Teratoma |
C00002903
|
| C536914 | Thyroid cancer, medullary |
C00002903
|
| D014062 | Tongue Neoplasms |
C00002903
|
| D002583 | Uterine Cervical Neoplasms |
C00002903
|
| D014594 | Uterine Neoplasms |
C00002903
|
| D018754 | Ventricular Dysfunction |
C00002903
|
| D018497 | Ventricular Dysfunction, Right |
C00002903
|
| D020257 | Ventricular Remodeling |
C00002903
|
| D008258 | Waldenstrom Macroglobulinemia |
C00002903
|
| D019282 | Wasting Syndrome |
C00002903
|
| D015430 | Weight Gain |
C00002903
|
| D015431 | Weight Loss |
C00002903
|
| D014898 | Werner Syndrome |
C00002903
|
| D014947 | Wounds and Injuries |
C00002903
|
| D000647 | Amnesia |
C00002776
|
| D006461 | Hemolysis |
C00002648
|
| D010212 | Papilloma |
C00002902
|
| D003556 | Cystitis |
C00000947
|
| D034381 | Hearing Loss |
C00000947
|
| D017114 | Liver Failure, Acute |
C00000947
|
| D008546 | Melanoma, Experimental |
C00002647
|
| D010182 | Pancreatic Diseases |
C00002647
|
| D000230 | Adenocarcinoma |
C00001180
|
| D009139 | Musculoskeletal Abnormalities |
C00001180
|
| D012021 | Reflex, Abnormal |
C00001180
|