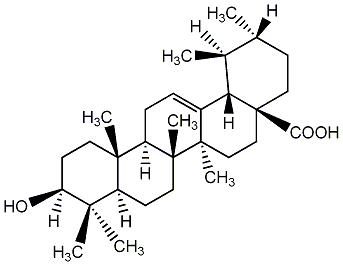
Metabolite
KNApSAcK Entry
| id | C00003558 |
|---|---|
| Name | Ursolic acid |
| CAS RN | 77-52-1 |
| Standard InChI | InChI=1S/C30H48O3/c1-18-10-15-30(25(32)33)17-16-28(6)20(24(30)19(18)2)8-9-22-27(5)13-12-23(31)26(3,4)21(27)11-14-29(22,28)7/h8,18-19,21-24,31H,9-17H2,1-7H3,(H,32,33)/t18-,19+,21+,22-,23+,24+,27+,28-,29-,30+/m1/s1 |
| Standard InChI (Main Layer) | InChI=1S/C30H48O3/c1-18-10-15-30(25(32)33)17-16-28(6)20(24(30)19(18)2)8-9-22-27(5)13-12-23(31)26(3,4)21(27)11-14-29(22,28)7/h8,18-19,21-24,31H,9-17H2,1-7H3,(H,32,33) |
Cluster
| Phytochemical cluster | No. 51 |
|---|---|
| KCF-S cluster | No. 13 |
Link
ChEMBL
| By standard InChI | CHEMBL169 |
|---|---|
| By standard InChI Main Layer | CHEMBL297810 CHEMBL56048 CHEMBL300594 CHEMBL169 CHEMBL176234 CHEMBL491715 CHEMBL1316667 CHEMBL1555307 CHEMBL1593360 CHEMBL1979720 |
KEGG
| By LinkDB | C08988 |
|---|
CTD
| By CAS RN | C005466 |
|---|
Species
Summary
Plant class
| class name | count |
|---|---|
| asterids | 26 |
| rosids | 14 |
| Euphyllophyta | 1 |
Family (Top 10)
| family name | count |
|---|---|
| Lamiaceae | 10 |
| Rosaceae | 6 |
| Rubiaceae | 6 |
| Moraceae | 2 |
| Myrtaceae | 2 |
| Ericaceae | 2 |
| Asteraceae | 2 |
| Nothofagaceae | 1 |
| Stilbaceae | 1 |
| Apocynaceae | 1 |
List (41)
* NCBI
Human Protein / Gene in interaction
47 ChEMBL Protein in interactions
| accession | description | class description | compound | assay ID (# of activities) |
# of diseases
(OMIM / KEGG) |
|---|---|---|---|---|---|
| P10635 | Cytochrome P450 2D6 | Cytochrome P450 2D6 | CHEMBL1316667 |
CHEMBL1741321
(1)
|
1 / 0 |
| P18031 | Tyrosine-protein phosphatase non-receptor type 1 | Tyr | CHEMBL169 |
CHEMBL904966
(1)
CHEMBL995832
(1)
CHEMBL972808 (1) CHEMBL1036591 (1) CHEMBL963010 (1) CHEMBL1036431 (1) CHEMBL1041807 (1) CHEMBL1041808 (2) CHEMBL1041717 (1) CHEMBL1101504 (1) CHEMBL1115822 (1) CHEMBL1781409 (1) CHEMBL1816250 (1) CHEMBL1828245 (1) CHEMBL2089751 (1) CHEMBL2149371 (1) CHEMBL2149372 (2) CHEMBL2149377 (1) CHEMBL2162374 (1) CHEMBL2210497 (1) CHEMBL2344762 (1) CHEMBL2344763 (1) |
0 / 0 |
| O60218 | Aldo-keto reductase family 1 member B10 | Enzyme | CHEMBL169 |
CHEMBL2168780
(1)
CHEMBL2168783
(1)
CHEMBL2169016 (1) |
0 / 0 |
| P35354 | Prostaglandin G/H synthase 2 | Oxidoreductase | CHEMBL169 |
CHEMBL1014032
(1)
CHEMBL1003453
(1)
CHEMBL1003458 (1) CHEMBL1003459 (1) |
0 / 3 |
| P06746 | DNA polymerase beta | Enzyme | CHEMBL169 |
CHEMBL1002742
(1)
|
0 / 0 |
| Q13547 | Histone deacetylase 1 | Hydrolase | CHEMBL169 |
CHEMBL1785663
(1)
|
0 / 0 |
| P24666 | Low molecular weight phosphotyrosine protein phosphatase | Tyr | CHEMBL169 |
CHEMBL2149373
(1)
CHEMBL2149374
(1)
|
0 / 0 |
| Q8TDU6 | G-protein coupled bile acid receptor 1 | Steroid-like ligand receptor | CHEMBL169 |
CHEMBL1041788
(1)
CHEMBL1041789
(1)
|
0 / 0 |
| O75604 | Ubiquitin carboxyl-terminal hydrolase 2 | Enzyme | CHEMBL1593360 |
CHEMBL1614331
(1)
|
0 / 0 |
| P02545 | Prelamin-A/C | Unclassified protein | CHEMBL1593360 |
CHEMBL1614544
(1)
|
11 / 10 |
| P08047 | Transcription factor Sp1 | Unclassified protein | CHEMBL169 |
CHEMBL1058704
(1)
|
0 / 0 |
| P10828 | Thyroid hormone receptor beta | NR1A2 | CHEMBL1316667 |
CHEMBL1613776
(1)
|
3 / 1 |
| P10586 | Receptor-type tyrosine-protein phosphatase F | Receptor tyrosine-protein phosphatase | CHEMBL169 |
CHEMBL963013
(1)
CHEMBL1828251
(1)
CHEMBL2149376 (1) |
0 / 0 |
| P11712 | Cytochrome P450 2C9 | Cytochrome P450 2C9 | CHEMBL1316667 |
CHEMBL1741325
(1)
|
0 / 1 |
| P23219 | Prostaglandin G/H synthase 1 | Oxidoreductase | CHEMBL169 |
CHEMBL1003454
(1)
CHEMBL1003456
(1)
CHEMBL1003457 (1) |
0 / 0 |
| P28845 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | Enzyme | CHEMBL169 |
CHEMBL1072762
(1)
CHEMBL1065225
(1)
|
1 / 1 |
| Q9UQL6 | Histone deacetylase 5 | Hydrolase | CHEMBL169 |
CHEMBL1785672
(1)
|
0 / 0 |
| Q92830 | Histone acetyltransferase KAT2A | Enzyme | CHEMBL169 |
CHEMBL1738606
(1)
|
0 / 0 |
| P15121 | Aldose reductase | Enzyme | CHEMBL169 |
CHEMBL2168781
(1)
|
0 / 0 |
| P49841 | Glycogen synthase kinase-3 beta | Gsk | CHEMBL169 |
CHEMBL1053705
(1)
|
0 / 0 |
| Q04206 | Transcription factor p65 | Transcription Factor | CHEMBL169 |
CHEMBL1109182
(1)
|
0 / 0 |
| P83916 | Chromobox protein homolog 1 | Unclassified protein | CHEMBL169 |
CHEMBL1794401
(1)
|
0 / 0 |
| O94782 | Ubiquitin carboxyl-terminal hydrolase 1 | Enzyme | CHEMBL1593360 |
CHEMBL1794467
(1)
|
0 / 0 |
| P11511 | Cytochrome P450 19A1 | Cytochrome P450 19A1 | CHEMBL169 |
CHEMBL978973
(1)
CHEMBL978974
(1)
CHEMBL978975 (1) |
2 / 2 |
| Q96RI1 | Bile acid receptor | NR1H4 | CHEMBL169 |
CHEMBL1041790
(1)
|
0 / 0 |
| O15379 | Histone deacetylase 3 | Hydrolase | CHEMBL169 |
CHEMBL1785662
(1)
|
0 / 0 |
| P18433 | Receptor-type tyrosine-protein phosphatase alpha | Receptor tyrosine-protein phosphatase | CHEMBL169 |
CHEMBL963014
(1)
CHEMBL1828250
(1)
|
0 / 0 |
| P05177 | Cytochrome P450 1A2 | Cytochrome P450 1A2 | CHEMBL1316667 |
CHEMBL1741322
(1)
|
0 / 0 |
| P56524 | Histone deacetylase 4 | Hydrolase | CHEMBL169 |
CHEMBL1785671
(1)
|
1 / 1 |
| P19838 | Nuclear factor NF-kappa-B p105 subunit | Transcription Factor | CHEMBL169 |
CHEMBL1109181
(1)
|
0 / 0 |
| P30305 | M-phase inducer phosphatase 2 | Ser_Thr_Tyr | CHEMBL169 |
CHEMBL1828247
(1)
|
0 / 0 |
| P33261 | Cytochrome P450 2C19 | Cytochrome P450 2C19 | CHEMBL1316667 |
CHEMBL1741323
(1)
|
1 / 1 |
| P08684 | Cytochrome P450 3A4 | Cytochrome P450 3A4 | CHEMBL1316667 |
CHEMBL1741324
(1)
|
0 / 1 |
| P27695 | DNA-(apurinic or apyrimidinic site) lyase | Enzyme | CHEMBL169 |
CHEMBL1614211
(1)
|
0 / 0 |
| Q07869 | Peroxisome proliferator-activated receptor alpha | NR1C1 | CHEMBL169 |
CHEMBL1837228
(1)
CHEMBL1837229
(2)
CHEMBL1837230 (1) CHEMBL1837231 (1) CHEMBL1837232 (1) CHEMBL1837233 (1) CHEMBL1839605 (1) CHEMBL1839606 (1) CHEMBL1839607 (1) CHEMBL1839608 (1) CHEMBL1839609 (1) CHEMBL1839610 (1) CHEMBL1839611 (1) CHEMBL1839612 (1) CHEMBL1839613 (1) CHEMBL1839614 (1) CHEMBL1839615 (1) |
0 / 0 |
| P17706 | Tyrosine-protein phosphatase non-receptor type 2 | Tyr | CHEMBL169 |
CHEMBL963011
(1)
CHEMBL1828246
(1)
CHEMBL2344761 (1) |
0 / 1 |
| Q9UBN7 | Histone deacetylase 6 | Hydrolase | CHEMBL169 |
CHEMBL1785670
(1)
|
0 / 0 |
| P29350 | Tyrosine-protein phosphatase non-receptor type 6 | Tyr | CHEMBL169 |
CHEMBL963012
(1)
CHEMBL1828248
(1)
|
0 / 0 |
| P80365 | Corticosteroid 11-beta-dehydrogenase isozyme 2 | Enzyme | CHEMBL169 |
CHEMBL1072765
(1)
|
1 / 1 |
| P23469 | Receptor-type tyrosine-protein phosphatase epsilon | Receptor tyrosine-protein phosphatase | CHEMBL169 |
CHEMBL963015
(1)
|
0 / 0 |
| P46063 | ATP-dependent DNA helicase Q1 | Enzyme | CHEMBL169 |
CHEMBL1613829
(1)
|
0 / 0 |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | Enzyme | CHEMBL169 |
CHEMBL2354311
(1)
|
1 / 0 |
| O00255 | Menin | Unclassified protein | CHEMBL169 CHEMBL1316667 |
CHEMBL1614257
(2)
CHEMBL1614531
(1)
|
2 / 5 |
| Q03164 | Histone-lysine N-methyltransferase 2A | Enzyme | CHEMBL169 CHEMBL1316667 |
CHEMBL1614257
(2)
CHEMBL1614531
(1)
|
1 / 3 |
| Q13951 | Core-binding factor subunit beta | Unclassified protein | CHEMBL1316667 |
CHEMBL1613933
(1)
|
0 / 1 |
| Q01196 | Runt-related transcription factor 1 | Unclassified protein | CHEMBL1316667 |
CHEMBL1613933
(1)
|
1 / 6 |
| O94925 | Glutaminase kidney isoform, mitochondrial | Enzyme | CHEMBL169 |
CHEMBL2114738
(1)
|
0 / 0 |
CTD interaction (20)
| compound | gene | gene name | gene description | interaction | interaction type | form |
reference
pmid |
|---|---|---|---|---|---|---|---|
| C005466 | 9131 |
AIFM1
AIF CMTX4 COWCK COXPD6 PDCD8 |
apoptosis-inducing factor, mitochondrion-associated, 1 | AIFM1 mutant form results in decreased susceptibility to ursolic acid |
decreases response to substance
|
mutant form |
20630652
|
| C005466 | 9131 |
AIFM1
AIF CMTX4 COWCK COXPD6 PDCD8 |
apoptosis-inducing factor, mitochondrion-associated, 1 | ursolic acid results in increased localization of AIFM1 protein |
increases localization
|
protein |
20630652
|
| C005466 | 578 |
BAK1
BAK BAK-LIKE BCL2L7 CDN1 |
BCL2-antagonist/killer 1 | ursolic acid results in increased activity of BAK1 protein |
increases activity
|
protein |
20630652
|
| C005466 | 581 |
BAX
BCL2L4 |
BCL2-associated X protein | ursolic acid results in increased expression of BAX protein |
increases expression
|
protein |
16012738
|
| C005466 | 596 |
BCL2
Bcl-2 PPP1R50 |
B-cell CLL/lymphoma 2 | ursolic acid results in decreased expression of BCL2 protein |
decreases expression
|
protein |
16012738
|
| C005466 | 836 |
CASP3
CPP32 CPP32B SCA-1 |
caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) | acetyl-aspartyl-glutamyl-valyl-aspartal inhibits the reaction [ursolic acid results in increased cleavage of and results in increased activity of CASP3 protein] |
decreases reaction
/ increases activity / increases cleavage |
protein |
16012738
|
| C005466 | 836 |
CASP3
CPP32 CPP32B SCA-1 |
caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) | ursolic acid results in increased activity of CASP3 protein |
increases activity
|
protein |
20005942
|
| C005466 | 836 |
CASP3
CPP32 CPP32B SCA-1 |
caspase 3, apoptosis-related cysteine peptidase (EC:3.4.22.56) | ursolic acid results in increased cleavage of and results in increased activity of CASP3 protein |
increases activity
/ increases cleavage |
protein |
16012738
|
| C005466 | 841 |
CASP8
ALPS2B CAP4 Casp-8 FLICE MACH MCH5 |
caspase 8, apoptosis-related cysteine peptidase (EC:3.4.22.61) | ursolic acid results in increased activity of CASP8 protein |
increases activity
|
protein |
20005942
|
| C005466 | 54205 |
CYCS
CYC HCS THC4 |
cytochrome c, somatic | ursolic acid affects the localization of CYCS protein |
affects localization
|
protein |
16012738
|
| C005466 | 1557 |
CYP2C19
CPCJ CYP2C P450C2C P450IIC19 |
cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) | ursolic acid inhibits the reaction [CYP2C19 protein results in increased metabolism of Mephenytoin] |
decreases reaction
/ increases metabolic processing |
protein |
15043991
|
| C005466 | 1557 |
CYP2C19
CPCJ CYP2C P450C2C P450IIC19 |
cytochrome P450, family 2, subfamily C, polypeptide 19 (EC:1.14.13.48 1.14.13.49 1.14.13.80) | ursolic acid results in decreased activity of CYP2C19 protein |
decreases activity
|
protein |
15043991
|
| C005466 | 3383 |
ICAM1
BB2 CD54 P3.58 |
intercellular adhesion molecule 1 | ursolic acid results in decreased expression of ICAM1 mRNA |
decreases expression
|
mRNA |
21539908
|
| C005466 | 3383 |
ICAM1
BB2 CD54 P3.58 |
intercellular adhesion molecule 1 | ursolic acid results in decreased expression of ICAM1 protein |
decreases expression
|
protein |
20005942
|
| C005466 | 4313 |
MMP2
CLG4 CLG4A MMP-II MONA TBE-1 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (EC:3.4.24.24) | ursolic acid results in decreased expression of MMP2 mRNA |
decreases expression
|
mRNA |
21539908
|
| C005466 | 4318 |
MMP9
CLG4B GELB MANDP2 MMP-9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (EC:3.4.24.35) | ursolic acid results in decreased expression of MMP9 mRNA |
decreases expression
|
mRNA |
21539908
|
| C005466 | 142 |
PARP1
ADPRT ADPRT_1 ADPRT1 ARTD1 PARP PARP-1 PPOL pADPRT-1 |
poly (ADP-ribose) polymerase 1 (EC:2.4.2.30) | ursolic acid results in increased cleavage of PARP1 protein |
increases cleavage
|
protein |
16012738
|
| C005466 | 7040 |
TGFB1
CED DPD1 LAP TGFB TGFbeta |
transforming growth factor, beta 1 | ursolic acid results in decreased expression of TGFB1 protein |
decreases expression
|
protein |
21539908
|
| C005466 | 7150 |
TOP1
TOPI |
topoisomerase (DNA) I (EC:5.99.1.2) | ursolic acid results in decreased activity of TOP1 protein |
decreases activity
|
protein |
10860928
|
| C005466 | 7153 |
TOP2A
TOP2 TP2A |
topoisomerase (DNA) II alpha 170kDa (EC:5.99.1.3) | ursolic acid results in decreased activity of TOP2A protein |
decreases activity
|
protein |
10860928
|
Related Disease
Diseases related to proteins in ChEMBL interactions
OMIM (26)
| OMIM | preferred title | UniProt |
|---|---|---|
| #218030 | Apparent mineralocorticoid excess; ame |
P80365
|
| #613546 | Aromatase deficiency |
P11511
|
| #139300 | Aromatase excess syndrome; aexs |
P11511
|
| #600430 | Brachydactyly-mental retardation syndrome; bdmr |
P56524
|
| #115200 | Cardiomyopathy, dilated, 1a; cmd1a |
P02545
|
| #212112 | Cardiomyopathy, dilated, with hypergonadotropic hypogonadism |
P02545
|
| #605588 | Charcot-marie-tooth disease, axonal, type 2b1; cmt2b1 |
P02545
|
| #604931 | Cortisone reductase deficiency 1; cortrd1 |
P28845
|
| #609535 | Drug metabolism, poor, cyp2c19-related |
P33261
|
| #608902 | Drug metabolism, poor, cyp2d6-related |
P10635
|
| #181350 | Emery-dreifuss muscular dystrophy 2, autosomal dominant; edmd2 |
P02545
|
| #137800 | Glioma susceptibility 1; glm1 |
O75874
|
| #605130 | Hairy elbows, short stature, facial dysmorphism, and developmental delay |
Q03164
|
| #610140 | Heart-hand syndrome, slovenian type |
P02545
|
| #176670 | Hutchinson-gilford progeria syndrome; hgps |
P02545
|
| #145000 | Hyperparathyroidism 1; hrpt1 |
O00255
|
| #151660 | Lipodystrophy, familial partial, type 2; fpld2 |
P02545
|
| #248370 | Mandibuloacral dysplasia with type a lipodystrophy; mada |
P02545
|
| #131100 | Multiple endocrine neoplasia, type i; men1 |
O00255
|
| #613205 | Muscular dystrophy, congenital, lmna-related |
P02545
|
| #159001 | Muscular dystrophy, limb-girdle, type 1b; lgmd1b |
P02545
|
| #601399 | Platelet disorder, familial, with associated myeloid malignancy |
Q01196
|
| #275210 | Restrictive dermopathy, lethal |
P02545
|
| #188570 | Thyroid hormone resistance, generalized, autosomal dominant; grth |
P10828
|
| #274300 | Thyroid hormone resistance, generalized, autosomal recessive; grth |
P10828
|
| #145650 | Thyroid hormone resistance, selective pituitary; prth |
P10828
|
KEGG DISEASE (33)
| KEGG | disease name | UniProt |
|---|---|---|
| H00033 | Adrenal carcinoma |
O00255
(related)
|
| H00034 | Carcinoid |
O00255
(related)
|
| H00045 | Malignant islet cell carcinoma |
O00255
(related)
|
| H00246 | Primary hyperparathyroidism |
O00255
(related)
|
| H01102 | Pituitary adenomas |
O00255
(related)
|
| H00264 | Charcot-Marie-Tooth disease (CMT) |
P02545
(related)
|
| H00294 | Dilated cardiomyopathy (DCM) |
P02545
(related)
|
| H00420 | Familial partial lipodystrophy (FPL) |
P02545
(related)
|
| H00563 | Emery-Dreifuss muscular dystrophy |
P02545
(related)
|
| H00590 | Congenital muscular dystrophies (CMD/MDC) |
P02545
(related)
|
| H00593 | Limb-girdle muscular dystrophy (LGMD) |
P02545
(related)
|
| H00601 | Hutchinson-Gilford progeria syndrome |
P02545
(related)
|
| H00663 | Restrictive dermopathy |
P02545
(related)
|
| H00665 | Mandibuloacral dysplasia |
P02545
(related)
|
| H01216 | Left ventricular noncompaction (LVNC) |
P02545
(related)
|
| H00036 | Osteosarcoma |
P08684
(marker)
|
| H00249 | Thyroid hormone resistance syndrome |
P10828
(related)
|
| H00599 | 46,XX disorders of sex development (Disorders related to androgen excess) |
P11511
(related)
|
| H00794 | Aromatase excess syndrome |
P11511
(related)
|
| H01205 | Coumarin resistance |
P11712
(related)
|
| H00408 | Type I diabetes mellitus |
P17706
(related)
|
| H01111 | Cortisone reductase deficiency (CRD) |
P28845
(related)
|
| H01171 | Poor drug metabolism (PM) |
P33261
(related)
|
| H00017 | Esophageal cancer |
P35354
(related)
|
| H00025 | Penile cancer |
P35354
(related)
|
| H00046 | Cholangiocarcinoma |
P35354
(related)
|
| H00561 | Brachydacytly-mental retardation syndrome and Smith-Magenis syndrome |
P56524
(related)
|
| H00259 | Apparent mineralocorticoid excess syndrome |
P80365
(related)
|
| H00001 | Acute lymphoblastic leukemia (ALL) (precursor B lymphoblastic leukemia) |
Q01196
(related)
Q01196 (marker) Q03164 (related) Q03164 (marker) |
| H00003 | Acute myeloid leukemia (AML) |
Q01196
(related)
Q01196 (marker) Q13951 (marker) |
| H00004 | Chronic myeloid leukemia (CML) |
Q01196
(related)
|
| H00978 | Thrombocytopenia (THC) |
Q01196
(related)
|
| H00002 | Acute lymphoblastic leukemia (ALL) (precursor T lymphoblastic leukemia) |
Q03164
(related)
|
Diseases related to CTD interactions
12 disease from interactions of metabolites
| MeSH disease | OMIM | compound | disease name | evidence type |
reference
pmid |
|---|---|---|---|---|---|
| D001284 | C005466 | Atrophy |
therapeutic
|
20869956
|
|
| D006528 | C005466 | Carcinoma, Hepatocellular |
therapeutic
|
20630652
|
|
| D002471 | C005466 | Cell Transformation, Neoplastic |
therapeutic
|
19329757
|
|
| D003921 | C005466 | Diabetes Mellitus, Experimental |
therapeutic
|
20869956
|
|
| D006943 | C005466 | Hyperglycemia |
therapeutic
|
20869956
|
|
| D006949 | C005466 | Hyperlipidemias |
therapeutic
|
8847885
|
|
| D006965 | C005466 | Hyperplasia |
therapeutic
|
19329757
|
|
| D007249 | C005466 | Inflammation |
therapeutic
|
8847885
|
|
| D008103 | C005466 | Liver Cirrhosis |
therapeutic
|
21172400
|
|
| D017202 | C005466 | Myocardial Ischemia |
therapeutic
|
17052790
20020988 |
|
| D009369 | C005466 | Neoplasms |
therapeutic
|
8847885
|
|
| D012878 | C005466 | Skin Neoplasms |
therapeutic
|
19329757
|